Background: Site-specific in vivo dynamics of histone acetylation have not been analyzed in a quantitative manner.
Results: Histone acetylation turnover varies depending on the histone residue and presence of neighboring modifications.
Conclusion: Acetylation of histones is a dynamic process that involves the dual action of HATs and HDACs to affect chromatin.
Significance: Acetylation turnover can be quantitatively measured in many cellular processes.
Keywords: Histone Acetylase, Histones, Kinetics, Mass Spectrometry (MS), Metabolism, Post-translational Modification, PTM, Acetylation, Proteomic, Quantitative
Abstract
Acetylation on the tails of histones plays an important role in controlling transcription initiation. Although the steady-state abundances of histone acetyl groups have been reported, the rate at which histones are acetylated and deacetylated on a residue-specific basis has not been quantitatively established. We added [13C]glucose to human cells and monitored the dynamic incorporation of 13C-labeled acetyl groups onto specific histone lysines with quantitative mass spectrometry. We determined the turnover of acetylation to be generally slower than phosphorylation, but fast relative to methylation, and that the rate varied depending on the histone, the residue modified, and also the neighboring modifications. Cells were also treated with a deacetylase inhibitor to determine the rate due to histone acetyltransferase activity alone and in the absence of deacetylase activity. Introduction of 13C-labeled glucose also resulted in the incorporation of 13C into alanine, which allowed us to partition histones into existing and newly synthesized protein categories. Newly synthesized histones were slower to accumulate histone modifications, especially modifications associated with silent chromatin. Finally, we applied our new approaches to find that quiescent fibroblasts exhibited lower levels of labeled acetyl accumulation compared with proliferating fibroblasts. This suggests that acetylation rates can be modulated in cells in different biological states and that these changes can be detected with the approach presented here. The methods we describe can be broadly applied to defining the turnover of histone acetylation in other cell states such as during cellular reprogramming and to quantify non-histone protein acetylation dynamics.
Introduction
Acetylation of histone tails has been demonstrated to be an important regulator of transcriptional activation in eukaryotic cells (1, 2). Enzymes that add acetyl groups to the tails of histones, histone acetyltransferases (HATs),4 are frequently associated with transcription factors, which facilitate their interaction with promoters or enhancers (3, 4). Experiments in which the localization of acetylated histones was determined with chromatin immunoprecipitation followed by detection of the associated DNA have indicated that the histones located upstream of actively transcribed genes are acetylated on the lysines of histones H3 and H4 (2, 5–7). Histone deacetylation is associated with gene repression, and histone deacetylases (HDACs) have been identified as components of complexes that repress transcription (8, 9). Recent studies suggest that complexes containing HATs and HDACs are actually present on the same promoters (10–12) and that a balance exists between the two complexes. That balance is shifted to activate or repress the neighboring gene.
The donor for histone acetylation is acetyl-CoA, in both yeast and metazoans. In yeast, the intracellular pool of acetyl-CoA fluctuates widely across the cell cycle, peaking near the start of a new cycle. Cell growth can be initiated by the addition of acetyl-CoA, which stimulates acetylation of key growth gene promoters via an acetyltransferase complex (13). Human cells do not undergo division in response to the addition of carbon sources but instead use cues from growth factors and hormones. Similar to yeast, acetylation is regulated by metabolic intermediates of glucose, and one major enzyme involved is ATP-citrate lyase, which converts citrate produced by the mitochondria into acetyl-CoA (14). Removal of glucose or a reduction of ATP-citrate lyase resulted in a loss of acetylation on several histones and reduced transcription of genes involved in glucose metabolism (14).
Although the steady-state levels of histone lysine acetylation can be determined by methods such as Western blotting, understanding the dynamics whereby acetyl groups are added and removed requires methods for monitoring the newly deposited acetyl groups independently of the previously deposited acetyl groups. One approach has been to monitor the incorporation of radiolabeled carbon atoms. Radiolabeled acetate has been used to monitor the presence of acetyl moieties and to estimate histone turnover (15). Using radiolabeled acetate to label bulk histones, a half-life of ∼15 min for deacetylation was estimated by averaging all histones and all acetylated residues (no residue specificity) (16). More recent approaches have also been developed to monitor acetylation dynamics in vitro and in vivo. In one study, NMR was used to monitor acetylation and deacetylation of reporter peptides. Recombinant HATs and HDACs were tested for the kinetics of acetyl exchange to determine specificity for particular lysines (17). In another study, a FRET probe was designed with an acetyl-binding domain (bromodomain) that specifically recognized H4K12Ac. The probe was used in live cells to monitor the accumulation of H4K12Ac following treatment with the HDAC inhibitor trichostatin A. Acetylation levels peaked after ∼3 h, suggesting that the half-maximal time for the addition of acetyl on H4K12 is <3 h. Another method used acid-urea gels to separate and quantify acetylated and non-acetylated H3 and also detected methylation marks on the acetylated histones (18, 19). It was discovered that acetylation accumulates faster on H3K4me3-containing histones than histones with other methylation marks in the presence of trichostatin A. Moreover, the transition from minimally acetylated to hyperacetylated histones with trichostatin A treatment was achieved after 15 min, which suggests that the half-maximal time for the addition of acetyl on H3K4me3-containing histones is <15 min (18).
Here, we describe a new method for monitoring the dynamics of histone acetylation using metabolic isotopic labeling of proteins with heavy 13C-labeled glucose in human cells paired with quantitative high-resolution mass spectrometry. We show that acetylation rates vary depending on the modification state as well as the presence of neighboring modifications. We further characterized the rate of histone acetylation in the presence of an HDAC inhibitor and found that alanine labeling from glucose can partition histones into existing and new pools, revealing that modifications accumulate on new histones at different rates than old histones. Finally, we measured the accumulation of labeled acetyl groups in proliferating and quiescent fibroblasts and found that quiescent cells have lower labeling efficiency, although both cell types have similar overall steady-state acetylation levels.
EXPERIMENTAL PROCEDURES
Isotope-labeling Experiments
Human embryonic kidney cells (HEK293) were grown in DMEM (Invitrogen) + 7.5% dialyzed fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA). Cells were washed twice with PBS and introduced into labeling medium. DMEM + 7.5% dialyzed fetal bovine serum + 10 mm [U-13C]sodium acetate (Cambridge Isotopes, Andover, MA) was used for acetate labeling experiments; DMEM without glutamine + 7.5% dialyzed fetal bovine serum + 4 mm [U-13C]glutamine (Cambridge Isotopes) was used for glutamine labeling experiments; and DMEM without glucose + 7.5% dialyzed fetal bovine serum + 4.5 g/liter [U-13C]glucose (Cambridge Isotopes) was used for glucose-labeling experiments. Cells were harvested at the indicated time points by washing with PBS followed by scraping into PBS, pelleting, and flash freezing for −80 °C storage until subsequent analysis.
Histone Preparation for MS
Histones were isolated as described previously using acid extraction (20). Histones were then derivatized with propionic anhydride to block unmodified lysines. Briefly, 100 μg of bulk histone was concentrated to 40 μl using a vacuum concentrator, and 20 μl of 3:1 anhydrous isopropyl alcohol to propionic anhydride (Sigma) was added. The solution was immediately neutralized by addition of a base. The mixture was incubated at 37 °C for 15 min and concentrated back to 40 μl, constituting one round of “propionylation.” One additional round of propionylation was performed, and the solution was reduced to near dryness. 85 μl of 100 mm ammonium bicarbonate (pH 8.0) was added, and histones were digested with trypsin (Promega, Madison, WI) at a ratio of 1:50 trypsin to histone for 6–8 h at 37 °C. The reaction was quenched by acidifying with glacial acetic acid (to < pH 5) and freezing at −80 °C. Newly exposed N termini were modified with two rounds of propionylation. Histone peptides were purified from salts with C18 STAGE-Tips constructed as described previously (21).
