Background: EpCAM is important for intestinal epithelial integrity and is also involved in tumorigenesis.
Results: EpCAM interacts with claudin-7 and claudin-1, protects them from lysosomal degradation, and alters their intercellular distribution.
Conclusion: EpCAM modifies tight junction composition and function by regulating amounts and locations of claudins.
Significance: Effects of EpCAM on epithelial physiology and tumorigenicity may be mediated via modulation of claudins and tight junctions.
Keywords: Cell Adhesion, Cell-Cell Interaction, Intestinal Epithelium, Protein-Protein Interactions, Tight Junctions, Barrier, EpCAM, Junctional Complexes, Claudin, Lysosomal Degradation
Abstract
Epithelial cell adhesion molecule (EpCAM) (CD326) is a surface glycoprotein expressed by invasive carcinomas and some epithelia. Herein, we report that EpCAM regulates the composition and function of tight junctions (TJ). EpCAM accumulated on the lateral interfaces of human colon carcinoma and normal intestinal epithelial cells but did not co-localize with TJ. Knockdown of EpCAM in T84 and Caco-2 cells using shRNAs led to changes in morphology and adhesiveness. TJ formed readily after EpCAM knockdown; the acquisition of trans-epithelial electroresistance was enhanced, and TJ showed increased resistance to disruption by calcium chelation. Preparative immunoprecipitation demonstrated that EpCAM bound tightly to claudin-7. Co-immunoprecipitation documented associations of EpCAM with claudin-7 and claudin-1 but not claudin-2 or claudin-4. Claudin-1 associated with claudin-7 in co-transfection experiments, and claudin-7 was required for association of claudin-1 with EpCAM. EpCAM knockdown resulted in decreases in claudin-7 and claudin-1 proteins that were reversed with lysosome inhibitors. Immunofluorescence microscopy revealed that claudin-7 and claudin-1 continually trafficked into lysosomes. Although EpCAM knockdown decreased claudin-1 and claudin-7 protein levels overall, accumulations of claudin-1 and claudin-7 in TJ increased. Physical interactions between EpCAM and claudins were required for claudin stabilization. These findings suggest that EpCAM modulates adhesion and TJ function by regulating intracellular localization and degradation of selected claudins.
Introduction
Epithelial cell adhesion molecule (EpCAM),2 also known as CD326, TACSTD1, and TROP-1, is a type I transmembrane glycoprotein that was first identified as a tumor-associated antigen (1). EpCAM is highly expressed by many carcinomas, and its possible involvement in tumor development, progression, and metastasis has been extensively studied. The therapeutic potential of anti-EpCAM antibodies is also being actively explored (2). In some settings, EpCAM has pro-oncogenic activity, promoting cell proliferation, motility, carcinogenesis, and metastasis. A recent report links pro-oncogenic effects of EpCAM to sequential proteolysis that releases a soluble intracellular fragment that enhances TCF/LEF1-dependent signaling, thereby promoting expression of downstream cell cycle regulators, including c-Myc (3). In contrast, in other tumors EpCAM appears to suppress cancer progression (2, 4). Mechanisms responsible for the divergent activities of EpCAM in different contexts have not yet been elucidated.
EpCAM is also widely expressed by developing epithelia in normal individuals, and high levels persist in some epithelia in adults (e.g. in the gastrointestinal tract). Although EpCAM has been less extensively studied in normal tissues and nontransformed cells than in cancer, there are indications that EpCAM influences epithelial homeostasis. Expression of EpCAM promotes aggregation of fibroblasts in suspension, suggesting that EpCAM can function as an intercellular adhesion molecule (5). It has been reported that EpCAM forms multimers within the plasma membrane and that adhesive interactions are homophilic and end-to-end. Binding of the short intracellular C terminus of EpCAM to the cytoskeleton is prerequisite for intercellular adhesion (6, 7).
In vivo studies also suggest a role for EpCAM in intercellular adhesion in epithelia. Zebrafish embryos with null mutations in EpCAM displayed defective epithelial morphogenesis with attenuated epiboly involving cells of the enveloping layer of primordial epidermis (8). In Xenopus, ectopic EpCAM expression induced tissue mixing during gastrulation (9). In humans, EpCAM mutations cause congenital tufting enteropathy, a rare diarrheal syndrome that results from severe intestinal epithelial dysplasia and loss of epithelial integrity (10). Finally, very recent studies indicate that EpCAM mutations in mice also cause major perturbations in intestinal epithelial homeostasis (11, 12).
The formation of apical junctional complexes (AJC) involving adjacent cells is required for the establishment and maintenance of normal epithelial structure and function. Typical AJC are composed of tight junctions (TJ) and adherens junctions, and AJC may also include gap junctions and desmosomes (13). TJ represent the most apical components of AJC, forming belt-like structures that encircle individual cells and closely approximate adjacent cells to each other. TJ comprise the principal components of the paracellular diffusion barrier that determines bidirectional epithelial permeability of small molecules and water, and TJ also restrict apical-basolateral diffusion of membrane components maintaining the structural and functional polarity of individual epithelial cells (14). Abrogation of barrier function in epithelia that interface with the environment is associated with a variety of gastrointestinal, renal, and cutaneous diseases (15).
TJ are composed of several transmembrane and membrane-associated proteins, including the tetraspan transmembrane proteins occludin and one or more of more than 20 structurally related claudins. These transmembrane proteins interact with each other and with additional membrane and nonmembrane proteins, including intracellular zonula occludens (ZO-1 and ZO-2) and other PDZ domain-containing proteins (16). Despite their elaborate morphology and substantial structural organization, AJC are highly dynamic with regard to composition and function (17). AJC (including TJ) are remodeled in physiological and pathological circumstances such as organogenesis and the epithelial-mesenchymal transition associated with cancer progression (18, 19). AJC composition may also be routinely adjusted to adapt to continuously changing microenvironments associated with different developmental stages, metabolic stress, and/or epithelial injury (20).
Many details regarding regulation of AJC composition and function remain to be determined. Recognizing that EpCAM has been implicated in intercellular adhesion in multiple settings and that EpCAM mutations disrupt intestinal epithelial homeostasis, we predicted that exploring EpCAM function in vitro in intestinal epithelial cells (IEC) with well characterized intercellular adhesive properties would be informative. Studies described herein reveal that EpCAM regulates TJ formation, stability, composition, and function in human colon cancer cells (T84 and Caco-2 cells) by interacting with several components of AJC, notably claudin-7 and claudin-1. Association of EpCAM with claudin-7 and claudin-1 promotes claudin stability and accumulation and influences claudin distribution in these polarized epithelial cells with functional consequences.
EXPERIMENTAL PROCEDURES
Cell Lines
T84 cells were kindly provided by Dr. Asma Nusrat (Emory University), and Caco-2 cells were obtained from Drs. Toni Antalis and Marguerite Buzza (University of Maryland). T84 cells were cultured in DMEM/F-12 supplemented with 6% fetal bovine serum (FBS), 15 mm HEPES (pH 7.4), 100 units/ml penicillin, and 100 mg/ml streptomycin. Caco-2 cells were grown in DMEM containing 10% FBS, 15 mm HEPES (pH 7.4), nonessential amino acids, 100 units/ml penicillin, and 100 mg/ml streptomycin. COS-7 cells were from American Type Culture Collection (Manassas, VA).
Antibodies
Polyclonal anti-EpCAM antibodies (Ab) used for Western blotting were raised in rabbits by immunization with a fusion protein composed of the extracellular domain of mouse EpCAM fused in-frame with the Fc portion from human IgG1 and used after affinity purification to eliminate reactivity with human IgG and enrich reactivity with EpCAM. Mouse monoclonal anti-EpCAM Ab (mAb, clones EBA-1 and 323/A3, used for immunoprecipitation and immunostaining) and anti-ubiquitin (P4D1) mAb were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and rabbit anti-EpCAM (1141-1) mAb used for immunostaining was from Epitomics (Burlingame, CA). Anti-occludin, anti-ZO-1, anti-claudin-1, -2, -4, -5, and -7, and mouse anti-E-cadherin mAb were from Invitrogen. Rat anti-CD26 mAb was purchased from Bender MedSystems; mouse anti-Lamp1 (H4A3) and mouse anti-β-catenin were from BD Biosciences (San Jose, CA), mouse anti-β-actin mAb and rabbit anti-myosin IIA were from Sigma, and rat anti-HA Ab was from Roche Applied Science. Mouse anti-desmoglein 2 mAb was a gift from Dr. Nusrat (Emory University), and rabbit anti-Na/K-ATPase Ab was kindly provided by Dr. Haruo Homareda (Kyorin University, Japan).
Construction of EpCAM shRNA-containing and Gene Expression Vectors
Two plasmids containing shRNA oligonucleotides targeting the EpCAM sequences (CTACAAGCTGGCCGTAAAC (EpCAM shRNA 1) and GGACGAAGACATCTTTGAA (EpCAM shRNA 2)) were constructed. Corresponding synthetic oligonucleotides were annealed and cloned into pSUPER.retro.puro vector (OligoEngine, Seattle) at BglII and XhoI sites. The resulting plasmids were verified by restriction enzyme digestion and DNA sequencing. pcDNA3-EpCAMHA-encoding plasmids were produced by cloning sequence-verified PCR fragments of human EpCAMHA into pcDNA3 (Invitrogen) that had been digested with BamHI and XhoI. Sequence-verified claudin-7HA PCR fragments were cloned into pcDNA3 at BamHI and EcoRI sites to produce pcDNA3-claudin-7HA. pTRIP-claudin-1 was kindly provided by Dr. Charles Rice (Rockefeller University). EpCAM mutations of interest were introduced into cDNA using a QuikChange kit (Stratagene, La Jolla, CA) following the manufacturer's instructions.
