Background: Some children with leukemia do not respond to an immunotoxin containing Pseudomonas exotoxin A.
Results: We isolated a resistant cell line and showed that a WDR85 gene deletion causes immunotoxin resistance by allowing formation of diphthamide containing an extra methyl group.
Conclusion: Low WDR85 expression can cause immunotoxin resistance in patients.
Significance: Analysis of EF2 may reveal why some patients fail therapy.
Keywords: ADP-ribosylation, Cancer Therapy, Drug Resistance, Leukemia, Translation Elongation Factors, CD22, DPH5, WDR85, Immunotoxin
Abstract
HA22 is a recombinant immunotoxin that kills CD22-expressing cells by ADP-ribosylating and inactivating elongation factor-2 (EF2). HA22 is composed of an Fv that binds to CD22 fused to a portion of Pseudomonas exotoxin A. HA22 is very active in drug-resistant hairy cell leukemia but is less active in children with acute lymphoblastic leukemia. To understand why some patients do not respond to HA22, we isolated an HA22-resistant lymphoma cell line and showed that resistance was due to the inability of HA22 to ADP-ribosylate and inactivate EF2. We analyzed the diphthamide synthesis genes and found that the WDR85 gene was deleted. We show that WDR85 knockdown conferred HA22 resistance to sensitive cells and that sensitivity was restored by introduction of a WDR85 cDNA into resistant cells. Analysis of EF2 in the mutant cells revealed a novel form of diphthamide with an additional methyl group that prevented ADP-ribosylation and inactivation of EF2. The abnormal methylation appeared to be catalyzed by DPH5. Inactivation of the WDR85 gene could be a mechanism of immunotoxin resistance in patients undergoing immunotoxin therapy.
Introduction
Acute lymphoblastic leukemia (ALL)2 is the most common pediatric malignancy, making up ∼25% of childhood cancer (1). Great advances in ALL treatment have been achieved in recent years. However, ALL still accounts for a large percentage of cancer-related mortality in pediatric patients, and survivors are at risk of many treatment-related side effects (2–4). Novel therapies that can improve prognosis and reduce nonspecific toxicities are needed.
CD22 is a B-lineage differentiation antigen that is an excellent target for the therapy of hairy cell leukemia, B-lineage ALL, and non-Hodgkin lymphoma (5, 6). We have developed a recombinant immunotoxin (HA22/moxetumomab pasudotox) that targets and kills CD22-expressing cells. It was constructed by fusing the Fv portion of an antibody that reacts with CD22 to a 38-kDa portion of Pseudomonas exotoxin A (7, 8). After binding to CD22, HA22 is internalized via receptor-mediated endocytosis. After processing by furin, which releases the Fv fragment from the toxin, the toxin portion is transferred to the endoplasmic reticulum, from which it is translocated to the cytosol, where it ADP-ribosylates and inactivates elongation factor-2 (EF2). Protein synthesis is arrested, and programmed cell death occurs (8). A modified histidine residue termed “diphthamide” is required for EF2 to be ADP-ribosylated by the catalytic domains of the toxin.
For many years, it was thought that only five enzymes (DPH1–5) were required to modify histidine 715 in EF2 and produce diphthamide (9). Recently, a sixth protein, WDR85 (WD repeat domain 85), was also found to be required for this modification process (10, 11). WDR85 is a WD repeat-containing protein. Cells that do not express WDR85 are resistant to killing by Diphtheria toxin and Pseudomonas exotoxin A (PE) due to impaired ADP-ribosylation of EF2 (10, 11).
A pediatric phase I trial of HA22 is ongoing (ClinicalTrials.gov identified NCT00659425), and the results are promising, with clinical activity seen in most patients and complete responses observed in ∼25% of children with chemotherapy-refractory disease (12). However, some patients do not respond, whereas others relapse after initially responding to the agent. We have undertaken studies to understand possible mechanisms that can lead to resistance and recently reported that silencing of the DPH4 gene in the HAL-01 ALL cell line results in immunotoxin resistance (13). Here, we have selected CA46 cells, which are immunotoxin-resistant, and observed that the resistant cells have a deletion of the WDR85 gene, which causes a novel modification of diphthamide in EF2 and immunotoxin resistance.