Liquid Chromatography Mass Spectrometry
An Agilent 1200 series HPLC configured to split the flow for a final output of ∼200 nl/min was used to resolve histone peptides using a 75-μm inner diameter fused silica column packed with 10–15 cm of 5-μm C18 (Michrom, Auburn, CA). Peptides were eluted with a gradient of 0.7–30% buffer B (buffer A, 0.1 m acetic acid; buffer B, 95% acetonitrile in 0.1 m acetic acid) for 35 min followed by 30–98% buffer B for 30 min and were ionized via electrospray ionization. Peptides were analyzed using an LTQ-Orbitrap-XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Full scans of m/z 290–1000 with a resolution of 30,000 were measured in the Orbitrap. The mass spectrometry data were then analyzed using software that has been described previously elsewhere (22–24). Briefly, for a given peptide with several possible modifications, an optimization-based model simultaneously considers the MS isotopic distribution, MS/MS fragment ions, and relative peptide hydrophobicity relationships to identify and quantify all PTM isoforms. All charge states for modified peptides were included in the analysis. Interferences in the MS isotopic distributions between co-eluting labeled and unlabeled peptides were deconvoluted. The resulting identifications were subsequently validated by manual inspection of the tandem mass spectra. The raw abundance of each modified histone peptide identified by the algorithm was computed by integrating its contribution to the experimentally observed isotopic distribution across the peak width. This raw abundance is then normalized across all observable labeled states within a specific modified state. For instance, both the 12C- and 13C-labeled forms for each acetylation that we measured were used to determine the label incorporation for that modification (relative distribution). This method of normalization controls for variable ionization efficiencies across various modified states of the same peptide sequence, which could potentially skew the analysis and interpretation of the data. The software is available for licensing on the Princeton University website.
Modeling and Half-maximal Calculations
The relative distribution of the labeled acetyl lysines across the time course were fitted (MATLAB, version 7.8) to the logarithmic function, y = A × (1 − e−t/B). This function was chosen to extrapolate both the asymptotic labeling efficiency as A and the time required to reach half of the asymptotic labeling efficiency as ln(2) × B for each modified histone peptide. This work will abbreviate the latter parameter as half-maximal time (t½max, minutes). Within a given time point for a given modified peptide, each replicate measurement was treated as a discrete datum, rather than collated with the other respective measurements into a single average, for the model to account directly for the variability across biological replicates and to increase the statistical significance of the optimizations.
Quiescence Model
Primary human fibroblasts were isolated from foreskins. The subcutaneous cells of a foreskin were removed, and the tissue was cut into 1-mm-wide strips. The strips were incubated for 2 h at 37 °C in 0.5% dispase (Invitrogen). Following removal of the epidermis, the dermis was cut into 1 × 1 mm squares and incubated for 1–2 h at 37 °C in 1000 units/ml collagenase (Invitrogen). Cells were filtered through a 70-μm mesh filter, washed with growth medium (DMEM + 8% FBS + 1 mm sodium pyruvate + 10 mm HEPES + 2 nm l-glutamine + 0.1 mm non-essential amino acids), and plated in growth medium. Cells were cultured for 3–4 days to ensure purity of fibroblasts. Fibroblasts were plated at ∼10,000 cells/cm2 and collected after 24 h for the proliferating state or after 14 days for the contact inhibition-induced quiescence state. Cells were cultured for 9 h in DMEM without glucose + 7.5% dialyzed fetal bovine serum + 4.5 g/liter [U-13C]glucose for labeling experiments.
RESULTS
Histone Acetyl Carbons Are Largely Derived from Glucose
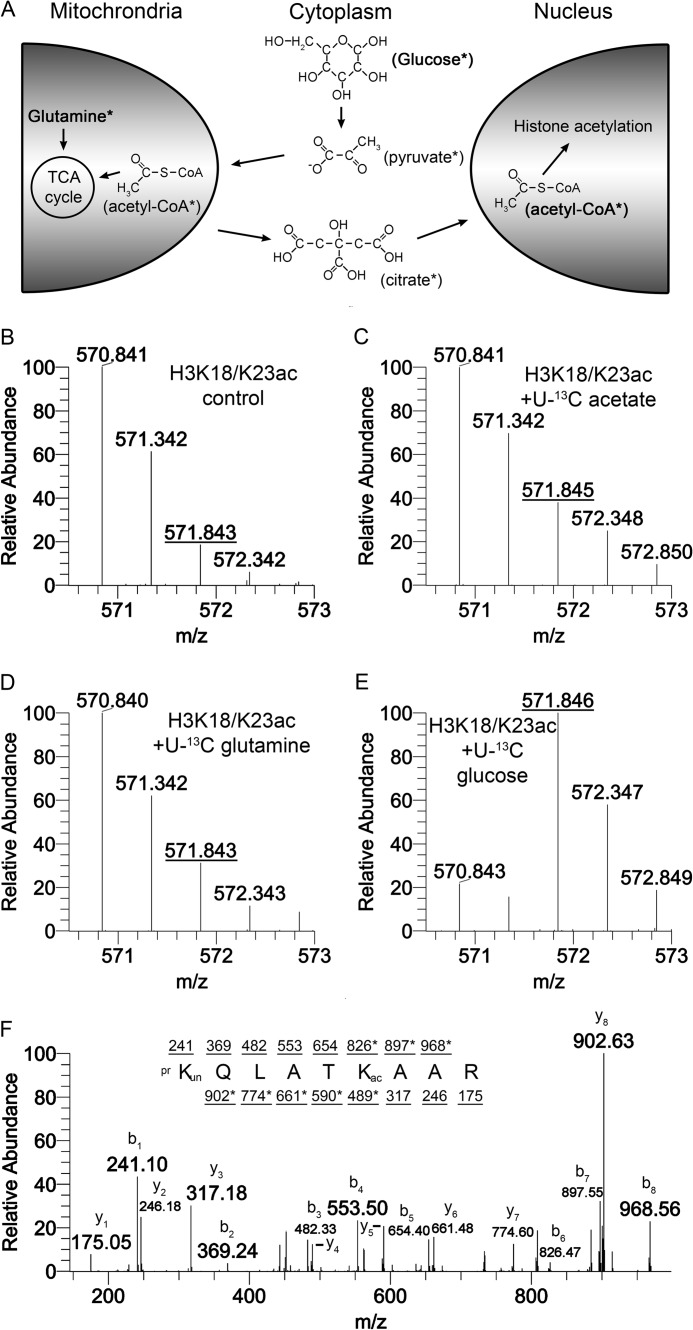
To quantitatively measure rates of acetylation, we used three approaches for labeling intermediates that provide the carbons for acetyl groups (Fig. 1A). In the first approach, we added 13C-labeled glucose because its metabolism can ultimately lead to production of acetyl-CoA, the donor substrate used by HATs to acetylate histones. In the second approach, we utilized 13C-labeled glutamine, which can contribute carbons to the pool of acetyl-CoA used by HATs via the TCA cycle. In the third approach, we bypassed central carbon metabolism and added 13C-labeled acetate directly to the cells as the substrate for acetylation. HEK293 cells were cultured with these three isotopically labeled components individually, and the histones were analyzed by nanoflow liquid chromatography-mass spectrometry (nanoLC-MS/MS). A control sample, in which no isotopically labeled components were added, served as a control that demonstrated the base-line isotopic distribution for the H3 peptide K18QLATK23(Ac)AAR (Fig. 1B). The species 570.84 m/z corresponds to the peptide in which Lys23 is acetylated and all atoms exist in their principal or most abundant isotopic state (monoisotopic state). The species 571.84 m/z is the acetylated peptide with two heavy isotopes, and this species exists due to naturally occurring isotopes that have been incorporated into the amino acids constituting the peptide. Labeling of the acetyl carbons with glucose, glutamine, or acetate caused a shift in abundance from the monoisotopic species (570.84) to the species with two heavy isotopes (571.84) because acetyl has two carbons that were 13C-labeled. The addition of 13C-labeled acetate or 13C-labeled glutamine for 24 h caused a modest amount of acetyl labeling, as indicated by a ∼20% increase in the relative abundance of 571.84 m/z (Fig. 1, C and D). 24 h of 13C-labeled glucose resulted in an increase in the relative abundance of the 571.84 species to ∼80% (Fig. 1E). The larger increase in the 13C labeling with glucose incubation compared with glutamine or acetate indicates that glucose is the major contributor of carbons to acetyl groups on human histones. Tandem mass spectrometry was used to confirm that the observed mass shift was in fact due to the heavy carbon isotopes incorporated into the acetyl group rather than the naturally occurring heavy isotopes within the peptide backbone. The H3 peptide K18QLATK23(Ac)AAR was fragmented in the mass spectrometer, and the fragments were detected. The masses of the fragments observed versus the theoretical masses revealed a 2-Da shift on the ions corresponding to the fragments that contain the acetylated lysine (Fig. 1F). Therefore, [13C]glucose is the preferred substrate to monitor the turnover of acetylation and is the predominant source of acetyl carbons in HEK293 cells.