Cell Transfection, Virus Infection, and Derivation of Stable Cell Lines
pSUPER.retro.puro or pSUPER.retro.puro plasmids containing EpCAM shRNA 1 or 2 were transfected into Phoenix-Ampho cells (from Dr. Gary Nolan, Stanford University) using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Retrovirus-containing supernatants were collected and used to infect T84 or Caco-2 cells. Following selection with puromycin for 10–14 days, stable EpCAM shRNA transductants were pooled and analyzed for EpCAM expression by immunoblotting. Single cell suspensions were prepared from pooled T84/EpCAM shRNA 2 and Caco-2/EpCAM shRNA 2 cells and replated at limiting dilutions to allow identification of single colonies to grow. Clones were isolated, expanded, and analyzed for EpCAM expression by flow cytometry. Clone 3 T84/shEpCAM 2 cells that exhibited dramatically reduced EpCAM expression were transiently transfected with pcDNA3 or pcDNA3 containing EpCAMHA or EpCAMmutHA (EpCAM(A279IG283I)HA) by electroporation (Gene Pulser, Bio-Rad) at 350 V for 20 s. pcDNA3 and pcDNA3 plasmids containing EpCAMHA or EpCAMmutHA were transfected into a clone of Caco-2/shEpCAM 2 cells that exhibited low EpCAM levels using Lipofectamine 2000. Plasmid-expressing cells were selected with 700 μg/ml G418 for 14 days and expanded, and cells that expressed high levels of EpCAM (comparable with levels in unmanipulated Caco-2 cells) were isolated via preparative flow cytometry.
Inhibition of Claudin Expression with siRNAs
Double-stranded RNA oligonucleotides containing claudin-1 siRNA that corresponded to a previously described sequence (21) were synthesized by Thermo Scientific Dharmacon (Lafayette, CO). Claudin-7 siRNA was purchased from Invitrogen. T84 or Caco-2 cells were transfected with claudin-7 siRNA with or without claudin-1 siRNA or negative control siRNA duplexes using electroporation (Gene Pulser, Bio-Rad) at 320 V (for T84) or 280 V (for Caco-2) for 20 s. Transfected cells were plated into 10-cm dishes, incubated for 24 h, and re-plated into Transwell chambers for monitoring TEER or as sources of cell lysates for monitoring protein levels by Western blotting.
Quantitative RT-PCR
Total RNA was prepared using RNeasy kits (Qiagen) and reverse-transcribed with SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen). Quantitative RT-PCR was performed using the following primers: claudin-7 (forward, 5′-TTTTCATCGTGGCAGGTCTTG-3′; reverse, 5′-CTTGCTCTCATTCCCAGGACAG-3′); claudin-1 (forward, 5′-TTGGTCAGGCTCTCTTCACTGG-3′; reverse, 5′-CTTGCTCTCATTCCCAGGACAG-3′), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (forward, 5′-ACAACTTTGGTATCGTGGAAGGAC-3′; reverse, 5′-AGGGATGATGTTCTGGAGAGCC-3′). PCR products were generated using a CFX96 real time PCR detection system (Bio-Rad) and quantified using SYBR Green PCR Master Mix (Applied Biosystems) in conjunction with CFX ManagerTM software version 1.5.
Flow Cytometry
T84 and Caco-2 cells were immunostained with mouse anti-EpCAM mAb, followed by FITC-conjugated anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA). Analytical flow cytometry was performed with a FACSCalibur (BD Biosciences), and flow cytometry sorting utilized a FACSAria II (BD Biosciences).
Cell Proliferation
T84 or Caco-2 cells were introduced into 96-well plates (3 × 103 cells/well) and cultured at 37 °C for various periods. Premixed WST-1 cell proliferation reagent (Clontech) was added, and incubations were continued for 1.5 h. Proliferation was assessed by measuring A450 in each well using an ELISA reader. A450 was directly proportional to viable cell number.
Cell Migration
Cell migration was evaluated using an in vitro wound healing assay. EpCAM knockdown or control Caco-2 cells (4.0 × 104) were plated into each well of 6-well culture plates that contained defect-creating ibidi inserts (Applied Biophysics Inc., Troy, NY). After overnight incubation, inserts were removed; media were replaced with low serum (0.1% FBS) or 10% FBS-containing media, and incubations were continued for an additional 24 h. Phase contrast images of “wounds” that were created by removing the inserts were acquired at identical locations at the beginning and conclusion of the assay, and residual wound areas were quantified using ImageJ.
Preparation of Epithelial Cell Monolayers and Measurement of Trans-epithelial Electrical Resistance
To establish polarized IEC monolayers, T84 cells or Caco-2 cells (6 × 105 and 3 × 105 cells per well, respectively) were plated into Transwell chambers with 12-mm diameter polycarbonate filters with pore sizes of 0.4 μm (Corning-Costar, Cambridge, MA). T84 and Caco-2 cells were cultured for 8–10 days or 21–28 days, respectively, with media changes every other day. TEER was monitored with an EVOM II electrovoltohmeter (World Precision Instruments, Sarasota, FL) and is expressed as megohms·cm2.
Immunoblotting, Immunoprecipitation, and Co-immunoprecipitation
Cells were lysed with Triton X-100 lysis buffer or RIPA lysis buffer as indicated and normalized for protein content using a Bradford protein assay (Bio-Rad) (22). EpCAM levels were assessed by immunoblotting with rabbit anti-EpCAM polyclonal Ab. To detect and quantify claudin-7 ubiquitination, cells were lysed in RIPA buffer, and the resulting cell lysates were normalized for protein concentration and immunoprecipitated with anti-claudin-7 prior to blotting with anti-ubiquitin Ab. Protein associations were assessed via co-immunoprecipitation of proteins from Triton X-100 lysates as described previously (22). For Western blotting, proteins in immunoprecipitates or cell lysates were resolved by SDS-PAGE and transferred onto nitrocellulose membranes that were subsequently incubated with the indicated Ab. Proteins of interest were visualized using horseradish peroxidase-conjugated secondary Ab (GE Healthcare) and enhanced chemiluminescence (Pierce).
Identification of EpCAM-associated Proteins via Mass Spectrometry
EpCAM-containing immunoprecipitates were resolved using SDS-PAGE and stained with colloidal Coomassie Blue (Invitrogen). After de-staining, protein bands were excised from gels, and protein characterization was performed by ProtTech, Inc. (Norristown, PA) using NanoLC-MS/MS peptide sequencing technology. Briefly, proteins from the gel bands were digested with modified sequencing grade trypsin (Promega, Madison, WI), and the resulting peptide mixture was subjected to tandem mass spectrometry to allow peptide sequencing. An ion trap mass spectrometer (Thermo, Palo Alto, CA) coupled with a high performance liquid chromatography system running a 75-μm inner diameter C18 column was used. MS/MS spectra were used to search recent nonredundant protein data bases from GenBankTM using ProtQuest software suite from ProtTech.
Immunofluorescence Microscopy and Image Analysis
Cells grown on coverslips were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 before immunostaining, whereas cells grown on Transwell filters were fixed with cold acetone/ethanol (3:1) or cold methanol. After blocking with 1% BSA, cells were stained with the indicated primary Ab or control IgG, followed by Alexa 488- or Alexa 568-conjugated secondary Ab (Invitrogen). Frozen sections of human small intestine were fixed with cold acetone/ethanol (3:1). After blocking with 3% dry milk (Bio-Rad), including 5% normal donkey serum or 5% normal goat serum (as appropriate) for 1 h at room temperature, sections were stained with the Ab of interest followed by Alexa 488- or Alexa 568-conjugated secondary Abs (Invitrogen). Stained cells or tissue sections were mounted in ProLong Gold antifade reagent containing 4,6-diamidino-2-phenylindole (DAPI) (Invitrogen) prior to visualization with an LSM510 confocal laser-scanning microscope (Zeiss) and analysis with LSM Image Browser 4.0. Quantification of co-localization of claudin-7 or claudin-1 with ZO-1 was accomplished using AIM software (Zeiss), and the results are presented as co-localization coefficients (claudin-associated pixels/ZO-1-associated pixels normalized to pixels above background; value = 1 indicates complete co-localization). TJ stability after calcium depletion was assessed by quantitative image analysis using AngioTool (23). The complexity of the network of TJ, as revealed by staining with an anti-occludin antibody, was evaluated by assessing the total length of the network and the number of junctions (locations where more than two segments of the network converge) per field.
Statistics
Probability values were calculated using the Student's t test.
RESULTS
EpCAM Localizes to the Lateral Interfaces of Polarized Epithelial Cells
Because EpCAM is expressed at high levels in intestinal epithelia and is important for intestinal epithelia architecture and function, we studied the human colon cancer cell lines T84 and Caco-2 to determine whether EpCAM regulated AJC composition or function. T84 and Caco-2 cells are useful tools for investigating intercellular adhesion and epithelial polarity because they retain characteristics of normal enterocytes and readily form monolayers that develop TEER. We first assessed EpCAM expression and localization in relationship to TJ structures that are key determinants of epithelial polarity. Consistent with previous reports (24), flow cytometry confirmed that T84 cells and Caco-2 cells expressed high levels of EpCAM on cell surfaces (data not shown). Characterization of EpCAM expression and localization in subconfluent T84 cells using confocal immunofluorescence microscopy revealed EpCAM on cell surfaces and in punctate intracellular accumulations as well (Fig. 1A). Confocal scanning coupled with image integration indicated that EpCAM localized to the lateral interfaces of polarized T84 cells in post-confluent monolayers characterized by robust formation of TJ manifesting as well as defined occludin and ZO-1-containing networks (Fig. 1, B and C). We observed that EpCAM was present below TJ and that EpCAM and TJ protein staining did not appear to co-localize. Importantly, analogous results were obtained in studies of EpCAM and TJ protein distribution in human small intestinal epithelial cells in vivo (Fig. 1D).