EXPERIMENTAL PROCEDURES
Reagents
Immunotoxins HA22 and HB21-PE40 were produced as described (14). MISSION lentiviral particles containing WDR85 shRNA (TRCN0000160958 and TRCN0000162819), DPH5 shRNA (TRCN0000152500), or METTL18 shRNA (TRCN0000143529) and non-target shRNA control transduction particles (SHC002V) were from Sigma. pCMV6-WDR85 (expressing WDR85) was from OriGene. Anti-actin and anti-EF2 antibodies were from Abcam, and anti-WDR85 antibody was from Sigma.
A fragment containing the WDR85 ORF was amplified from pCMV6-WDR85 using the following primer: sense, 5′-AGTCACAGATCTCACCATGATGGGCTGTTTCGCCCT-3′; and antisense, 5′-AGTCACGAATTCGGTTCCCCTCCCACTCCCAGA-3′. The amplified fragment, which did not contain the stop codon, was digested with BglII and EcoRI endonucleases and then cloned into the BglII and EcoRI sites of pEGFP-N1 (Clontech). The constructed vector, pEGFP-N1-WDR85-GFP, expressed WDR85-GFP fusion protein.
Establishment of Resistant Cell Line
The cell line CA46 was maintained in RPMI 1640 medium with 10% FBS. To isolate resistant cells, 2 × 107 cells were seeded in 10 ml of RPMI 1640 medium with HA22 at 100 ng/ml and incubated for 72 h. Residual viable cells were expanded for 4 weeks without HA22. A second round of selection was performed in a similar manner, and the resistant cells were expanded. A water-soluble tetrazolium salt (WST) assay was performed to confirm immunotoxin resistance, and aliquots were frozen in liquid nitrogen.
Antigen Expression and Internalization of HA22
Quantitation of CD22 surface expression and HA22 internalization was performed as described (15, 16) after excluding debris and dead cells with forward scatter and side scatter gating strategy.
Protein Synthesis Inhibition Assay
Protein synthesis inhibition was performed as described (17).
Toxin-induced ADP-ribosylation of EF2
Cells were lysed in 0.3 ml of radioimmune precipitation assay buffer with protease inhibitors, and 0.01 ml of cell lysate (30 μg) was incubated with 100 ng of HA22 in ADP-ribosylation buffer (20 mm Tris-HCl (pH 7.4), 1 mm EDTA, and 50 mm DTT) with 5 μm 6-biotin-17-NAD (Trevigen) for 60 min at 25 °C. Samples were subjected to SDS-PAGE, followed by Western blotting with HRP-conjugated streptavidin (Invitrogen) to detect biotin-ADP-ribose-EF2.
Real-time RT-PCR
Measurement of mRNA levels was performed by real-time RT-PCR as described (18). See supplemental “Experimental Procedures” for all primer sequences.
Immunoblots
Cells were collected, washed twice with cold Dulbecco's PBS, and solubilized in lysis buffer (25 nm Tris-HCl (pH 7.5), 150 nm NaCl, 1.0% Nonidet P-40, and 0.5% sodium deoxycholate) with membrane-blocking agent (GE Healthcare). Detection of proteins was performed as described (19).
Lentiviral Infections
CA46 or CA46-R cells were seeded at a density of 1 × 105 cells/well in 500 μl of medium containing 1 μg/ml Polybrene in a 24-well plate and infected with various lentiviral particles (listed above under “Reagents”). The plate was centrifuged at 1200 × g for 90 min at room temperature and placed in a 37 °C incubator. One day after infection, 1 ml of medium was added to each well, and puromycin was added at 4 μg/ml after 72 h. Puromycin was refreshed every 3 days. The transduced cells were maintained in puromycin and taken out of the puromycin for 3 days before each experiment.
Electrotransfection of CA46-R Cells
CA46-R cells were transfected with the pEGFP-N1-WDR85-GFP or pEGFP-N1 plasmid using the Amaxa Cell Line Nucleofector Kit C and Amaxa Nucleofector (Lonza). First, 2 × 106 CA46-R cells were harvested and suspended in 100 μl of Nucleofector Solution C, and the cell suspension was then mixed with 2 μg of plasmid DNA. The mixture was electrotransfected using Nucleofector Program R-028 in the Amaxa Nucleofector. The transfected cells were cultured in medium, and 750 μg/ml G418 was added 48 h after transfection. Three weeks after selection, GFP-expressing cells were isolated by cell sorting, and these cells used for all studies.