FIGURE 1.
Glucose is the major substrate for histone acetylation. A, metabolic pathways were utilized to label new acetyl groups with 13C. The subsequent accumulation of new acetyl groups on histone residues was monitored with mass spectrometry. All species that are or become labeled are marked with an asterisk. Carbons lacking four bonds should be assumed to have hydrogen at those positions. B–E, HEK293 cells were incubated in medium containing unlabeled glucose (B), [U-13C]acetate (C), [U-13C]glutamine (D), or [U-13C]glucose (E) for 24 h. The U refers to universally labeled, where all carbons are 13C. Cells were harvested, and histone modifications were analyzed by mass spectrometry. The isotopic distribution of the H3 peptide K18QLATK23AAR where Lys23 is acetylated is shown for each condition. An increase in the m/z 571.84 species indicates an increase in new, 13C-labeled acetyls. F, the MS/MS spectra for K18QLATK23AAR where Lys23 is acetylated shows the appropriate fragmentation ions corresponding to a +2 Da shift on the second lysine residue. b ions are the fragments starting on the N terminus of the peptide and y ions are fragments starting on the C terminus of the peptide. Fragments that contain a +2 Da shift are marked with an asterisk. B–E, represent [M + 2H]+2 spectra.
Acetylation Rates Are Residue-specific
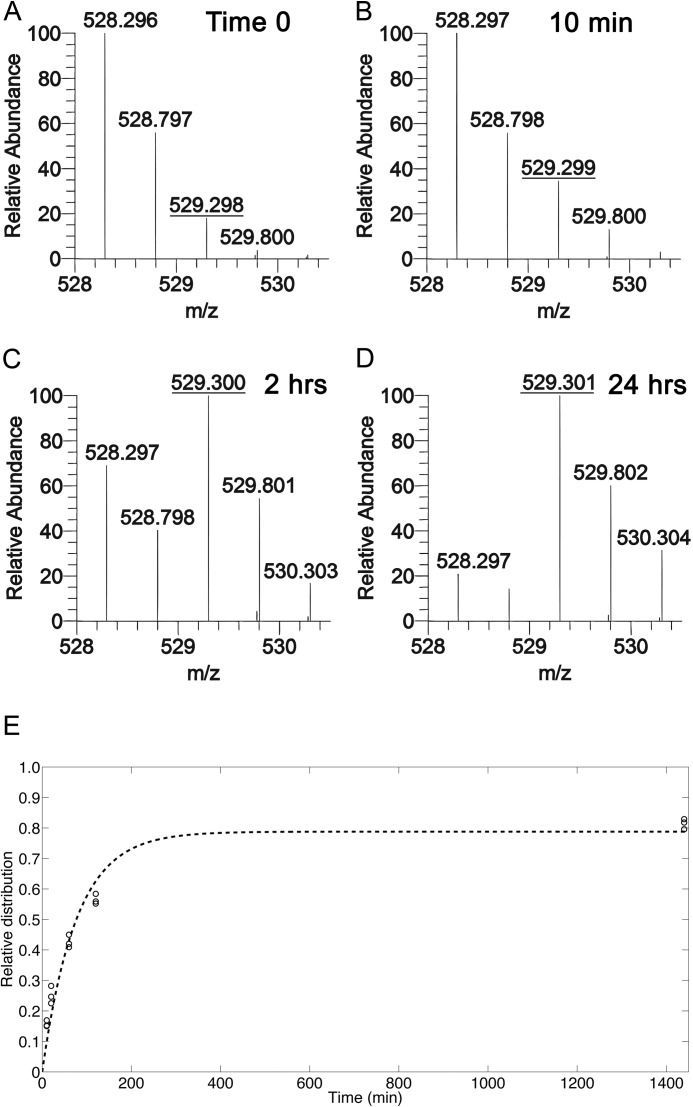
We extended our analysis using [13C]glucose to understand the dynamics of acetylation on several lysines on H3 and one acetylation on H4. HEK293 cells were cultured with 13C-labeled glucose and harvested at 10, 20, 60, 120, and 1440 min. Histones were acid-extracted and processed for nanoLC-MS/MS. Mass spectrometry analysis showed that after 10 min in the presence of labeled glucose, a shift was observed in the mass (529.3 m/z) corresponding to a labeled acetyl on the H3 peptide K9STGGK14APR in which either Lys9 or Lys14 is acetylated (Fig. 2, A and B). After 2 h of incubation with [13C]glucose, the abundance of the species with a labeled acetyl exceeded the abundance of the acetyl species with no labeled carbons (Fig. 2C). After incubation with 13C-labeled glucose for 24 h, the conversion of the unlabeled acetyl to the labeled form reached a maximum (Fig. 2D). Approximately 20% of the acetyl species was unlabeled even after 24 h and remained unlabeled after 48 h (data not shown). This is likely due to some contribution to acetylation from other unlabeled carbon sources such as glutamine supplied in the medium or recycling of metabolic or protein components.
FIGURE 2.
Metabolic incorporation of heavy glucose provided labeled acetyl groups. HEK293 cells were incubated in medium containing unlabeled glucose (A) or [13C]glucose. Cells were harvested at multiple time points, and the histones were analyzed by nanoLC-MS/MS. The relative abundances of isotopes for the H3 peptide K9STGGK14APR in which Lys9 or Lys14 is acetylated are shown for cells incubated for 0 min (A), 10 min (B), 2 h (C), or 24 h (D) with [13C]glucose. A shift in the relative abundance of the m/z 529.3 species indicates turnover from old acetyls to new acetyls. E, the fraction of labeled acetyl species, where 1.0 is 100% labeled, is plotted over time, and an exponential function was fitted to the observed abundances. A–D represent [M + 2H]+2 spectra.
Our method for processing histones produced several small peptides containing one or two lysines. These lysines could be unmodified, methylated, or acetylated in the cell, and the modification status was determined via their predicted mass and predicted elution properties from the HPLC used to separate the peptides prior to mass spectrometry analysis. Using custom software to identify and quantify these modified histone peptides, we determined the relative abundance of each of the modified forms for a given peptide. For the acetylated species, we then found the fraction that was unlabeled (old acetyl) versus labeled (new acetyl), and plotted the percentage of the labeled form over time (Fig. 2E). The data were fit to a logarithmic curve to generate a time constant used in calculating a half-maximum. Half-maximums were determined for six different acetylated forms on histone H3 and one acetylation on H4 (Table 1). Half-maximal times ranged from 53–87 min, which was much faster than the average cell cycle, and varied depending on the lysine modified as well as the presence of neighboring modifications. The half-maximum was fastest when Lys9 or Lys14 was acetylated, with no adjacent modifications on Lys9 or Lys14, or when Lys14 was acetylated and Lys9 was monomethylated. This is consistent with the absence of silencing marks near the monitored acetyls, at least on the observed peptide. The half-maximum of K14Ac was slower when measured on peptides that also contained Lys9 di- or trimethyl, suggesting that histone tails that contain modifications classified as “silencing marks” exhibit slower rates of acetylation on Lys14 than tails in which Lys14 is not near a silencing mark. The turnover of both Lys9 and Lys14 acetyl on the same peptide is 20 min slower than for a single acetylation event on Lys9 or Lys14, indicating that it is more time consuming to replace acetyls on two residues versus only one residue. The half-maximum of K18Ac or K23Ac was similar to that of K9me2K14Ac and K9me3K14Ac. The slowest half-maximum was on H4 in which Lys5, Lys8, Lys12, or Lys16 was acetylated. These data indicate that the rate of acetylation turnover varies as a function of the site of acetylation as well as the presence of nearby modifications on the same histone tail.