FIGURE 1.
EpCAM localizes to lateral interfaces of human intestinal epithelial cells below tight junctions. A, human carcinoma (T84) cells growing as colonies on coverslips were stained with anti-EpCAM and examined using confocal laser immunofluorescence microscopy. B and C, post-confluent T84 cell monolayers growing on membranes in Transwell chambers were fixed, and double-stained with anti-EpCAM and anti-occludin (B, XZ image) or anti-EpCAM and anti-ZO-1 (C, three-dimensional image). D, lightly fixed frozen sections of human small intestine were stained with anti-EpCAM and anti-occludin and analyzed via confocal microscopy. Scale bars, 10 μm (A–C), or 20 μm (D).
Previous studies suggested that EpCAM regulated cadherin-mediated intercellular adhesion (25), and our initial experiments indicated that EpCAM and E-cadherin both accumulated on lateral interfaces of IEC. However, confocal microscopy revealed that these two proteins did not precisely co-localize in T84 cell monolayers (supplemental Fig. 1A). Co-immunoprecipitation studies also did not indicate that EpCAM and E-cadherin were tightly associated in T84 cells (supplemental Fig. 1B).
Reductions in EpCAM Expression Change Intestinal Epithelial Cell Morphology and Physiology
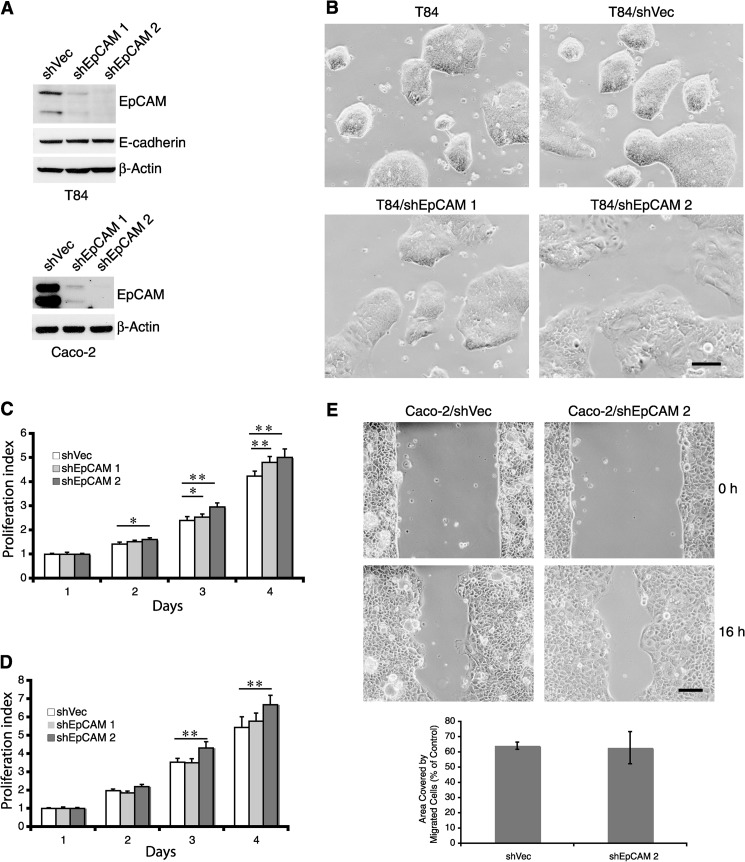
We utilized two different shRNAs to inhibit EpCAM expression by T84 and Caco-2 cells to begin to assess the functional significance of EpCAM expression by IEC. Western blotting revealed that shEpCAM 2 reduced EpCAM expression more efficiently than shEpCAM 1 in both T84 and Caco-2 cells (Fig. 2A). Unmanipulated subconfluent T84 cells formed tight dome-shaped colonies that exhibited compaction (apparent loss of intercellular borders). Although transduction with control retrovirus did not alter colony morphology, introduction of shEpCAM 1, and especially shEpCAM 2, caused modest flattening of colonies that were composed of cells whose borders could be discerned (Fig. 2B), consistent with the concept that EpCAM influences intercellular adhesion.
FIGURE 2.
Reduction of EpCAM expression changes the morphology and physiology of colon cancer cells. T84 and Caco-2 cells were infected with retroviruses expressing control or EpCAM shRNAs, and stable transductants were selected using puromycin. A, cell lysates from transduced T84 (upper panel) or Caco-2 (lower panel) cells were assessed for EpCAM content using Western blotting. B, phase contrast images reveal morphologies of unmanipulated as well as control vector- or EpCAM shRNA-transduced T84 cells. Cells were plated in 10-cm dishes and cultured for 6 days prior to imaging. C and D, control vector- or EpCAM shRNA-transduced T84 (C) or Caco-2 (D) cells were assessed for proliferative activity using the WST-1 assay. Bars represent daily A450 determinants (means ± S.D.) for each cell line normalized to the day 1 A450 for that cell line. E, abilities of control vector-transduced and EpCAM shRNA 2-expressing Caco-2 cells to migrate were studied using a wound healing assay. Defects in monolayers were created by adding cells into 6-well plates containing inserts in each well. After incubation for 24 h in medium containing 0.1% FBS, inserts were removed, and phase contrast photomicrographs were obtained at the indicated times. Representative images from n = 3 experiments are shown. Residual wound areas (means ± S.E.) after 16 h were determined using ImageJ. Scale bars, 200 μm. *, p < 0.05; **, p < 0.01.
Because several prior reports related EpCAM expression to proliferation and migration (3, 26, 27), we measured these parameters in control and EpCAM knockdown T84 and Caco-2 cells. Somewhat surprisingly, in both cell lines, knockdown of EpCAM expression led to 10–20% increases in cell proliferation that were inversely correlated with EpCAM levels (Fig. 2, C and D). These results are consistent with a report indicating that overexpression of EpCAM in invasive colon cancer cell lines down-regulated cell proliferation (28). T84 cells did not migrate appreciably, independent of levels of EpCAM (data not shown). Characterization of Caco-2 cell migration in an in vitro wound healing assay conducted in low serum-containing media that mitigated effects of EpCAM on cell proliferation (supplemental Fig. 2A) revealed that EpCAM reduction did not appreciably alter “wound healing” (Fig. 2E).
EpCAM Regulates Tight Junction Function
To additionally explore the role of EpCAM in intercellular adhesion, we characterized TJ structure and function in control and EpCAM knockdown T84 and Caco-2 cells. Confocal laser microscopy of post-confluent T84 monolayers demonstrated continuous networks of ZO-1-containing TJ that were well defined in both EpCAM knockdown and control vector-transduced cells (data not shown, also see Fig. 9D). Barrier function of TJ can be quantified by measuring TEER. Thus, we made serial measurements of TEER exhibited by monolayers of T84 and Caco-2 cells expressing usual or reduced levels of EpCAM. As reported previously, T84 monolayers acquired higher levels of TEER than Caco-2 monolayers and also acquired TEER more quickly (29, 30). Reductions of EpCAM led to significant alterations in TEER acquisition in both cell lines. In T84 monolayers, EpCAM reduction caused TEER to increase more rapidly than in control cells, but maximal levels of TEER exhibited by monolayers of T84 cells with normal and reduced levels of EpCAM were not different (Fig. 3A). Rates of TEER acquisition in post-confluent monolayers did not correlate with increases in cell numbers (supplemental Fig. 2B), suggesting that this difference is not due to altered cell proliferation. Interestingly, EpCAM reduction in Caco-2 cells resulted in acquisition of maximal TEER that was more than 2-fold higher than in monolayers composed of cells with high levels of EpCAM (Fig. 3B).
FIGURE 9.
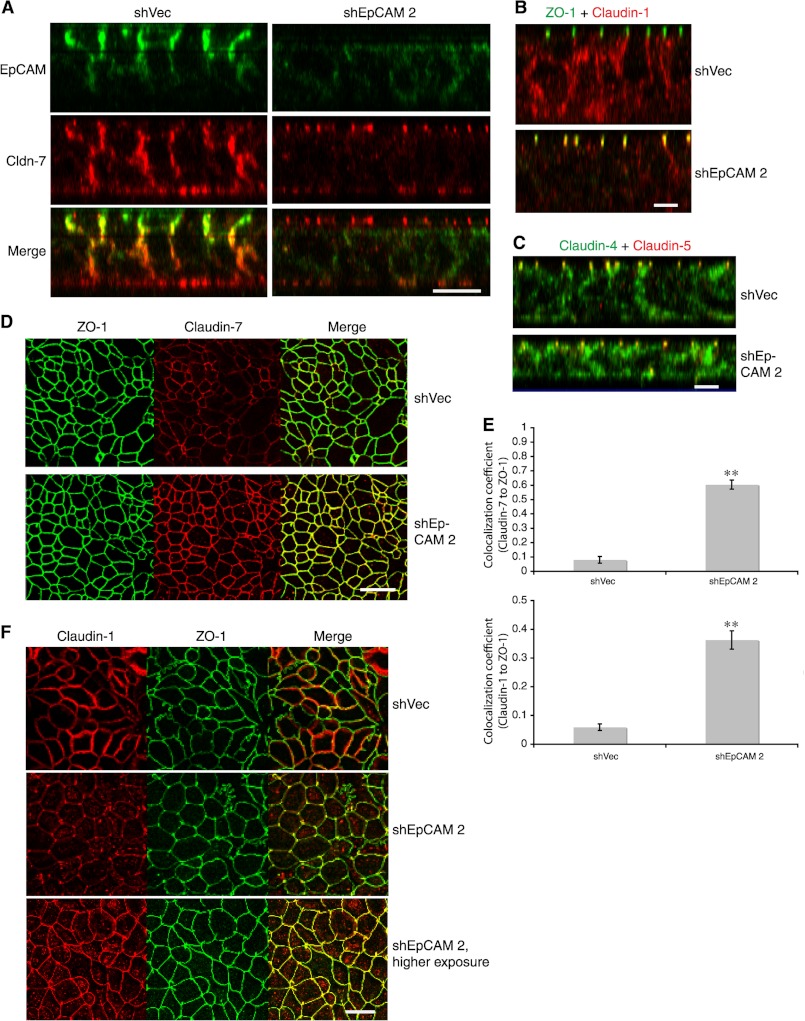
Reduction of EpCAM levels promotes accumulation of claudin-7 and claudin-1 in tight junctions. A and B, shVector- or shEpCAM-transduced T84 cell monolayers were grown in Transwells for 9 days to allow acquisition of maximal TEER, fixed with cold acetone/ethanol (3:1), stained for EpCAM and claudin-7 (A), ZO-1 and claudin-1 (B), or claudin-4 and claudin-5 (C) and analyzed using confocal microscopy (XZ images). D, ZO-1 and claudin-7 distributions were assessed in T84 cells using an analogous approach (en face images). Each panel depicts representative data from three to four experiments. Scale bars, 10 μm (A–C) or 20 μm (D). E, shVector- or shEpCAM-transduced T84 cell monolayers grown in Transwells were fixed and stained for ZO-1 and claudin-1 or claudin-7 as in B and D. Ten randomly selected XZ images as shown in B corresponding to each condition were analyzed to determine co-localization of claudin-7 (upper panel) or claudin-1 (lower panel) with ZO-1 signals. Co-localization coefficients were shown. **, p < 0.01. F, Caco-2 cell monolayers with maximal TEER acquired after culturing for 23 days were fixed with cold methanol and then stained for claudin-1 and ZO-1 (en face images). Representative data from one of three experiments are shown. Scale bar, 10 μm.