EF2 Purification, In-gel Digest, and MALDI-TOF-MS Analysis
EF2 was purified from 5 × 106 cells by ion exchange chromatography on TOYOPEARL DEAE-650 (Tosoh Bioscience) and SDS gel electrophoresis, the EF2 gel band was digested with trypsin, and peptides were fractionated by reverse phase HPLC on Vydac C18 columns (5 μm, 250 × 2.1 mm). Fractions were analyzed by MALDI-TOF-MS on an Ultraflex III TOF/TOF mass spectrometer (Bruker Daltonics, Billerica, MA). Fractions containing diphthamide-modified peptides were further analyzed. See supplemental “Experimental Procedures” for a detailed description of the procedures and mass spectrometry data analysis.
High Resolution SNP Array Analysis
High resolution copy number variation/SNP analysis of CA46 and CA46-R cells was performed using the Illumina 5.0M BeadChip platform following the manufacturer's guidelines. GenomeStudio software was used to analyze the data, and the final report containing the log relative ratio and the B-allele frequency were exported (Illumina).
Statistical Analysis
The data are expressed as the mean ± S.D. Statistical analysis was performed using Student's t test for comparison between two groups.
RESULTS
Establishment and Characterization of HA22-resistant Cell Line
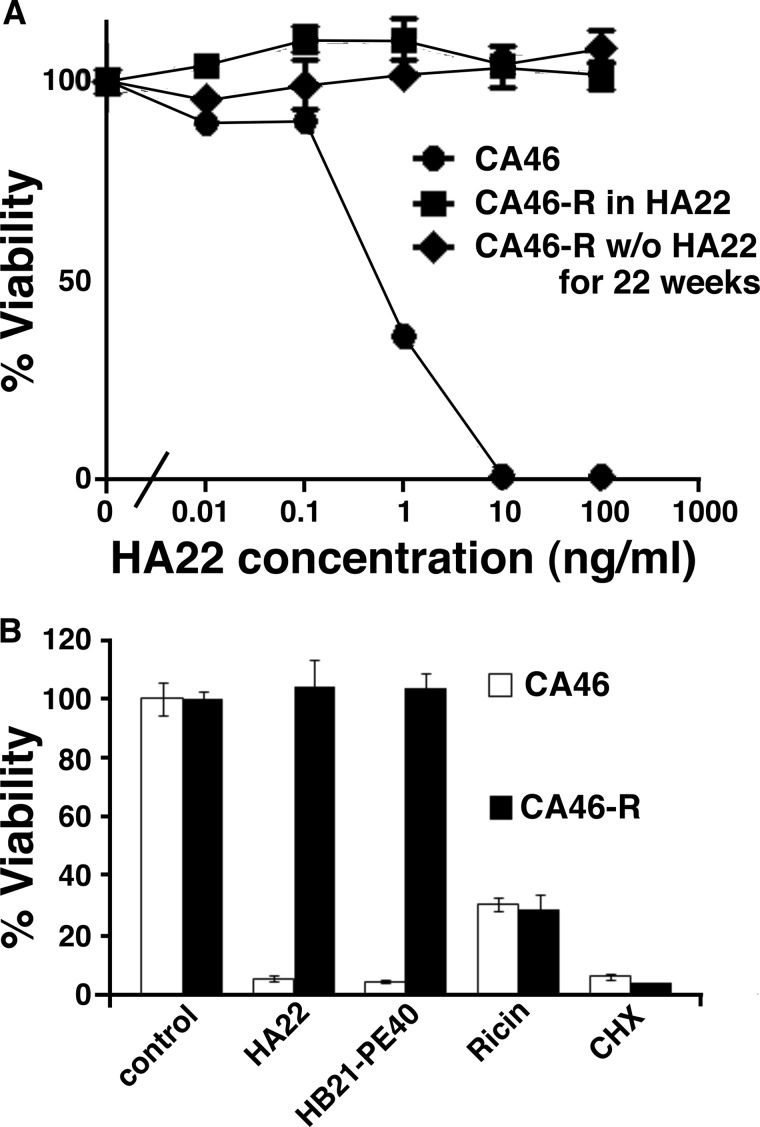
A cell line that was resistant to HA22 (CA46-R) was isolated by repeated incubation of the parental cell line (CA46) with 100 ng/ml HA22 as described under “Experimental Procedures.” Fig. 1A shows that the sensitive cells had an IC50 of 0.6 ng/ml, whereas the resistant cells were not killed by 100 ng/ml. To determine whether the resistance was stable, we cultured the cells without HA22 for 22 weeks and measured their response to HA22. We found that the cells were still fully resistant to HA22 after 22 weeks (Fig. 1A). We measured the growth rate of the cells and found that the doubling time of CA46 cells was 1.7 days and that CA46-R cells was 2.3 days.