TABLE 1.
Half-maximal (t½, minutes) values for acetylated residues on H3 and H4
The half-maximum was calculated as described in the methods for each modification state, with and without sodium butyrate treatment. A range of half-maximal values shows the mean and the upper and lower values for 5% error. Ratios are shown representing the half-maximum with butyrate treatment over the half-maximum without butyrate treatment.
| Modification | [13C]Glucose (t½ maximum) | [13C]Glucose + Butyrate (t½ maximum) | Ratio of butyrate treated to glucose alone |
|---|---|---|---|
| min | min | ||
| H3K9Ac or K14Ac | 49.2–52.7–56.4 | 91.3–102.0–114.4 | 1.9 |
| H3K9me1K14Ac | 50.5–54.2–58.0 | 93.6–102.7–113.0 | 1.9 |
| H3K9me2K14Ac | 64.4–66.8–69.5 | 88.4—96.2–105.0 | 1.4 |
| H3K9me3K14Ac | 58.6–62.5–66.6 | 88.2–97.2–107.4 | 1.6 |
| H3K9AcK14Ac | 68.9–75.4–82.7 | 177.0–185.3–194.4 | 2.5 |
| H3K18Ac or K23Ac | 63.7–66.1–68.6 | 84.2–90.6–97.7 | 1.4 |
| H4-1Ac | 80.0–86.7–94.1 | 86.5–93.5–101.2 | 1.1 |
Estimation of Acetylation On-rates using HDAC Inhibition
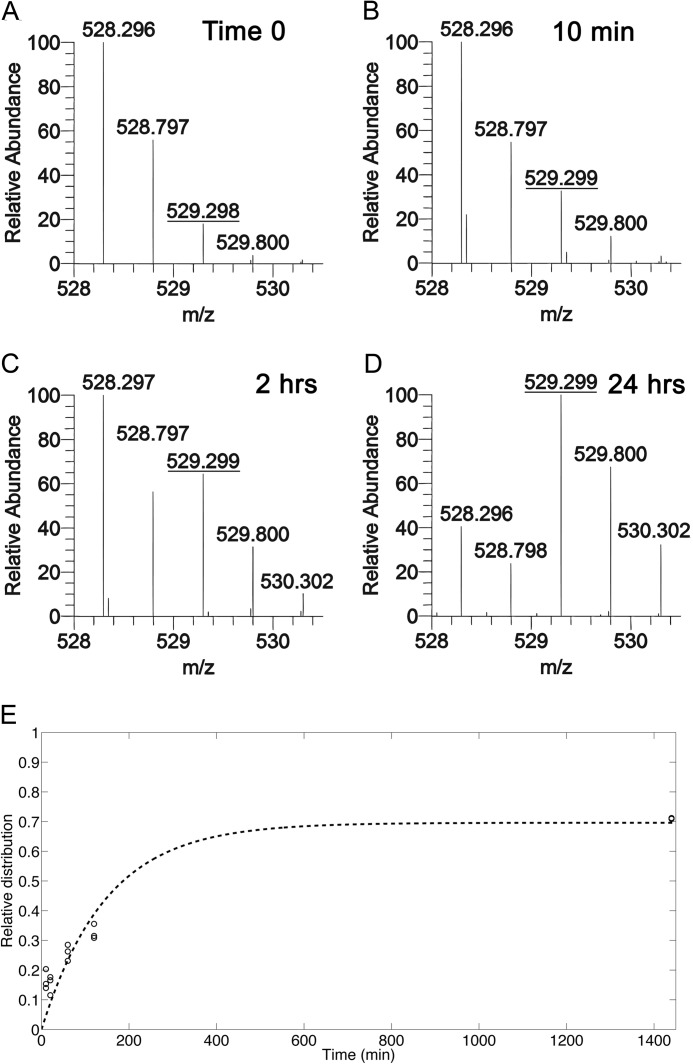
To decouple the HATs and HDACs that contribute to the overall observed acetylation turnover, we cultured HEK293 cells in [13C]glucose medium that contained 10 mm sodium butyrate, a potent HDAC inhibitor. We did not, however, assume that butyrate targeted all HDACs similarly in our experiments, and therefore, some HDACs may still have been active even in the presence of butyrate. Cells were harvested at 10, 20, 60, 120, and 1440 min and processed for nanoLC-MS/MS. A shift in the relative abundance of the unlabeled acetyl to the 13C-labeled acetyl on either Lys9 or Lys14 of the H3 peptide K9STGGK14APR was observed after 10 min of incubation in [13C]glucose and butyrate (Fig. 3, A and B). After 2 h, the turnover approached 50% but was noticeably slower than without butyrate (Figs. 2C and 3C). After 24 h of labeling, the turnover reached a maximum that was lower than that observed with [13]glucose alone (Figs. 2D and 3D). The labeled fraction at each time point was plotted and fit to a logarithmic curve (Fig. 3E). Half-maximums were determined for six different acetylated forms on H3 and one acetyl on H4 (Table 1). Half-maximums were similar for H3K9Ac or K14Ac, K9me1K14Ac, K9me2K14Ac, K9me3K14Ac, K18Ac or K23Ac, and H4-1Ac. However, the ratio of the half-maximum with butyrate to without butyrate was different. A ratio of 2 would suggest that butyrate was targeting the relevant HDACs with high specificity and that turnover of the acetyl group is equally dependent on HATs and HDACs, instead of new histone synthesis or degradation. The ratio with to without butyrate was 1.9 for H3K9Ac or K14Ac and K9me1K14Ac, suggesting that the HAT(s) and HDAC(s) that modify these sites are contributing equally and that butyrate is an efficient inhibitor of the HDAC(s). The ratio for H3K9me2K14Ac, K9me3K14Ac, and K18Ac or K23Ac was ∼1.5, indicating that the HAT(s) and HDAC(s) that target these sites are not contributing equally or butyrate is not an efficient inhibitor of the HDAC(s). The ratio for H4-1Ac was the lowest at 1.1, suggesting that HDACs are less involved in the turnover of this modification, or that butyrate does not target the HDAC(s). In fact, the majority of this modification is on H4K16, which is deacetylated by sirtuin 1, and not a target of butyrate (25). The ratio for H3K9AcK14Ac was the slowest at 2.5, suggesting that deacetylation is a large contributor to the rate of turnover for these modifications. The addition of H3K9acK14ac may only occur at specific sites in the genome that must be initially deacetylated. Loss of deacetylation would limit production of these unmodified sites and may contribute to the slow half-maximum observed from our data. Overall, the half-maximums of acetyl groups were slower when cells were treated with butyrate, and the decrease in turnover rates varied based on the particular acetyl site and neighboring modifications.
FIGURE 3.
HDAC inhibition leads to slower turnover by blocking removal of old acetyl groups. HEK293 cells were incubated in medium containing unlabeled glucose (A) or [13C]glucose + 10 mm sodium butyrate. Cells were harvested at multiple time points, and the isolated histones were analyzed by nanoLC-MS/MS. The relative abundance of isotopes for the H3 peptide K9STGGK14APR in which Lys9 or Lys14 is acetylated is shown for cells incubated for 0 min (A), 10 min (B), 2 h (C), or 24 h (D) with [13C]glucose + 10 mm sodium butyrate. A shift in the relative abundance of the m/z 529.3 species indicates an increased fraction of the peptide that has a labeled acetyl. E, the fraction of labeled acetyl species, where 1.0 is 100% labeled, is plotted over time, and an exponential function was fitted to the observed abundances. A–D represent [M + 2H]+2 spectra.