FIGURE 3.
Regulation of TEER and tight junction stability by EpCAM. A and B, monolayers of control vector- or EpCAM shRNA-transduced T84 (A) or Caco-2 (B) cells were cultured in Transwells, and TEERs were determined daily for T84 or every other day for Caco-2 cells. Mean TEERs ± S.E. are depicted. Representative results from one of three experiments are shown. C, T84 cell monolayers were grown and monitored for acquisition of TEER. Maximal TEERs were obtained after 9 days of incubation, and monolayers were subsequently incubated in EGTA-containing (calcium-depleted) or regular medium for 40 min, fixed, and stained with anti-occludin. D, digital images corresponding to 10 random fields of anti-occludin-stained retrovirus-transduced T84 monolayers that had been calcium-depleted were subjected to quantitative analysis using AngioTool. Aggregate lengths of tight junction networks and numbers of network junctions per field (means ± S.E.) are shown. Representative results from one of three experiments are presented. Scale bars, 10 μm (C, upper panel) or 20 μm (C, lower panel). *, p < 0.05; **, p < 0.01.
We also determined the impact of calcium depletion on TJ integrity. Control vector- and shEpCAM-treated T84 cells were allowed to form monolayers with maximal TEER; extracellular calcium was removed via exposure to EGTA (31), and TJ were visualized via occludin staining of fixed cell monolayers 40 min later. TJ were significantly more resistant to disruption by calcium depletion after EpCAM knockdown, and the degree of resistance was inversely proportional to the levels of EpCAM that were expressed (Fig. 3, C and D). In aggregate, these results suggest that EpCAM regulates TJ formation, maintenance, and function.
It has been reported that EpCAM modulates adhesion mediated by E-cadherin (25), a critical component of adherens junctions in epithelia. Adherens junctions are also required for TJ formation (32). Thus, we compared E-cadherin as well as β-catenin expression and localization in T84 cells expressing the usual high levels of EpCAM and EpCAM knockdown cells. We also examined desmoglein 2 expression and localization as a surrogate for desmosomes. Immunofluorescence staining of control and EpCAM knockdown T84 cell monolayers indicated that reduction in EpCAM expression did not change E-cadherin and β-catenin expression or localization (Fig. 4, A and B). This is consistent with the results of co-localization studies and reciprocal co-immunoprecipitation experiments described above (see supplemental Fig. 1). EpCAM knockdown also did not lead to obvious alterations in desmoglein 2 expression or distribution (Fig. 4C). We also examined the distributions of cell surface proteins that are indicative of intestinal epithelial cell polarity. EpCAM knockdown did not change in expression or distribution of CD26 (on apical membrane surfaces), Na/K-ATPase (on basolateral surfaces) (Fig. 4D), or the subapical localizations myosin IIA (Fig. 4E). We conclude that effects of EpCAM on TJ are selective, do not cause gross abnormalities in cell polarity, and are not secondary to changes in E-cadherin expression or disruption of microfilament networks.
FIGURE 4.
EpCAM depletion does not markedly alter the distribution of adherens junction- or desmosome-associated proteins or epithelial polarity. Stable control vector- or shEpCAM 2-transduced T84 cells were grown in Transwells for 8–10 days to allow development of maximal TEER and then fixed and stained for ZO-1 and E-cadherin (E-cad) (A), occludin (Occl) and β-catenin (β-cat) (B), ZO-1 and desmoglein 2 (Dsg2) (C), for Na/K-ATPase (red) and CD26 (green) (D), or for myosin IIA (red) and EpCAM (green) (E). Results shown are representative of those observed in three experiments. Scale bars, 10 μm (A–C, left panels, and D and E, XZ images), or 20 μm (right panels, XY images).
EpCAM Interacts with Claudin-7 and Claudin-1
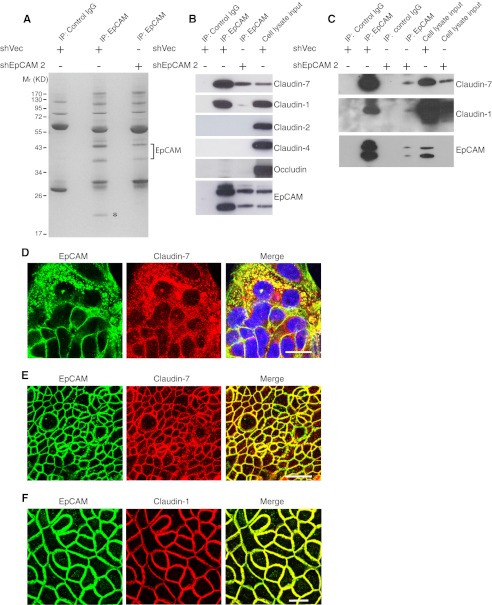
To gain insights into mechanisms by which EpCAM might influence TJ function and IEC physiology, we characterized proteins that associated with EpCAM. To accomplish this, we carried out preparative immunoprecipitation of detergent-solubilized control and EpCAM knockdown T84 cell lysates with anti-EpCAM mAb in the absence of cross-linking reagents and identified SDS-PAGE-resolved proteins that were more abundant in immunoprecipitates using specific mAb and control cells. We also screened for proteins that were less abundant in immunoprecipitates from EpCAM knockdown cells. Differentially represented proteins were eluted from gels, and peptides generated by exhaustive trypsinization were characterized by mass spectrometry. As anticipated, anti-EpCAM mAb precipitated 43- and 37-kDa species that corresponded to EpCAM (Fig. 5A). Strikingly, there was a single ∼20-kDa band in EpCAM immunoprecipitates from vector-transduced cells that was markedly less abundant in immunoprecipitates from T84 cells with reduced EpCAM expression (see asterisk in Fig. 5A). Mass spectrometry indicated that this 20-kDa protein corresponded to claudin-7.
FIGURE 5.
EpCAM interacts with claudin-7 and claudin-1. A, control vector-transduced or EpCAM knockdown T84 cells were solubilized in Triton X-100-containing buffer, and lysates were immunoprecipitated (IP) with control IgG or anti-EpCAM Ab. Immunoprecipitates were resolved via SDS-PAGE, and gels were stained with Coomassie Blue. Differentially represented bands/proteins were characterized using mass spectrometry. B and C, immunoprecipitates from vector-transduced or EpCAM knockdown T84 cells (B) or Caco-2 cells (C) acquired as in A were fractionated using SDS-PAGE and immunoblotted with anti-claudin-7, -1, -2 or -4 or anti-occludin Abs. D–F, co-localization of EpCAM with claudin-7 and claudin-1 detected using confocal scanning laser microscopy. D, T84 cells were grown on coverslips for 2 days, fixed with paraformaldehyde, and permeabilized with 0.5% Triton X-100. E, T84 monolayers were cultured on Transwell filters for 8 days, fixed with cold acetone/ethanol (3:1), and stained with anti-EpCAM and anti-claudin-7 Abs. A mid-level optical section (below TJ) is displayed. F, Caco-2 monolayers were grown on Transwell filters for 23 days, fixed with cold methanol, and stained for EpCAM and claudin-1. A mid-level optical section (below TJ) is displayed. Scale bars, 20 μm.
To confirm that EpCAM associated with claudin-7 in T84 cells, we prepared immunoprecipitates with anti-EpCAM mAb and probed for claudin-7 in immunoprecipitates by Western blotting. Claudin-7 was readily detected in anti-EpCAM immunoprecipitates but not in those prepared with control IgG (Fig. 5B). Because the claudin family is composed of ∼25 members, we also determined whether EpCAM associated with other claudin-7-related proteins. In addition to claudin-7, EpCAM co-immunoprecipitated with closely related claudin-1 (33), but not claudin-2, claudin-4 or the unrelated TJ protein occludin (Fig. 5B). We subsequently determined that Caco-2 cells also expressed claudin-7 and claudin-1 (supplemental Fig. 3) and that EpCAM also associated with these proteins in Caco-2 cells (Fig. 5C). Note, however, that EpCAM and claudin-7 were much more abundant in T84 cells than in Caco-2 cells (supplemental Fig. 3). We also assessed EpCAM-claudin-7/claudin-1 interactions in subconfluent (Fig. 5D) and polarized T84 (Fig. 5E) and Caco-2 (Fig. 5F) cell monolayers using confocal microscopy. EpCAM co-localized with claudin-7 and claudin-1 in all of these settings. Interestingly, results obtained with subconfluent T84 cells suggested that EpCAM and claudin-7 were associated in intracellular compartments as well as on cell surfaces (Fig. 5D).