FIGURE 1.
A, CA46-R cells are still resistant to HA22 after being removed from HA22 for 9 weeks. CA46-R cells were cultured in normal medium without HA22 for the indicated number of days, followed by treatment with the indicated concentrations of HA22 for 3 days. B, CA46-R cells are cross-resistant to HB21 but are still sensitive to cycloheximide (CHX) and ricin as determined by the WST-8 assay. CA46 and CA46-R cells were treated for 3 days with HA22 (100 ng/ml), HB21-PE40 (100 ng/ml), cycloheximide (10 μg/ml), or ricin (10 ng/ml). Sensitivity to drugs was assessed by the WST-8 assay.
To assess if the resistance was specific for a PE-based immunotoxin or extended to other cytotoxic agents, we examined resistance to a PE-containing immunotoxin targeted at the transferrin receptor (HB21-PE40), as well as resistance to ricin and cycloheximide. We found that the CA46-R cells were resistant to HB21-PE40, but not to the two other protein synthesis inhibitors, cycloheximide and ricin (Fig. 1B). These results show that the resistance is not due to a general defect related to protein synthesis.
Resistance Is Due to Impaired ADP-ribosylation of EF2
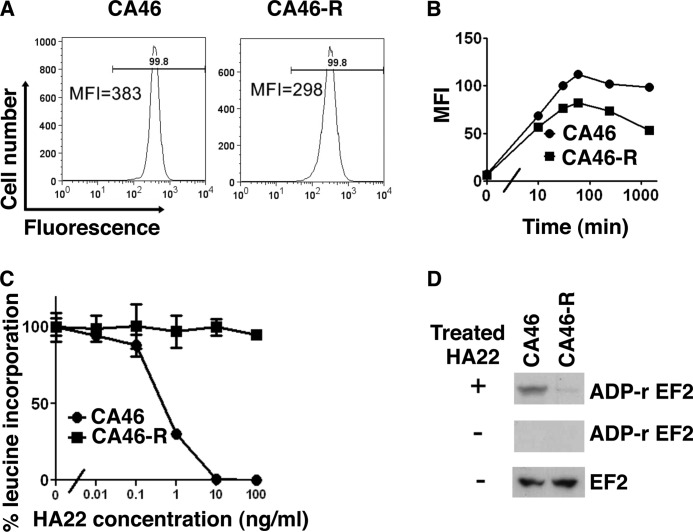
There are several steps in the immunotoxin pathway that could be modified and account for the resistance (8). The first step is binding to CD22. The number of CD22-binding sites on the cell surface was evaluated by flow cytometry; there was a small decrease in CD22-binding sites that could not account for the resistance (Fig. 2A). To explore whether decreased internalization accounted for resistance, we measured HA22 internalization by flow cytometry using HA22 labeled with Alexa Fluor 647 and found the total amount of HA22 internalized was only slightly decreased in the resistant cell line (Fig. 2B). These data indicate that decreased expression and internalization do not account for the resistance.
FIGURE 2.