Alanine Becomes Labeled in Newly Synthesized Histones
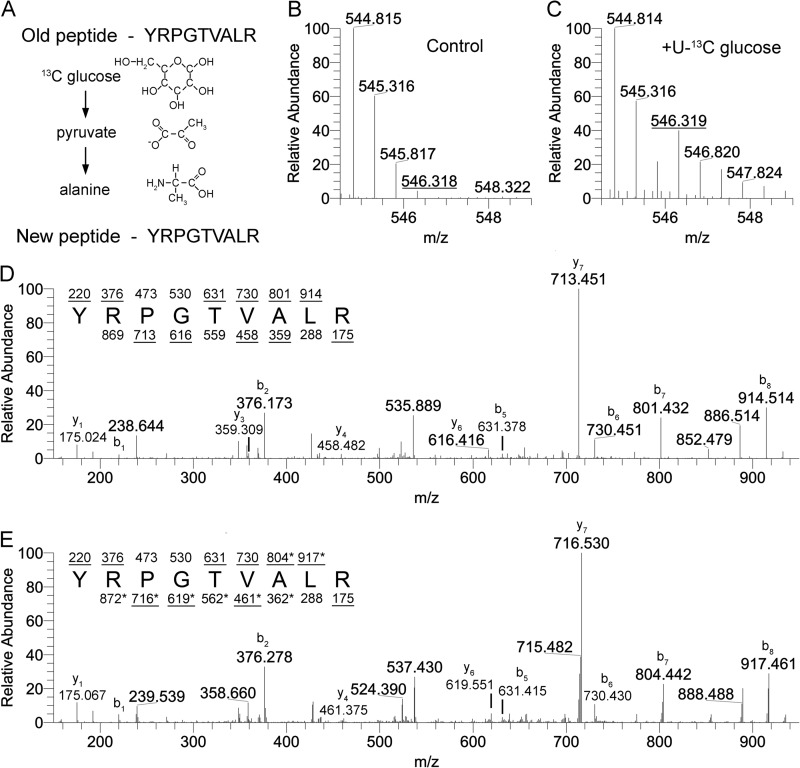
Alanine can be synthesized from pyruvate, a metabolite of glycolysis, as well as branched chain amino acids (Fig. 4A) (26, 27). Alanine is a nonessential amino acid and was not present in the DMEM medium used for culturing cells herein, so newly synthesized proteins contained endogenously synthesized alanines, some of which were derived from glucose. Many histone peptides and all of the peptides that we analyzed contain at least one alanine residue. Mass spectrometry analysis of a control peptide that contained an alanine but no known modifications (data not shown) revealed that after 24 h in the presence of [13C]glucose, a shift was observed that corresponded to +3 Da, or a fully 13C-labeled alanine (Fig. 4, B and C). This H3 peptide is represented in H3.1, H3.2, and H3.3, making it an ideal peptide for measuring new protein synthesis, both from replication dependent and independent histones. Fragmentation of the unlabeled, monoisotopic species (m/z 544.81) produced fragments that correspond to the predicted amino acid sequence (Fig. 4D). Fragmentation of the alanine-labeled species (m/z 546.32) produced fragments that were shifted 3 Da when alanine was present on the fragment (Fig. 4E). No shift was observed for fragments that contain portions of the sequence lacking alanine. The fraction of the peptide that was labeled would be ∼50% after 24 h if every alanine was derived from glucose, and the cell cycle for HEK293 cells was 24 h long. We observed ∼35% turnover of unlabeled alanine to labeled alanine. Thus, although some new proteins contain unlabeled alanines (Fig. 4, B and C), most newly synthesized proteins contain labeled alanines derived from [13C]glucose. This allowed us to discriminate between old and new histones based on the presence or absence of an isotopic shift.
FIGURE 4.
Labeled carbons derived from exogenously provided glucose can be metabolized to form 13C-labeled alanine. A, alanine can be synthesized directly from pyruvate, allowing for labeled carbons from glucose to be incorporated into newly synthesized alanine. Carbons lacking four bonds should be assumed to have hydrogen at those positions. B and C, the 41–49-amino acid peptide from H3 that has no known modification sites shows a shift of a +1.5 m/z species (+3 Da) after 24 h of [13C]glucose treatment. MS/MS analysis of the control peptide treated with unlabeled glucose (D) or [13C]glucose (E) identifies the +1.5 m/z shift as corresponding to alanine based on fragmentation ions. b ions are the fragments starting at the N terminus of the peptide, and y ions are fragments starting on the C terminus of the peptide. Fragments that contain a +3 Da shift are marked with an asterisk. B and C represent [M + 2H]+2 spectra.
Newly Synthesized Histones Are Modified at Different Rates
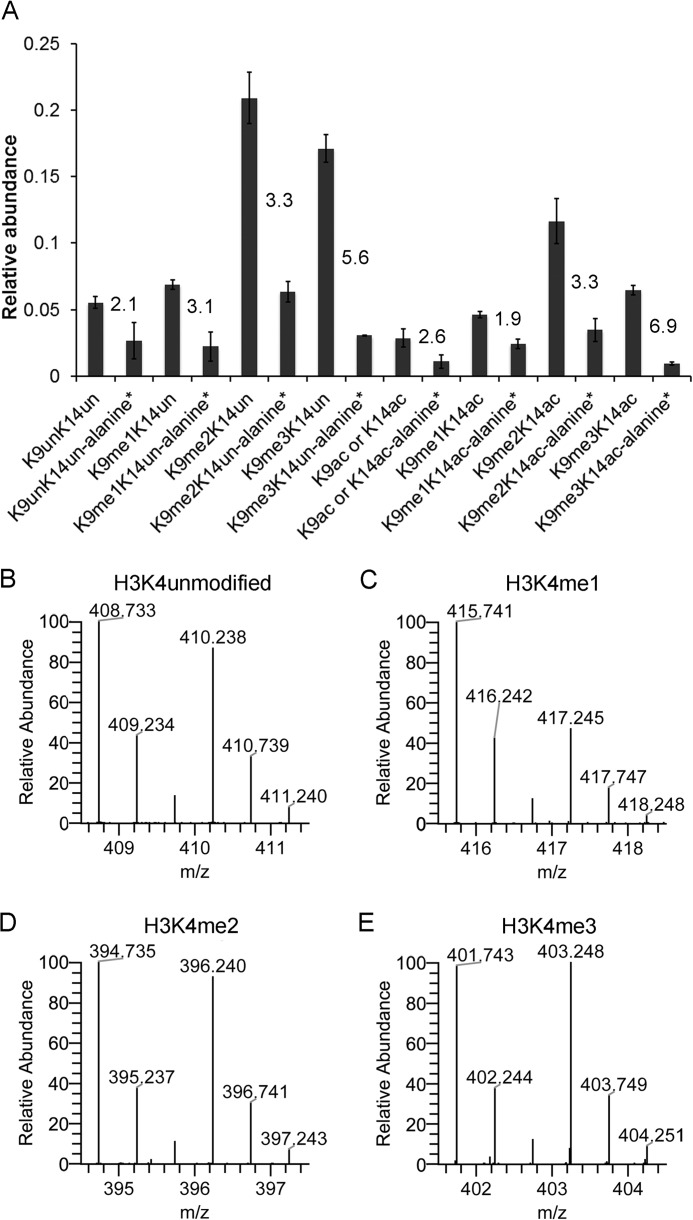
Based on our determination that the presence of 13C in alanine can serve to distinguish most old proteins from new proteins, we analyzed the relative abundance of selected modifications on old histones and new histones after 24 h in the presence of [13C]glucose. Using an estimated turnover of ∼35% for alanine-labeled histones (Fig. 4, B and C), we would expect that the ratio of old to new histone peptides would be ∼1.85:1. The unmodified form of the peptide K9STGGK14APR was present at a ratio of 2.1:1, indicating that most newly synthesized histones that are unlabeled at Lys9 and Lys14 are synthesized at a rate similar to that expected for general protein synthesis (Fig. 5A). However, the ratio of old to new histones increased as Lys9 became more modified; the ratio was 3.1:1 for K9me1K14un, 3.3:1 for K9me2K14un, and 5.6:1 for K9me3K14un. This indicates that these modifications are not added to new histones immediately but are instead added after a delay and may take a full cell cycle to become fully modified. A similar trend was observed when we analyzed the relative abundance of acetylated species on the histone H3 peptide K9STGGK14APR, with the exception that K9me1K14Ac was produced faster than K9Ac or K14Ac (Fig. 5A). The slowest accumulation of alanine-labeled species occurred on K9me2K14Ac and K9me3K14Ac. The higher ratio of old histone peptides to new peptides correlated with previous observations that higher methylation states are slower to accumulate after S phase (28).