Claudin-7 Associates with Claudin-1 and Facilitates Incorporation of Claudin-1 into EpCAM-containing Complexes
Claudin-7 and claudin-1 are closely related (∼50% homologous at the amino acid level (33)), suggesting that they might be similar with regard to their ability to interact with EpCAM. Although both claudin-7 and claudin-1 co-immunoprecipitate with EpCAM, preferential association of EpCAM with claudin-7 as compared with claudin-1 (Fig. 5, B, 2nd and 4th lanes, and C, 2nd and 5th lanes) suggested that EpCAM-claudin-7 associations were of higher avidity than EpCAM-claudin-1 associations. These results are consistent with the observation that mass spectrometry only identified claudin-7 in EpCAM immunoprecipitates (Fig. 5A). Because claudin family members can interact with each other, it was possible that claudin-7 and claudin-1 might interact with each other and that EpCAM-claudin-1 associations might be mediated by claudin-7. To assess this possibility, we examined EpCAM-claudin-1 association in T84 cells with reduced claudin-7 levels. As shown in Fig. 6A, the presence of claudin-1 in EpCAM immunoprecipitates was markedly decreased after claudin-7 knockdown with specific siRNA. This suggested that claudin-1 might bind to claudin-7 that, in turn, interacted with EpCAM. Consistent with this conclusion, claudin-7 in T84 cell lysates co-immunoprecipitated with claudin-1 (Fig. 6B). Because claudin-1 and claudin-7 are homologous, it was important to exclude antibody cross-reactivity as a confounder in these experiments. To address this, HA-tagged claudin-7 and nontagged claudin-1 were ectopically expressed in EpCAM-negative (data not shown) COS-7 fibroblasts, and detergent lysates were immunoprecipitated with anti-HA antibodies. We observed claudin-1 to claudin-7 co-immunoprecipitation and confirmed that anti-claudin-1 and anti-claudin-7 antibodies were not cross-reactive in Western blots (Fig. 6C). The simplest (but not exclusive) explanation for this result is that claudin-1 and claudin-7 interact directly.
FIGURE 6.
Claudin-7 mediates EpCAM-claudin-1 interactions by associating with claudin-1. A, T84 cells transfected with negative control (siNeg) or claudin-7 siRNA (siC7) were solubilized, and cell lysates were immunoprecipitated (IP) with control IgG or anti-EpCAM Ab as in Fig. 5A. Immunoprecipitates and cell lysates were fractionated using SDS-PAGE and immunoblotted with anti-claudin-1, anti-claudin-7, or anti-EpCAM. B, anti-claudin-1 immunoprecipitates of T84 cell lysates acquired as in A were fractionated using SDS-PAGE and immunoblotted with anti-claudin-7 or anti-claudin-1. C, COS-7 cells were co-transfected with pTRIP-claudin-1 and pcDNA3-claudin-7HA or control empty vectors (Vec) using Lipofectamine. Cells were lysed in Triton X-100-containing buffer 24 h after transfection, and cell lysates were immunoprecipitated with anti-HA antibody. SDS-PAGE-resolved immunoprecipitates were blotted with anti-claudin-1 or anti-claudin-7.
EpCAM Protects Claudin-7 and Claudin-1 from Degradation in Lysosomes
While conducting the immunoprecipitation and Western blotting experiments described above, we observed that claudin-7 and claudin-1 levels were selectively and dramatically decreased in Triton X-100 lysates from T84 cells (Fig. 7A) and RIPA buffer lysates from T84 (Fig. 7B, left panel) and Caco-2 (Fig. 7B, right panel) cells that had been treated with viruses encoding two different EpCAM shRNAs. Note that the amounts of claudin-7 and claudin-1 in virus-treated cells were directly proportional to the amounts of EpCAM that were present and that levels of claudin-2, claudin-4, occludin, ZO-1, and E-cadherin did not correlate with EpCAM levels.
FIGURE 7.
EpCAM protects claudin-7 and claudin-1 from degradation via a lysosome-dependent pathway. A, shVector- or shEpCAM-transduced T84 cells were lysed in Triton X-100-containing buffer, and normalized amounts of solubilized proteins were quantified by immunoblotting (IB). B, shVector- or shEpCAM-transduced T84 cells or Caco-2 cells were lysed with RIPA buffer, and supernatants were cleared via centrifugation at 12,000 × g for 15 min. Normalized amounts of cell lysate proteins were resolved using SDS-PAGE and immunoblotted for the indicated proteins. β-Actin was used as a loading control. C, T84 cell total RNA was analyzed for claudin-7 and claudin-1 mRNA content using real time RT-PCR. Claudin mRNA levels in each sample are expressed relative to GAPDH mRNA levels and were normalized such that expression levels in control (vector-transduced) cells = 1. D and E, T84 cells were plated for 40 h and subsequently treated with the lysosome inhibitor chloroquine for 24 h (D) or the proteasome inhibitor lactacystin for 20 h (E) (or appropriate diluent controls) at the indicated concentrations, and RIPA buffer lysates were prepared. Cell lysates were normalized for protein concentrations; lysate proteins were resolved via SDS-PAGE, and EpCAM, claudin-7, and claudin-1 were detected via immunoblotting. β-Actin was used as a loading control. F, vector-transduced control and EpCAM shRNA 2-expressing T84 cells were treated with, or without, 100 μm chloroquine for 24 h and lysed with RIPA buffer. Cell lysates were normalized for protein concentrations and immunoprecipitated with anti-claudin-7 antibody. Immunoprecipitates were blotted with anti-ubiquitin (Ub) and anti-claudin-7.
To address the possibility that these results reflected differential solubilization of junction-associated proteins, we lysed T84 cells directly in SDS sample buffer followed by boiling. Claudin-1 and especially claudin-7 levels were also decreased in EpCAM knockdown lysates prepared with SDS sample buffer (data not shown). We also sought to determine whether claudin down-regulation could be attributed to changes in mRNA levels, and we performed real time RT-PCR experiments. Claudin-7 and claudin-1 mRNA levels were not decreased in T84 EpCAM knockdown cells (Fig. 7C). Thus, decreases in claudin levels that were associated with reductions in EpCAM expression reflect post-transcriptional, and probably post-translational, alterations.
Having observed that EpCAM and claudin-7 and claudin-1 co-localized in intracellular vesicular compartments as well as on cell surfaces (Fig. 5D), we reasoned that EpCAM might regulate claudin levels by influencing susceptibility to lysosomal degradation, and we hypothesized that inhibition of lysosomal enzymes might rescue expression of claudins in EpCAM knockdown cells. Indeed, exposure of T84 cells to increasing concentrations of chloroquine resulted in almost complete restoration of claudin-7 and claudin-1 levels without dramatic changes in EpCAM expression (Fig. 7D). We also explored the possible role of ubiquitination and proteosomal degradation in regulation of claudin stability by treating T84 cells with control or shEpCAM-encoding viruses and increasing concentrations of lactacystin (Fig. 7E) or MG132 (data not shown). Inhibition of proteosomes also led to increased accumulation of claudin-7 and claudin-1 in EpCAM knockdown cells, but the changes observed were less marked than with chloroquine. That there may be a relationship between ubiquitination and lysosomal degradation is suggested by the observations that ubiquitinylated claudin-7 can be detected in T84 cells with reduced levels of EpCAM and that treatment with EpCAM knockdown cells with chloroquine led to marked accumulation of this species (Fig. 7F).
Based on these results and the observation of punctate intracellular staining of EpCAM by ourselves and others (34), we sought to determine whether claudin-7, claudin-1, and EpCAM turnover occurred via lysosomes, and whether EpCAM might protect claudin-7 and claudin-1 from lysosomal degradation. To accomplish this, we localized claudin-7 (or claudin-1) and Lamp1, a late endosome/lysosome marker, in Caco-2 cells using confocal laser microscopy. In addition to cell surface expression, we observed similar punctate intracellular staining of claudin-7 and claudin-1 in subconfluent cells that co-localized with Lamp1 (Fig. 8 and supplemental Fig. 5). As reported previously (34), EpCAM also exhibited strong perinuclear staining in nonconfluent Caco-2 cells, and much of this co-localized with claudin-1 (supplemental Fig. 4). Co-staining with Lamp1 revealed that EpCAM was indeed present in lysosomal compartments (Fig. 8B). Immunofluorescence microscopy indicated that EpCAM knockdown led to dramatic decreases in cytoplasmic claudin-7 and claudin-1. Chloroquine treatment resulted in accumulation of claudin-7 and claudin-1 in lysosomes in EpCAM knockdown cells as in vector-transduced cells (Fig. 8 and supplemental Fig. 5). In aggregate, these results strongly suggest that, in Caco-2 cells, claudin-7 and claudin-1 are continually transported into and degraded in lysosomes and indicate that EpCAM may enhance claudin-7 and claudin-1 stability by sequestering these proteins.
FIGURE 8.
Intracellular claudin-7, claudin-1, and EpCAM localize to endosomal/lysosomal compartments. A and B, Caco-2 cells grown on chamber slides were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and co-stained for claudin-1 and Lamp1 (A) or EpCAM and Lamp1 (B). C, stable shVector- or shEpCAM-transduced Caco-2 cells were treated with or without 100 μm chloroquine for 24 h and fixed with 4% paraformaldehyde. Fixed cells were permeabilized with 0.5% Triton and co-stained with anti-claudin-7 and anti-Lamp1. Stained cells were imaged using confocal laser immunofluorescence microscopy. Scale bars, 50 μm.