A, all CA46-R cells are CD22-positive, but the intensity is slightly decreased. Cells were incubated on ice with PE-labeled anti-CD22 antibody for 45 min and analyzed by FACS. MFI, geometric mean of fluorescence intensity. B, the internalization rate is also decreased in CA46-R cells, and it is possibly because of lower CD22 expression. Cells were incubated with Alexa Fluor 647-labeled HA22 at 37 °C for 10 and 30 min and 1, 4, and 24 h. Internalized immunotoxin is shown as the geometric mean fluorescence. C, HA22 cannot inhibit leucine incorporation in CA46-R cells. Cells were treated with HA22 for 20 h. Protein synthesis was measured by incorporation of [3H]leucine. Mean triplicate values are shown. D, HA22 is not able to ADP-ribosylate (ADP-r) EF2 in CA46-R cells. Cell lysate (30 μg) was incubated with or without 100 ng of HA22 for 60 min at 25 °C. Samples were analyzed by Western blotting with HRP-conjugated streptavidin to detect biotin-ADP-ribose-EF2. EF2 is shown as a loading control.
The next step we measured is the amount of [3H]leucine incorporated into protein. We found that [3H]leucine incorporation was not reduced by HA22 in the resistant cells (Fig. 2C). This result indicates either that the toxin did not reach the cytosol, where EF2 is located, or that the toxin could not inactivate EF2. To evaluate if EF2 could be ADP-ribosylated by the immunotoxin, HA22 and 6-biotin-17-NAD were added to cell-free extracts prepared from sensitive or resistant cells; the proteins were resolved by SDS-PAGE; and after transfer to a membrane, the amount of biotin-NAD incorporated into EF2 was assessed using peroxidase-conjugated streptavidin. As shown in Fig. 2D, we found that HA22 catalyzed the incorporation of ADP-ribose into EF2 in extracts of sensitive CA46 cells but not CA46-R cells and that the EF2 protein levels were similar in both cell lines. These results indicate that EF2 probably does not contain a functional diphthamide residue, which is required for ADP-ribosylation of EF2, and that the resistance is due to a defect in diphthamide biosynthesis.
Identification of a New Modification of the Diphthamide Residue in CA46-R Cells
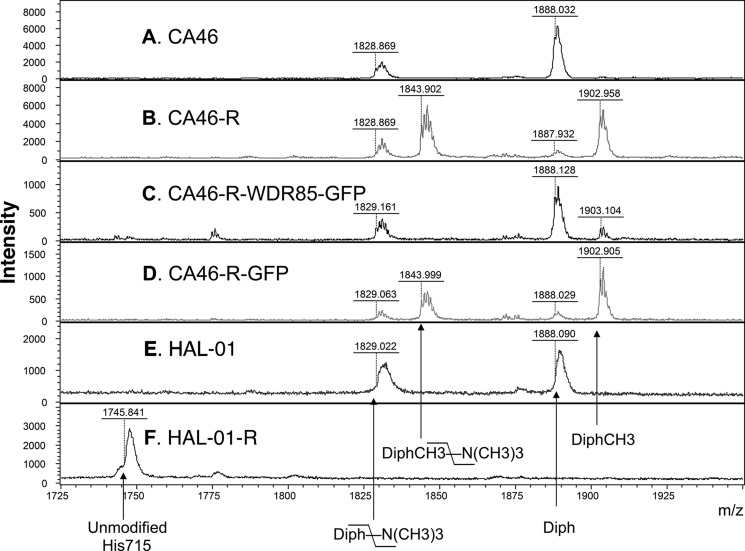
To evaluate the nature of the possible diphthamide modifications, EF2 from CA46 and CA46-R cells was purified and digested by trypsin. For a comprehensive comparison of diphthamide modifications, EF2 from HAL-01 cells, which have normal diphthamide in EF2, and EF2 from HAL-01-R cells, which have unmodified EF2 due to reduced expression of DPH4 (13), were also purified and digested. The tryptic peptides from the four samples were fractionated by reverse phase HPLC (supplemental Fig. S1A). MALDI-TOF-MS analysis of fractions containing peptide FDVHDVTLHADAIHR (residues 702–716) modified and not modified by diphthamide revealed significant differences among these cells (Fig. 3 and supplemental Fig. S1). We detected unmodified His-715 (m/z 1745) in HAL-01-R cells (Fig. 3F). EF2 from parental CA46 and HAL-01 cells yielded peptides with “normal” intact diphthamide (m/z 1888) and a form that had lost a trimethylamino group (m/z 1828) (Fig. 3, A and E). These correspond to peptides containing normal diphthamide (Fig. 4A). CA46-R cells (Fig. 3B) also had small amounts of these peptides, but their relative amounts were significantly lower.