FIGURE 5.
Estimating the lag between histone synthesis and lysine modification. HEK293 cells were cultured with [13C]glucose for 24 h, and the histones were analyzed by nanoLC-MS. The intensities of each modified form of the peptide K9STGGK14APR were calculated for unlabeled alanine and labeled alanine (+3 Da). A, intensity values were used to create a relative abundance (1 = 100%) of both unlabeled and labeled modified forms. The numbers on the graph indicate the ratio of unlabeled to labeled. The relative abundance of unlabeled and labeled modifications on H3K4 (B–E) is plotted. B–E represent [M + 2H]+2 spectra.
In contrast, for H3K4, peptides with more methyl groups were generated faster than peptides with fewer methyl groups. Alanine-labeled peptides of H3K4me1 accumulated more slowly than H3K4 that was di- or trimethylated (Fig. 5, C–E). H3K4 unmodified also had high amounts of alanine-labeled species, potentially due to demethylation of H3K4me3 (Fig. 5B). Turnover of the H3K4-methylated species is higher than the control peptide in Fig. 4C, likely due to these modifications existing in higher frequencies on replication independent variants such as H3.3. Replication-independent variants can be exchanged more rapidly throughout the cell cycle, allowing for some modifications to turnover faster than expected, if only cell cycle dynamics are considered. Additionally, Henikoff and co-workers (29) demonstrated using a metabolic/genomic-based approach that active gene regions (trithorax group protein-bound, Lys4 methylation) of the genome have faster turnover rates than silenced regions (polycomb protein-bound, Lys27 methylation) in Drosophila, which would be consistent with our mass spectrometry results.
Quiescent Fibroblasts Exhibit Altered Incorporation of Labeled Acetyl Groups
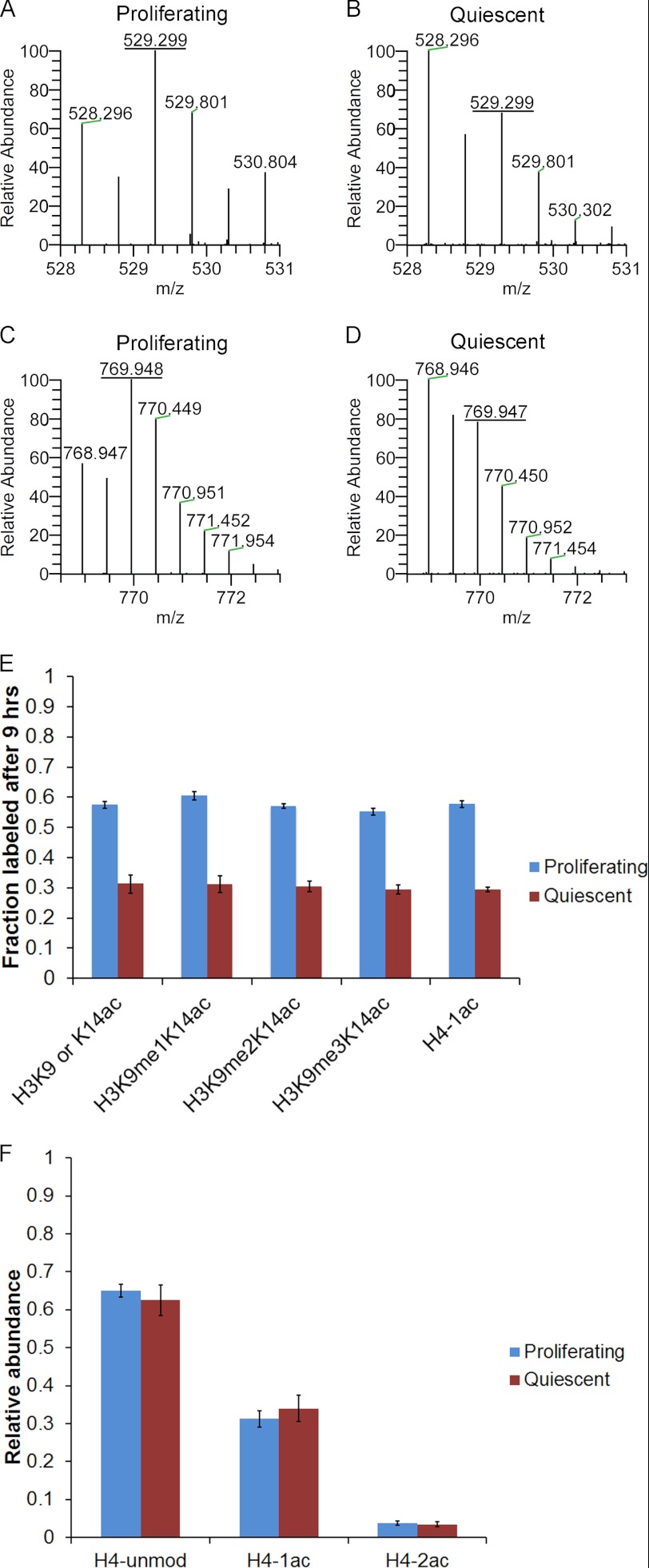
To determine whether our method for monitoring acetylation turnover can be used to identify differences in acetylation dynamics between cells in different biological states, we analyzed primary human foreskin fibroblasts either in proliferating or quiescent conditions. Cells were cultured for 9 h in medium containing [13C]glucose and analyzed by nanoLC-MS. A mass spectrum of the H3 peptide K9STGGK14APR acetylated on Lys9 or Lys14 revealed that proliferating fibroblasts contained a higher amount of labeled acetyl groups after 9 h than quiescent fibroblasts (Fig. 6, A and B). These differential labeling rates were not unique to H3K9 or H3K14 and were observed on H4 in which the histone was singly acetylated (Fig. 6, C and D). The fraction of acetyl groups that were labeled in proliferating and quiescent fibroblasts was determined for five modification states (Fig. 6E). All modifications showed a highly reproducible level of incorporation for all five sites, and the incorporation for quiescent cells was approximately half of that for proliferating cells. The incorporation was less than that observed for HEK293 cells, which had ∼80% incorporation after 9 h (Fig. 2E). The steady-state level of acetylation was similar between proliferating and quiescent cells as shown for histone H4 (Fig. 6F). Therefore, we observed a higher incorporation of labeled acetyl groups in proliferating than quiescent fibroblasts, even though the steady-state levels were similar, suggesting that proliferating cells are more rapidly exchanging acetyl groups on histones or that quiescent cells utilize different pathways for acetyl carbons.
FIGURE 6.
Quiescent fibroblasts incorporate less labeled acetyl. Fibroblasts were cultured in [13C]glucose medium for 9 h and harvested while proliferating or after contact inhibition-induced quiescence. The relative abundance of isotopes for the H3 peptide K9STGGK14APR where Lys9 or Lys14 is acetylated is shown for proliferating (A) and quiescent (B) fibroblasts. The relative abundance of isotopes for the H4 peptide GRGK5GGK8GLGK12GGAK16R in which one lysine is acetylated is shown for proliferating (C) and quiescent (D) fibroblasts. E, the fraction of labeled acetyl groups after 9 h of incubation in [13C]glucose is plotted for several acetylations for proliferating and quiescent fibroblasts. F, the steady-state relative abundances (1 = 100%) for the H4 peptide (residues 4–17) when unmodified (unmod), singly acetylated, or doubly acetylated are plotted for proliferating and quiescent fibroblasts. Three independent experiments were performed, and the error bars represent S.E. A–D represent [M + 2H]+2 spectra.