EpCAM Regulates Tight Junction Composition
TJ properties are determined, in part, by TJ composition. Although understanding is incomplete, TJ function can be influenced by incorporation of different claudin family members into TJ. Thus, both levels of expression and distribution of claudins in relationship to AJC may be important. To determine whether EpCAM regulated claudin distribution in IEC, we analyzed post-confluent T84 monolayers that had acquired maximal TEER using confocal laser microscopy. XZ projections revealed that whereas claudin-7 and claudin-1 were distributed over much of the lateral interfaces of control virus-treated T84 cells in regions below TJ, the small amount of claudin-7 and claudin-1 that remained in EpCAM knockdown T84 cells was preferentially TJ-associated (Fig. 9, A, B, and E). Importantly, EpCAM knockdown did not result in obvious redistribution of claudin-4 and claudin-5 (Fig. 9C). The enhanced incorporation of claudin-7 and claudin-1 into TJ in EpCAM knockdown T84 cells was confirmed by co-localization of the claudins with TJ-associated ZO-1 visualized in XY projections of corresponding T84 monolayers (Fig. 9D), and similar observations were made in Caco-2 monolayers that exhibited maximal TEER (Fig. 9F). We conclude that EpCAM knockdown promotes accumulation of claudin-7 and claudin-1 in TJ.
EpCAM-induced Changes in TJ Function Are Dependent on Claudin-7 and Claudin-1
We sought to determine whether regulation of claudin-7 and claudin-1 dynamics by EpCAM was related to physical interactions of EpCAM with these proteins and to determine whether EpCAM acted via these claudins to regulate TJ function. Previous studies demonstrated that binding of rat EpCAM to claudin-7 could be attenuated by mutating two amino acids in the EpCAM transmembrane domain (35). We prepared cDNAs encoding the corresponding human protein, and we tested the ability of C-terminal HA-tagged wild type and mutant (A279IG283I) EpCAM to associate with claudin-7 and claudin-1. Expression plasmids encoding the relevant proteins or control plasmids were transiently introduced into T84 cells; lysates were prepared; EpCAM was immunoprecipitated with anti-HA Ab, and immunoprecipitates were assessed for claudin content via Western blotting. Claudin-7 and claudin-1 co-immunoprecipitated with wild type EpCAM, whereas neither claudin co-immunoprecipitated with the mutant (Fig. 10A).
FIGURE 10.
Regulation of claudin levels by EpCAM requires physical interactions involving EpCAM and claudins, and claudin redistribution is responsible for the modulation of TJ function by EpCAM. A, T84 cell clone that had markedly reduced EpCAM expression after transduction with shEpCAM 2 was transfected with pcDNA3 or pcDNA3 containing HA-tagged EpCAM or EpCAM(A279IG283I) (EpCAMmut) using electroporation. Cells were lysed in Triton X-100-containing buffer 48 h after transfection, and cell lysates that had been normalized for protein content were immunoprecipitated with anti-HA antibody. SDS-PAGE-resolved immunoprecipitates were blotted with anti-claudin-7, anti-claudin-1, and anti-EpCAM. B, cloned EpCAM knockdown T84 cells were transfected as in A with pcDNA3 or pcDNA3 containing HA-tagged EpCAM or EpCAM(A279IG283I). Cell lysates normalized for protein content were obtained 48 h after transfection, and SDS-PAGE-resolved proteins were immunoblotted with anti-claudin-7 and anti-EpCAM. C, Caco-2 clone with dramatically reduced EpCAM expression subsequent to transduction with shEpCAM 2 was transfected with pcDNA3 or plasmid encoding HA-tagged EpCAM or EpCAM(A279IG283I). After selection with G418 for several weeks, cells with EpCAM expression comparable with the endogenous levels were isolated via preparative flow cytometry. Sorted cells were plated into Transwells and cultured for 21 days and stained for HA (EpCAM) and claudin-1 after fixation with cold methanol. Scale bar, 10 μm. D and E, stable shVector- or shEpCAM 2-transduced Caco-2 cells were transfected with control siRNA (siNeg) or claudin-7 siRNA and claudin-1 siRNA (siC7C1) duplexes using electroporation. Twenty four h later, 5 × 105 cells were plated into Transwell chambers with 12-mm diameter polyester filters, and TEERs were determined daily thereafter. RIPA buffer cell lysates were collected over the course of the experiment and examined for claudin-7 and claudin-1 expression using Western blotting (D). Mean TEERs ± S.E. are depicted (E). **, p < 0.01, compared with Caco-2/shEpCAM 2 cells transfected with siC7C1 or compared with Caco-2/shVector cells transfected with siNeg. Representative results from one of three experiments are shown. Vec, vector.
To determine whether the ability of EpCAM to associate with claudins was linked to claudin stabilization, we attempted to rescue claudin expression in EpCAM knockdown cells with HA-tagged wild type and mutant EpCAM. Transient transfection of EpCAM knockdown T84 cells with plasmids encoding HA-tagged wild type EpCAM increased claudin-7 levels, whereas expression of mutant EpCAM was less effective (Fig. 10B). Technical difficulties (including low transfection efficiencies) precluded the use of T84 cells in efforts to prepare stable transfectants and also limited the success of experiments with Caco-2 cells. Nonetheless, transfection of Caco-2 cells that expressed very low levels of EpCAM with plasmids encoding wild type or mutant EpCAM followed by isolation of G418-resistant cells and enrichment of cells expressing EpCAM using preparative flow cytometry resulted in mixtures of cells in which claudin expression and distribution could be assessed at the single cell level. Introduction of wild type HA-tagged EpCAM led to dramatic recovery of claudin-1 expression exclusively in EpCAM-expressing cells, and EpCAM and claudin-1 co-localized in these cells (Fig. 10C). Expression of similar levels of mutant EpCAM did not result in accumulation of claudin-1 to the same extent. The ability of EpCAMmutHA to stabilize claudin-7 and claudin-1 to some extent (Fig. 10, B and C) suggests that this protein retains some claudin-binding activity that is not evident in the immunoprecipitation experiments or that additional mechanisms may also regulate stabilization of claudins by EpCAM.
To assess the involvement of claudin-7 and claudin-1 in EpCAM-induced alterations in TJ function, we transiently decreased expression of these claudins in Caco-2 cells expressing usual and reduced levels of EpCAM. After verifying that claudin-7 and claudin-1 siRNA decreased claudin-7 and claudin-1 expression in Caco-2 cells for at least 7 days (Fig. 10D), we quantified TJ function by measuring TEER developed by Caco-2 cells during a 1-week culture period. As expected, knockdown of EpCAM expression in Caco-2 monolayers led to increased TEER (Fig. 10E). Increased acquisition of TEER by Caco-2 monolayers with decreased EpCAM expression was abrogated by treatment with siRNA directed against claudin-7 and claudin-1. Treatment of EpCAM-sufficient Caco-2 cells with the same siRNA had no effect on TEER. These results are consistent with the observation that claudin-7 and claudin-1 are not abundant in TJ in EpCAM-containing cell monolayers (Fig. 9), and the concept that EpCAM-related changes in TJ function result from increased accumulation of claudin-7 and claudin-1 in TJ in the absence of EpCAM.
DISCUSSION
Although the acronym EpCAM (epithelial cell adhesion molecule) has gained wide acceptance, the extent to which and the mechanism by which EpCAM influences intercellular adhesion are incompletely understood. Initial studies indicated that EpCAM promoted adhesion (6), but it was subsequently suggested that EpCAM was a negative modulator of cadherin-mediated interactions (25). In this study, we systematically explored the relationship between EpCAM expression and intercellular adhesion in two human colon cancer cell lines that are frequently used to study AJCs and TJ. We showed that EpCAM did not accumulate in TJ. Although EpCAM and E-cadherin co-localized at lateral interfaces of polarized epithelial cells, they did not bind tightly to each other, and EpCAM and E-cadherin expression was not coordinately regulated. However, EpCAM did physically associate with claudin-7 and claudin-1, cell surface proteins that are important components of TJ. The avidities of these interactions were sufficient to allow wash-resistant co-immunoprecipitation in the absence of cross-linking agents. We confirmed that EpCAM also associated with claudin-1 but not claudin-2 or claudin-4, and we demonstrated that EpCAM-claudin-1 association was claudin-7-dependent. EpCAM and claudin-7 bands in SDS gels of resolved immunoprecipitates were of similar intensities and additional specifically immunoprecipitated proteins were not detected (Fig. 5A). These observations, coupled with the finding that claudin-7 and claudin-1 co-immunoprecipitated from EpCAM-negative, claudin-expressing fibroblast lysates, suggest that claudin-7 and claudin-1 bind to each other. Consistent with this, these two closely related claudins are similarly distributed in subconfluent Caco-2 cells (Fig. 8 and supplemental Figs. 4 and 5) and they may act coordinately to regulate cell-cell adhesion and TJ function.
Interaction of EpCAM with claudin-7 and claudin-1 in IEC had several consequences. First, EpCAM-claudin-7/claudin-1 binding resulted in accumulation of these claudins at intercellular interfaces in regions that were distinct from TJ. Second, in the absence of EpCAM, levels of cellular claudin-7 and claudin-1 decreased dramatically as these proteins were degraded by lysosome-dependent and, to some extent, proteosome-dependent mechanisms. Third, claudin-7 and claudin-1 that remained in cells that did not express EpCAM preferentially accumulated in TJ. Inhibition of claudin-7 and claudin-1 expression in Caco-2 cells with siRNA prevented claudin redistribution in EpCAM knockdown cells and obviated the increased TEER acquisition that otherwise occurred. Changes in TJ function have previously been related to differential incorporation of individual claudin family members into TJ (36). Our results suggest that EpCAM is an important regulator of TJ function that acts by sequestering claudin-7 and claudin-1 at lateral interfaces of IEC in regions that are distinct from TJ. Interestingly, claudin-7 also redistributed from basolateral surfaces to TJ in HT-29 human colon carcinoma cells, although its levels were reduced secondary to Sox-9 overexpression (37). It is not known if EpCAM has a role in this modulation.