FIGURE 3.
Cells with a deletion of the WDR85 promoter have an additional modification on diphthamide at m/z 1902. Shown are the results from MALDI-TOF-MS of diphthamide-containing peptides purified from different cells. MS spectra were collected in linear mode to prevent detection of metastable forms of diphthamide peptides (see supplemental “Experimental Procedures”). A, CA46 cells. B, CA46-R cells. C, CA46-R-WDR85-GFP cells. WDR85-GFP protein was stably transfected and expressed in CA46-R cells. D, CA46-R-GFP cells. GFP protein was stably transfected and expressed in CA46-R cells. E, HAL-01 cells. HAL-01 is an ALL cell line. F, HAL-01-R cells. HAL-01-R is an HA22-resistant cell line derived from HAL-01 cells. DPH4 expression was reduced and His-715 in EF2 was not modified in HAL-01-R cells (13). Unmodified His715, peptide FDVHDVTLHADAIHR with unmodified His-715; Diph-\-N(CH3)3, peptide with diphthamide that lost the trimethylamino group; DiphCH3-\-N(CH3)3, peptide with methylated diphthamide that lost the trimethylamino group; Diph, peptide with diphthamide; DiphCH3, peptide with methylated diphthamide.
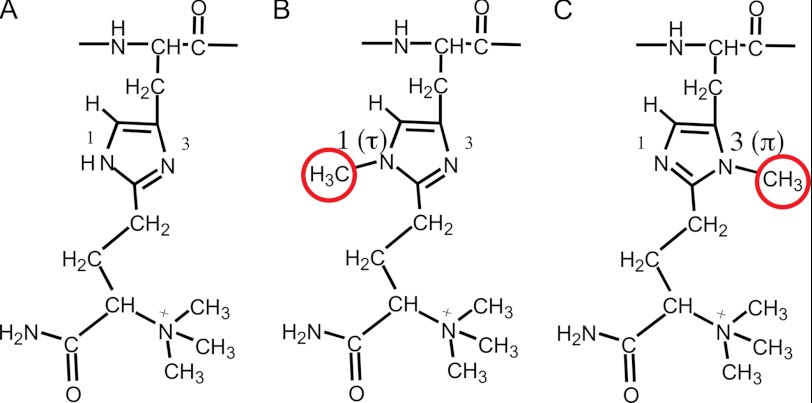
FIGURE 4.
Two potential methylation sites in diphthamide. A, normal diphthamide. B, methylation at N1 (Nτ). C, methylation at N3 (Nπ).
In contrast, EF2 from CA46-R cells contained two major peptide forms (m/z 1902 and 1843), indicating an additional modification on diphthamide. Detailed mass spectrometry analyses indicated that the peptides represent diphthamide containing an additional methyl group at N1 (Nτ) (Fig. 4B) or chemically almost equivalent N3 (Nπ) of the imidazole ring (Fig. 4C).