DISCUSSION
Deriving accurate estimates of the rate of histone acetylation has been hampered by the large number of different types of acetylation events and the fact that the rate of acetylation can depend on the presence of nearby lysine modifications. The application of mass spectrometry to this problem has the potential to solve these issues because it can provide highly quantitative information on the presence of specific modifications. In this study, we measured the dynamics of histone acetylation for several lysines on H3 and H4 using quantitative mass spectrometry. Our data indicate that the half-maximal values for acetylation are fast relative to reports of histone methylation (30, 31) but likely slower than reports of non-histone phosphorylation in both the cytoplasm and the mitochondria (32, 33). Our data also indicate that the rate of new acetyl accumulation is dependent on the histone, the lysine, and neighboring modifications. Furthermore, by distinguishing newly synthesized from preexisting histones, we show evidence that the modifications on newly deposited histones differ from those that preexist in the chromatin. Finally, acetylation dynamics are not uniform among different cell states, and this may be an area of future exploration in processes such as reprogramming and differentiation.
Several approaches have been used to measure the rate of acetylation in model organisms. Early approaches did not have the resolution to distinguish one lysine from another and reported half-lives that were averages across all lysines of a particular histone (19, 34, 35). The half-life values ranged from 12–200 min in chicken erythrocytes to 4–15 min in Saccharomyces cerevisiae, depending on the histone. More recent approaches have been used to measure acetylation rates on particular lysines and even to take into account neighboring modifications. In vitro analysis of acetylation using reporter peptides and recombinant HATs revealed that full acetylation of the reporter was achieved within 4 min for H4K12 and H4K20 (17). FRET can be used to monitor H4K12 acetylation dynamics, and acid-urea gels in combination with Western blotting can be used to determine acetylation turnover for H3K4me3- or H3K9me3-containing histones (18, 36). We are able to estimate the half-maximum of acetylation in a residue-specific manner and incorporate information about neighboring residues. We show that the half-max of acetylation ranges from 53 to 87 min depending on the lysine monitored and the nearby modifications. Our values are slightly higher than other estimates, which likely reflect the fact that each method focuses on different collections of acetylation events. Middle-down mass spectrometry analysis of entire histone tails (37) would help to account for all additional modifications and the interdependency of modifications on acetylation dynamics.
We found varying effects of butyrate treatment on the half-maximum of different modification states. Some residues had an increase in half-maximal time by a factor of 1.9 relative to no butyrate treatment, suggesting that incorporation of [13C]glucose in the absence of butyrate is dependent equally on acetylation and deacetylation. Other modification states showed smaller increases in the half-maximal time, and this indicates that butyrate is not an efficient inhibitor of deacetylation for these modifications. The exact HDACs inhibited by butyrate are not known with certainty, although reports suggest that class I HDACs and some class II HDACs are targets (38). Sirtuins are not known to be targets of butyrate. Lysine 16 acetylation, the predominant form that we measured on H4, is removed via sirtuins (25, 39). This may explain why the half-maximum did not increase for H4 acetylation when treated with butyrate. It also opens up the possibility of using this technique to screen existing or new HDAC inhibitors for their specificity.
Our results indicate that glucose provides the major source of carbons for acetylation, at least in HEK293 cells. Labeled glutamine and sodium acetate treatment resulted in small shifts in labeled acetyl groups, whereas labeled glucose treatment resulted in near complete turnover. These results are consistent with other reports showing that glucose or metabolic intermediates are involved in histone acetylation (13, 14, 40, 41). Our method is capable of testing whether changes in the flux from glucose to nuclear acetyl-CoA can influence acetylation turnover, and therefore transcription activity, or whether downstream enzymes such as ATP-citrate lyase act as master regulators to maintain pools of acetyl-CoA for acetylation.
We show that proliferating and quiescent fibroblasts have differential labeling of acetyl groups. The disparity in labeled acetyl accumulation may be due to the rate of glucose uptake between the two cell states, Indeed, proliferating and quiescent cells may consume different amounts of glucose (42). This would support the idea that glucose is the major factor contributing not only to steady-state acetylation but also the rate of acetylation turnover. The difference in labeled acetyl accumulation may also be due to fundamental differences in the proliferating and quiescent states reflecting unique transcriptional programs that are regulated via HAT and HDAC activity (43). The approach described to define the dynamics of acetylation could be highly useful in reprogramming studies or differentiation models that require extensive transcriptional and chromatin changes on an acute dynamic or epigenetic time-scale.
Acetylation of non-histone proteins is becoming more widely appreciated, and large-scale proteomic screens have revealed the existence of thousands of acetylated proteins in various cell types (44, 45). Some of the more common acetylated protein examples other than histones include tubulin and p53 (46–48). In the case of p53, p300 or p300/CBP-associated factor can catalyze the acetylation event that facilitates the transcription factor activity of p53 (49). Sirt1 acts as the deacetylase for p53 and can reverse the ability of p53 to induce processes such as apoptosis (50). The methods presented here can easily be extended and coupled with existing immunoaffinity enrichment of global acetylated peptides for investigating the stability of these modifications under a variety of cellular conditions. Understanding the rates at which these protein acetylation reversals occur, or whether acetylation is constantly being turned over, will add to the understanding of how this important protein modification is regulated in normal physiology and disease situations.
Footnotes
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase
- nanoLC
- nanoflow liquid chromatography.