In addition to regulating epithelial permeability and polarity, TJ likely constrain movement of cells in epithelia. Physiological processes, including epiboly (8), shedding of apoptotic cells from tips of intestinal villi (38), and movement of leukocytes within the epidermis (39) require that cells in epithelia change positions with respect to each other in a carefully orchestrated fashion without disrupting barrier function. This may involve modulation of TJ composition and/or function, and EpCAM may be an important contributor by attenuating TJ avidity. Although EpCAM is expressed at high levels in only a few tissues in adults, EpCAM is widely expressed in developing epithelia during embryogenesis, and it has been suggested that EpCAM plays an important role in organogenesis (40). EpCAM is also abundant in a number of carcinomas, and EpCAM expression has been correlated with increased cancer invasiveness and poor prognosis. Abrogation of TJ function appears to be a prerequisite for metastasis (41), and EpCAM may promote metastasis by modulating TJ.
TJ are dynamic structures, and turnover and trafficking of individual TJ components is regulated via different mechanisms. Some TJ proteins (e.g. ZO-1 and occludin) rapidly exchange between TJ and extra-junctional pools, whereas others (e.g. claudins) do not (42). Recent studies demonstrated that, at least in some cells, claudins (including claudin-1) are continuously removed from cell surfaces via an endosomal sorting complex required for a transport-dependent process after ubiquitination by LNX1p80 (42, 43). Internalization into endosomes can lead to recycling to cell surfaces or ultimately to lysosomal degradation. Importantly, intracellular claudin trafficking is linked to regulation of TJ barrier strength and movement of adjacent epithelial cells within cellular sheets (42, 44). Herein, we demonstrate that claudin-7 and claudin-1 continually traffic into lysosomes where they are degraded. However, in EpCAM-expressing cells, there is a pool of claudin molecules on cell surfaces that is EpCAM-associated, protected from lysosomal degradation, and not available to be incorporated into TJ.
Several prior studies characterized EpCAM-associated proteins. Co-immunoprecipitation experiments utilizing rat pancreatic adenocarcinoma cells and bifunctional cross-linking reagents indicated that EpCAM and claudin-7 were physically associated (45). Assessment of larger macromolecular complexes in human colorectal cancer cells via differential ultracentrifugation revealed that EpCAM and claudin-7 were concentrated in tetraspanin-enriched microdomains that also included CD9, CD81, β2-containing integrins, and CD44v (46). EpCAM may bind to CD9 and claudin-7 directly, and claudin-7 may also associate with β2 integrins and CD44v (47). Thus, selected claudins may participate in the organization of subcellular signaling platforms in addition to being essential components of TJ. Whether or not these tetraspanin-enriched microdomains correspond to the accumulations of EpCAM and claudins that we visualized at lateral interfaces of IEC remains to be determined.
It is striking that, in the presence of EpCAM, most claudin-7 and claudin-1 proteins are localized at lateral intercellular interfaces distinctly below TJ that form in T84 and Caco-2 cell monolayers. We (data not shown) and others (47, 48) have observed that claudin-7 and claudin-1 are similarly localized in murine IEC in situ. Little is known about the function of extra-junctional claudins. The distinctive non-TJ localization of claudin-7 and claudin-1 in IEC in vitro and in vivo suggests that studies of claudin function in the intestine may be particularly informative. Indeed, a recent study revealed that claudin-7-deficient mice display severe defects in intestinal mucosal architecture, without obvious disruption of TJ and epithelial polarity (47), and it was suggested that claudin-7 regulation of intestinal epithelial integrity was unrelated to effects on TJ (47).
Relevant to this study, two very recent reports describe new lines of EpCAM null mutant mice. Interestingly, in both papers, EpCAM knock-out pups exhibited phenotypes that share features with that seen in claudin-7 knock-out mice as well as humans with congenital tufting enteropathy that is caused by EpCAM mutations (11, 12). Lei et al. (11) attributed the phenotype of EpCAM knock-out mice to alterations of TJ and barrier function that related to differential recruitment of claudins to TJ, whereas Guerra et al. (12) ascribed the phenotype that they observed to dysregulation of E-cadherin and/or β-catenin localization or function. Well controlled in vitro studies are important complements to those that are conducted using the knock-out animals in which intestinal epithelial architecture is seriously disrupted to differentiate changes in cellular physiology that are causally linked to decreased EpCAM expression from those that are indirectly related to epithelial homeostasis or other alterations. Although our report of enhancement of intestinal epithelial TJ function by EpCAM reduction appears to not be in accord with the results of studies of EpCAM null mice, it is consistent with in vivo observations regarding zebrafish epiboly reported by Slanchev et al. (8).
In conclusion, demonstration that claudin stability in IEC is determined by EpCAM expression that regulates susceptibility of claudins to lysosomal degradation and influences TJ composition and function provides mechanistic insight into the phenotypes of EpCAM- and claudin-7-deficient mice and a rare but devastating human disease. Our study has also identified a valuable experimental platform that will facilitate future investigations.
Acknowledgments
We thank Barbara Taylor (Center for Cancer Research, FACS Core Laboratory) and Nga Voong (Experimental Transplantation and Immunology Branch, NCI, National Institutes of Health) for help with cell sorting. We also thank the following: Drs. Su-Young Kim and Michael Weiger (Center for Cancer Research) for help with cell migration assay; Dr. Enrique Zudaire (Center for Cancer Research) for quantitation of tight junction staining; Dr. Xu Feng for constructing the plasmid encoding the EpCAM-IgG fusion protein used for producing EpCAM polyclonal Ab; Susan Garfield (Laboratory of Experimental Carcinogenesis, NCI, National Institutes of Health) for providing confocal imaging expertise; the Laboratory of Pathology Tissue Processing and Procurement Facility for providing human small intestinal tissue, and Drs. James Anderson and Christina Van Itallie (National Institutes of Health) for helpful discussions. Dr. Asma Nusrat (Emory University) provided valuable advice and access to critical cells and reagents during the early phases of this project.
This work was supported, in whole or in part, by National Institutes of Health grants from the Intramural Research Program, Center for Cancer Research, and NCI.
This article was selected as a Paper of the Week.

This article contains supplemental Figs. 1–5.
- EpCAM
- epithelial cell adhesion molecule
- AJC
- apical junctional complex
- IEC
- intestinal epithelial cell
- TEER
- trans-epithelial electrical resistance
- TJ
- tight junction
- Ab
- antibody.
REFERENCES
- 1. Baeuerle P. A., Gires O. (2007) EpCAM (CD326) finding its role in cancer. Br. J. Cancer 96, 417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van der Gun B. T., Melchers L. J., Ruiters M. H., de Leij L. F., McLaughlin P. M., Rots M. G. (2010) EpCAM in carcinogenesis: the good, the bad or the ugly. Carcinogenesis 31, 1913–1921 [DOI] [PubMed] [Google Scholar]
- 3. Maetzel D., Denzel S., Mack B., Canis M., Went P., Benk M., Kieu C., Papior P., Baeuerle P. A., Munz M., Gires O. (2009) Nuclear signalling by tumour-associated antigen EpCAM. Nat. Cell Biol. 11, 162–171 [DOI] [PubMed] [Google Scholar]
- 4. Akita H., Nagano H., Takeda Y., Eguchi H., Wada H., Kobayashi S., Marubashi S., Tanemura M., Takahashi H., Ohigashi H., Tomita Y., Ishikawa O., Mori M., Doki Y. (2011) Ep-CAM is a significant prognostic factor in pancreatic cancer patients by suppressing cell activity. Oncogene 30, 3468–3476 [DOI] [PubMed] [Google Scholar]
- 5. Litvinov S. V., Bakker H. A., Gourevitch M. M., Velders M. P., Warnaar S. O. (1994) Evidence for a role of the epithelial glycoprotein 40 (Ep-CAM) in epithelial cell-cell adhesion. Cell Adhes. Commun. 2, 417–428 [DOI] [PubMed] [Google Scholar]
- 6. Litvinov S. V., Velders M. P., Bakker H. A., Fleuren G. J., Warnaar S. O. (1994) Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J. Cell Biol. 125, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balzar M., Bakker H. A., Briaire-de-Bruijn I. H., Fleuren G. J., Warnaar S. O., Litvinov S. V. (1998) Cytoplasmic tail regulates the intercellular adhesion function of the epithelial cell adhesion molecule. Mol. Cell. Biol. 18, 4833–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Slanchev K., Carney T. J., Stemmler M. P., Koschorz B., Amsterdam A., Schwarz H., Hammerschmidt M. (2009) The epithelial cell adhesion molecule EpCAM is required for epithelial morphogenesis and integrity during zebrafish epiboly and skin development. PLoS Genet. 5, e1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maghzal N., Vogt E., Reintsch W., Fraser J. S., Fagotto F. (2010) The tumor-associated EpCAM regulates morphogenetic movements through intracellular signaling. J. Cell Biol. 191, 645–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sivagnanam M., Mueller J. L., Lee H., Chen Z., Nelson S. F., Turner D., Zlotkin S. H., Pencharz P. B., Ngan B. Y., Libiger O., Schork N. J., Lavine J. E., Taylor S., Newbury R. O., Kolodner R. D., Hoffman H. M. (2008) Identification of EpCAM as the gene for congenital tufting enteropathy. Gastroenterology 135, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lei Z., Maeda T., Tamura A., Nakamura T., Yamazaki Y., Shiratori H., Yashiro K., Tsukita S., Hamada H. (2012) EpCAM contributes to formation of functional tight junction in the intestinal epithelium by recruiting claudin proteins. Dev. Biol. 371, 136–145 [DOI] [PubMed] [Google Scholar]
- 12. Guerra E., Lattanzio R., La Sorda R., Dini F., Tiboni G. M., Piantelli M., Alberti S. (2012) mTrop1/Epcam knockout mice develop congenital tufting enteropathy through dysregulation of intestinal E-cadherin/β-catenin. PLoS One 7, e49302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koch S., Nusrat A. (2009) Dynamic regulation of epithelial cell fate and barrier function by intercellular junctions. Ann. N.Y. Acad. Sci. 1165, 220–227 [DOI] [PubMed] [Google Scholar]
- 14. Matter K., Balda M. S. (2003) Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 4, 225–236 [DOI] [PubMed] [Google Scholar]
- 15. Förster C. (2008) Tight junctions and the modulation of barrier function in disease. Histochem. Cell Biol. 130, 55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paris L., Tonutti L., Vannini C., Bazzoni G. (2008) Structural organization of the tight junctions. Biochim. Biophys. Acta 1778, 646–659 [DOI] [PubMed] [Google Scholar]
- 17. Steed E., Balda M. S., Matter K. (2010) Dynamics and functions of tight junctions. Trends Cell Biol. 20, 142–149 [DOI] [PubMed] [Google Scholar]
- 18. Lui W. Y., Lee W. M. (2006) Regulation of junction dynamics in the testis–transcriptional and post-translational regulations of cell junction proteins. Mol. Cell. Endocrinol. 250, 25–35 [DOI] [PubMed] [Google Scholar]
- 19. Grünert S., Jechlinger M., Beug H. (2003) Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat. Rev. Mol. Cell Biol. 4, 657–665 [DOI] [PubMed] [Google Scholar]
- 20. Utech M., Brüwer M., Nusrat A. (2006) Tight junctions and cell-cell interactions. Methods Mol. Biol. 341, 185–195 [DOI] [PubMed] [Google Scholar]
- 21. Evans M. J., von Hahn T., Tscherne D. M., Syder A. J., Panis M., Wölk B., Hatziioannou T., McKeating J. A., Bieniasz P. D., Rice C. M. (2007) Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446, 801–805 [DOI] [PubMed] [Google Scholar]
- 22. Wu C. J., Conze D. B., Li T., Srinivasula S. M., Ashwell J. D. (2006) Sensing of Lys-63-linked polyubiquitination by NEMO is a key event in NF-κB activation (corrected). Nat. Cell Biol. 8, 398–406 [DOI] [PubMed] [Google Scholar]
- 23. Zudaire E., Gambardella L., Kurcz C., Vermeren S. (2011) A computational tool for quantitative analysis of vascular networks. PLoS One 6, e27385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gelderman K. A., Kuppen P. J., Bruin W., Fleuren G. J., Gorter A. (2002) Enhancement of the complement activating capacity of 17-1A mAb to overcome the effect of membrane-bound complement regulatory proteins on colorectal carcinoma. Eur. J. Immunol. 32, 128–135 [DOI] [PubMed] [Google Scholar]
- 25. Litvinov S. V., Balzar M., Winter M. J., Bakker H. A., Briaire-de Bruijn I. H., Prins F., Fleuren G. J., Warnaar S. O. (1997) Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J. Cell Biol. 139, 1337–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Osta W. A., Chen Y., Mikhitarian K., Mitas M., Salem M., Hannun Y. A., Cole D. J., Gillanders W. E. (2004) EpCAM is overexpressed in breast cancer and is a potential target for breast cancer gene therapy. Cancer Res. 64, 5818–5824 [DOI] [PubMed] [Google Scholar]
- 27. Zanna P., Trerotola M., Vacca G., Bonasera V., Palombo B., Guerra E., Rossi C., Lattanzio R., Piantelli M., Alberti S. (2007) Trop-1 are conserved growth stimulatory molecules that mark early stages of tumor progression. Cancer 110, 452–464 [DOI] [PubMed] [Google Scholar]
- 28. Basak S., Speicher D., Eck S., Wunner W., Maul G., Simmons M. S., Herlyn D. (1998) Colorectal carcinoma invasion inhibition by CO17-1A/GA733 antigen and its murine homologue. J. Natl. Cancer Inst. 90, 691–697 [DOI] [PubMed] [Google Scholar]
- 29. Tirumalasetty P. P., Eley J. G. (2006) Permeability enhancing effects of the alkylglycoside, octylglucoside, on insulin permeation across epithelial membrane in vitro. J. Pharm. Pharm. Sci. 9, 32–39 [PubMed] [Google Scholar]
- 30. Buzza M. S., Netzel-Arnett S., Shea-Donohue T., Zhao A., Lin C. Y., List K., Szabo R., Fasano A., Bugge T. H., Antalis T. M. (2010) Membrane-anchored serine protease matriptase regulates epithelial barrier formation and permeability in the intestine. Proc. Natl. Acad. Sci. U.S.A. 107, 4200–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Samarin S. N., Ivanov A. I., Flatau G., Parkos C. A., Nusrat A. (2007) Rho/Rho-associated kinase-II signaling mediates disassembly of epithelial apical junctions. Mol. Biol. Cell 18, 3429–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tunggal J. A., Helfrich I., Schmitz A., Schwarz H., Günzel D., Fromm M., Kemler R., Krieg T., Niessen C. M. (2005) E-cadherin is essential for in vivo epidermal barrier function by regulating tight junctions. EMBO J. 24, 1146–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hewitt K. J., Agarwal R., Morin P. J. (2006) The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer 6, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Balzar M., Briaire-de Bruijn I. H., Rees-Bakker H. A., Prins F. A., Helfrich W., de Leij L., Riethmüller G., Alberti S., Warnaar S. O., Fleuren G. J., Litvinov S. V. (2001) Epidermal growth factor-like repeats mediate lateral and reciprocal interactions of Ep-CAM molecules in homophilic adhesions. Mol. Cell. Biol. 21, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nübel T., Preobraschenski J., Tuncay H., Weiss T., Kuhn S., Ladwein M., Langbein L., Zöller M. (2009) Claudin-7 regulates EpCAM-mediated functions in tumor progression. Mol. Cancer Res. 7, 285–299 [DOI] [PubMed] [Google Scholar]
- 36. Yamazaki Y., Tokumasu R., Kimura H., Tsukita S. (2011) Role of claudin species-specific dynamics in reconstitution and remodeling of the zonula occludens. Mol. Biol. Cell 22, 1495–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Darido C., Buchert M., Pannequin J., Bastide P., Zalzali H., Mantamadiotis T., Bourgaux J. F., Garambois V., Jay P., Blache P., Joubert D., Hollande F. (2008) Defective claudin-7 regulation by Tcf-4 and Sox-9 disrupts the polarity and increases the tumorigenicity of colorectal cancer cells. Cancer Res. 68, 4258–4268 [DOI] [PubMed] [Google Scholar]
- 38. Wilson A. J., Gibson P. R. (1997) Epithelial migration in the colon: filling in the gaps. Clin. Sci. 93, 97–108 [DOI] [PubMed] [Google Scholar]
- 39. Kubo A., Nagao K., Yokouchi M., Sasaki H., Amagai M. (2009) External antigen uptake by Langerhans cells with reorganization of epidermal tight junction barriers. J. Exp. Med. 206, 2937–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trzpis M., Bremer E., McLaughlin P. M., de Leij L. F., Harmsen M. C. (2008) EpCAM in morphogenesis. Front. Biosci. 13, 5050–5055 [DOI] [PubMed] [Google Scholar]
- 41. Martin T. A., Mason M. D., Jiang W. G. (2011) Tight junctions in cancer metastasis. Front. Biosci. 16, 898–936 [DOI] [PubMed] [Google Scholar]
- 42. Dukes J. D., Fish L., Richardson J. D., Blaikley E., Burns S., Caunt C. J., Chalmers A. D., Whitley P. (2011) Functional ESCRT machinery is required for constitutive recycling of claudin-1 and maintenance of polarity in vertebrate epithelial cells. Mol. Biol. Cell 22, 3192–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Takahashi S., Iwamoto N., Sasaki H., Ohashi M., Oda Y., Tsukita S., Furuse M. (2009) The E3 ubiquitin ligase LNX1p80 promotes the removal of claudins from tight junctions in MDCK cells. J. Cell Sci. 122, 985–994 [DOI] [PubMed] [Google Scholar]
- 44. Matsuda M., Kubo A., Furuse M., Tsukita S. (2004) A peculiar internalization of claudins, tight junction-specific adhesion molecules, during the intercellular movement of epithelial cells. J. Cell Sci. 117, 1247–1257 [DOI] [PubMed] [Google Scholar]
- 45. Ladwein M., Pape U. F., Schmidt D. S., Schnölzer M., Fiedler S., Langbein L., Franke W. W., Moldenhauer G., Zöller M. (2005) The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Exp. Cell Res. 309, 345–357 [DOI] [PubMed] [Google Scholar]
- 46. Kuhn S., Koch M., Nübel T., Ladwein M., Antolovic D., Klingbeil P., Hildebrand D., Moldenhauer G., Langbein L., Franke W. W., Weitz J., Zöller M. (2007) A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol. Cancer Res. 5, 553–567 [DOI] [PubMed] [Google Scholar]
- 47. Ding L., Lu Z., Foreman O., Tatum R., Lu Q., Renegar R., Cao J., Chen Y. H. (2012) Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology 142, 303–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fujita H., Chiba H., Yokozaki H., Sakai N., Sugimoto K., Wada T., Kojima T., Yamashita T., Sawada N. (2006) Differential expression and subcellular localization of claudin-7, -8, -12, -13, and -15 along the mouse intestine. J. Histochem. Cytochem. 54, 933–944 [DOI] [PubMed] [Google Scholar]