REFERENCES
- 1. Braunstein M., Rose A. B., Holmes S. G., Allis C. D., Broach J. R. (1993) Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7, 592–604 [DOI] [PubMed] [Google Scholar]
- 2. Wang Z., Zang C., Rosenfeld J. A., Schones D. E., Barski A., Cuddapah S., Cui K., Roh T. Y., Peng W., Zhang M. Q., Zhao K. (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40, 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grant P. A., Duggan L., Côté J., Roberts S. M., Brownell J. E., Candau R., Ohba R., Owen-Hughes T., Allis C. D., Winston F., Berger S. L., Workman J. L. (1997) Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11, 1640–1650 [DOI] [PubMed] [Google Scholar]
- 4. Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., Nakatani Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 [DOI] [PubMed] [Google Scholar]
- 5. Schübeler D., MacAlpine D. M., Scalzo D., Wirbelauer C., Kooperberg C., van Leeuwen F., Gottschling D. E., O'Neill L. P., Turner B. M., Delrow J., Bell S. P., Groudine M. (2004) The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 18, 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roh T. Y., Cuddapah S., Cui K., Zhao K. (2006) The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. U.S.A. 103, 15782–15787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sinha I., Buchanan L., Rönnerblad M., Bonilla C., Durand-Dubief M., Shevchenko A., Grunstein M., Stewart A. F., Ekwall K. (2010) Genome-wide mapping of histone modifications and mass spectrometry reveal H4 acetylation bias and H3K36 methylation at gene promoters in fission yeast. Epigenomics 2, 377–393 [DOI] [PubMed] [Google Scholar]
- 8. Davie J. K., Edmondson D. G., Coco C. B., Dent S. Y. (2003) Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J. Biol. Chem. 278, 50158–50162 [DOI] [PubMed] [Google Scholar]
- 9. van der Vlag J., Otte A. P. (1999) Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23, 474–478 [DOI] [PubMed] [Google Scholar]
- 10. Simone C., Stiegler P., Forcales S. V., Bagella L., De Luca A., Sartorelli V., Giordano A., Puri P. L. (2004) Deacetylase recruitment by the C/H3 domain of the acetyltransferase p300. Oncogene 23, 2177–2187 [DOI] [PubMed] [Google Scholar]
- 11. Johnsson A., Durand-Dubief M., Xue-Franzén Y., Rönnerblad M., Ekwall K., Wright A. (2009) HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep. 10, 1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Z., Zang C., Cui K., Schones D. E., Barski A., Peng W., Zhao K. (2009) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cai L., Sutter B. M., Li B., Tu B. P. (2011) Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 42, 426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Allfrey V. G., Pogo B. G., Littau V. C., Gershey E. L., Mirsky A. E. (1968) Histone acetylation in insect chromosomes. Science 159, 314–316 [DOI] [PubMed] [Google Scholar]
- 16. Chestier A., Yaniv M. (1979) Rapid turnover of acetyl groups in the four core histones of simian virus 40 minichromosomes. Proc. Natl. Acad. Sci. U.S.A. 76, 46–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dose A., Liokatis S., Theillet F. X., Selenko P., Schwarzer D. (2011) NMR profiling of histone deacetylase and acetyl-transferase activities in real time. ACS Chem Biol 6, 419–424 [DOI] [PubMed] [Google Scholar]
- 18. Crump N. T., Hazzalin C. A., Bowers E. M., Alani R. M., Cole P. A., Mahadevan L. C. (2011) Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc. Natl. Acad. Sci. U.S.A. 108, 7814–7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Waterborg J. H. (2001) Dynamics of histone acetylation in Saccharomyces cerevisiae. Biochemistry 40, 2599–2605 [DOI] [PubMed] [Google Scholar]
- 20. Shechter D., Dormann H. L., Allis C. D., Hake S. B. (2007) Extraction, purification and analysis of histones. Nat. Protoc. 2, 1445–1457 [DOI] [PubMed] [Google Scholar]
- 21. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 22. DiMaggio P. A., Jr., Young N. L., Baliban R. C., Garcia B. A., Floudas C. A. (2009) A mixed integer linear optimization framework for the identification and quantification of targeted post-translational modifications of highly modified proteins using multiplexed electron transfer dissociation tandem mass spectrometry. Mol. Cell Proteomics 8, 2527–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Baliban R. C., DiMaggio P. A., Plazas-Mayorca M. D., Young N. L., Garcia B. A., Floudas C. A. (2010) A novel approach for untargeted post-translational modification identification using integer linear optimization and tandem mass spectrometry. Mol. Cell Proteomics 9, 764–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu Y., DiMaggio P. A., Jr., Perlman D. H., Zakian V. A., Garcia B. A. (2013) Novel phosphorylation sites in the S. cerevisiae Cdc13 protein reveal new targets for telomere length regulation. J. Proteome Res. 12, 316–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hajji N., Wallenborg K., Vlachos P., Füllgrabe J., Hermanson O., Joseph B. (2010) Opposing effects of hMOF and SIRT1 on H4K16 acetylation and the sensitivity to the topoisomerase II inhibitor etoposide. Oncogene 29, 2192–2204 [DOI] [PubMed] [Google Scholar]
- 26. Odessey R., Khairallah E. A., Goldberg A. L. (1974) Origin and possible significance of alanine production by skeletal muscle. J. Biol. Chem. 249, 7623–7629 [PubMed] [Google Scholar]
- 27. Perriello G., Jorde R., Nurjhan N., Stumvoll M., Dailey G., Jenssen T., Bier D. M., Gerich J. E. (1995) Estimation of glucose-alanine-lactate-glutamine cycles in postabsorptive humans: role of skeletal muscle. Am. J. Physiol. 269, E443–450 [DOI] [PubMed] [Google Scholar]
- 28. Zee B. M., Britton L. M., Wolle D., Haberman D. M., Garcia B. A. (2012) Origins and formation of histone methylation across the human cell cycle. Mol. Cell. Biol. 32, 2503–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deal R. B., Henikoff J. G., Henikoff S. (2010) Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328, 1161–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zee B. M., Levin R. S., Xu B., LeRoy G., Wingreen N. S., Garcia B. A. (2010) In vivo residue-specific histone methylation dynamics. J. Biol. Chem. 285, 3341–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zee B. M., Levin R. S., Dimaggio P. A., Garcia B. A. (2010) Global turnover of histone post-translational modifications and variants in human cells. Epigenetics Chromatin 3, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aponte A. M., Phillips D., Hopper R. K., Johnson D. T., Harris R. A., Blinova K., Boja E. S., French S., Balaban R. S. (2009) Use of (32)P to study dynamics of the mitochondrial phosphoproteome. J. Proteome Res. 8, 2679–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bennetzen M. V., Larsen D. H., Bunkenborg J., Bartek J., Lukas J., Andersen J. S. (2010) Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol. Cell Proteomics 9, 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Waterborg J. H. (1998) Dynamics of histone acetylation in Chlamydomonas reinhardtii. J. Biol. Chem. 273, 27602–27609 [DOI] [PubMed] [Google Scholar]
- 35. Zhang D. E., Nelson D. A. (1988) Histone acetylation in chicken erythrocytes. Rates of acetylation and evidence that histones in both active and potentially active chromatin are rapidly modified. Biochem. J. 250, 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ito T., Umehara T., Sasaki K., Nakamura Y., Nishino N., Terada T., Shirouzu M., Padmanabhan B., Yokoyama S., Ito A., Yoshida M. (2011) Real-time imaging of histone H4K12-specific acetylation determines the modes of action of histone deacetylase and bromodomain inhibitors. Chem. Biol. 18, 495–507 [DOI] [PubMed] [Google Scholar]
- 37. Young N. L., DiMaggio P. A., Plazas-Mayorca M. D., Baliban R. C., Floudas C. A., Garcia B. A. (2009) High throughput characterization of combinatorial histone codes. Mol. Cell Proteomics 8, 2266–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu W. S., Parmigiani R. B., Marks P. A. (2007) Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene 26, 5541–5552 [DOI] [PubMed] [Google Scholar]
- 39. Vaquero A., Sternglanz R., Reinberg D. (2007) NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene 26, 5505–5520 [DOI] [PubMed] [Google Scholar]
- 40. Morrish F., Noonan J., Perez-Olsen C., Gafken P. R., Fitzgibbon M., Kelleher J., VanGilst M., Hockenbery D. (2010) Myc-dependent mitochondrial generation of acetyl-CoA contributes to fatty acid biosynthesis and histone acetylation during cell cycle entry. J. Biol. Chem. 285, 36267–36274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mosley A. L., Ozcan S. (2003) Glucose regulates insulin gene transcription by hyperacetylation of histone h4. J. Biol. Chem. 278, 19660–19666 [DOI] [PubMed] [Google Scholar]
- 42. Lemons J. M., Feng X. J., Bennett B. D., Legesse-Miller A., Johnson E. L., Raitman I., Pollina E. A., Rabitz H. A., Rabinowitz J. D., Coller H. A. (2010) Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 8, e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Coller H. A., Sang L., Roberts J. M. (2006) A new description of cellular quiescence. PLoS Biol. 4, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 45. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 46. L'Hernault S. W., Rosenbaum J. L. (1985) Chlamydomonas α-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry 24, 473–478 [DOI] [PubMed] [Google Scholar]
- 47. Maruta H., Greer K., Rosenbaum J. L. (1986) The acetylation of α-tubulin and its relationship to the assembly and disassembly of microtubules. J. Cell Biol. 103, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sakaguchi K., Herrera J. E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C. W., Appella E. (1998) DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12, 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dornan D., Shimizu H., Perkins N. D., Hupp T. R. (2003) DNA-dependent acetylation of p53 by the transcription coactivator p300. J. Biol. Chem. 278, 13431–13441 [DOI] [PubMed] [Google Scholar]
- 50. Busch F., Mobasheri A., Shayan P., Stahlmann R., Shakibaei M. (2012) Sirt-1 is required for the inhibition of apoptosis and inflammatory responses in human tenocytes. J. Biol. Chem. 287, 25770–25781 [DOI] [PMC free article] [PubMed] [Google Scholar]