Reduced WDR85 Expression in the CA46-R Cell Line Is Responsible for Immunotoxin Resistance
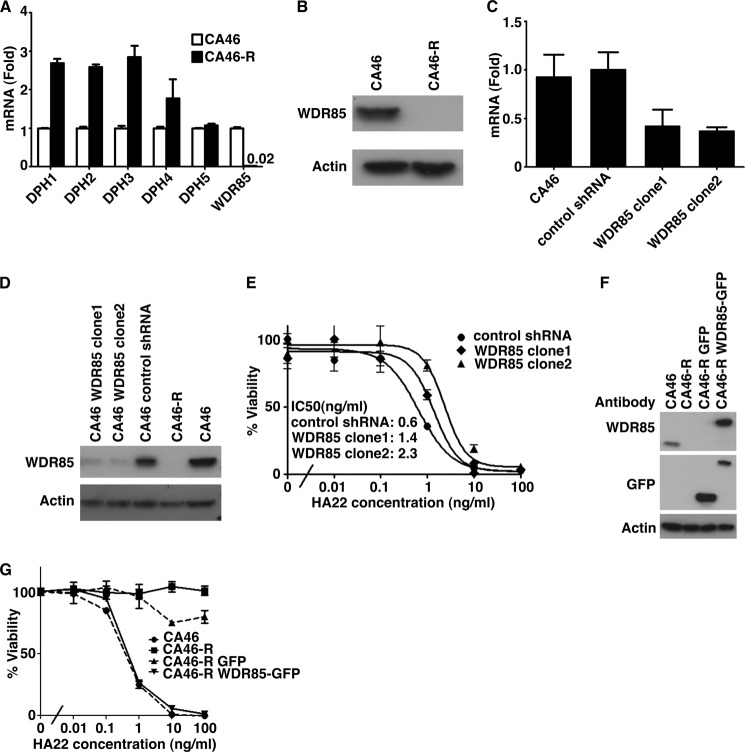
There are six proteins required for diphthamide biosynthesis: DPH1–5 and WDR85 (10, 11, 20). Using RT-PCR, we measured the level of the mRNA for each of these proteins and found that the level of WDR85 mRNA was extremely low in CA46-R cells (Fig. 5A and supplemental Table S1). The levels of the mRNAs for DPH1–4 were actually increased, whereas DPH5 mRNA was similar in both cell types (Fig. 5A). The increase in DPH1–4 was observed in three separate experiments. These data suggest that WDR85 may control the expression of DPH1–4 through a feedback mechanism. We also measured the level of WDR85 protein by immunoblotting and observed that it could not be detected (Fig. 5B). These data indicate that the absence of WDR85 is responsible for the altered diphthamide modification and resistance to HA22.
FIGURE 5.
A, DPH1–5 and WDR85 mRNA levels. mRNA levels were analyzed by quantitative RT-PCR, and the data represent the average of three samples. Only WDR85 mRNA decreased very much in CA46-R cells. B, WDR85 protein levels. Equal amounts of cell lysates from sensitive and resistant cells were subjected to SDS-PAGE, followed by Western blotting with anti-WDR85 and anti-actin antibodies. C–E, knockdown of WDR85 by shRNA. Two different shRNAs from Sigma were used: TRCN0000162819 (clone1) and TRCN0000160958 (clone2). C, quantitative RT-PCR showing efficient suppression of WDR85 mRNA in CA46 cells by shRNA. D, WDR85 down-regulation by shRNA was confirmed by Western blotting. E, the sensitivity to HA22 of control and WDR85 knockdown CA46 cells was assessed by WST assays and is expressed as a percentage of surviving cells relative to the untreated controls. The IC50 values are listed in the inset. F and G, rescue expression of WDR85 in CA46-R cells restores sensitivity. An empty vector was used as a control. F, WDR85-GFP fusion protein expression was confirmed by Western blotting. G, the sensitivity to HA22 of CA46-R cells after electrotransfection was assessed by WST assays and is expressed as a percentage of surviving cells relative to the untreated controls. CA46-R GFP, pEGFP-N1-transfected CA46-R cells; CA46-R WDR85-GFP, pEGFP-N1-WDR85-GFP-transfected CA46-R cells.
To address whether reduced expression of WDR85 is sufficient by itself to produce toxin resistance, we used a lentivirus to introduce shRNA targeting WDR85 into CA46 cells and isolated transduced cells with reduced WDR85 mRNA and protein levels (Fig. 5, C and D). We tested the cells for sensitivity to HA22 and found that the IC50 was increased by ∼4-fold from 0.6 to 2.3 ng/ml (Fig. 5E).
We introduced a WDR85-GFP fusion protein into CA46-R cells to determine whether the phenotype could be restored. Fig. 5F shows that the transfected cells expressed WDR85-GFP fusion protein, and Fig. 5G shows that sensitivity to HA22 was completely restored. We analyzed EF2 from cells transfected with WDR85 and found that the modified diphthamide form of EF2 was absent and replaced by EF2 with normal diphthamide (Fig. 3C).
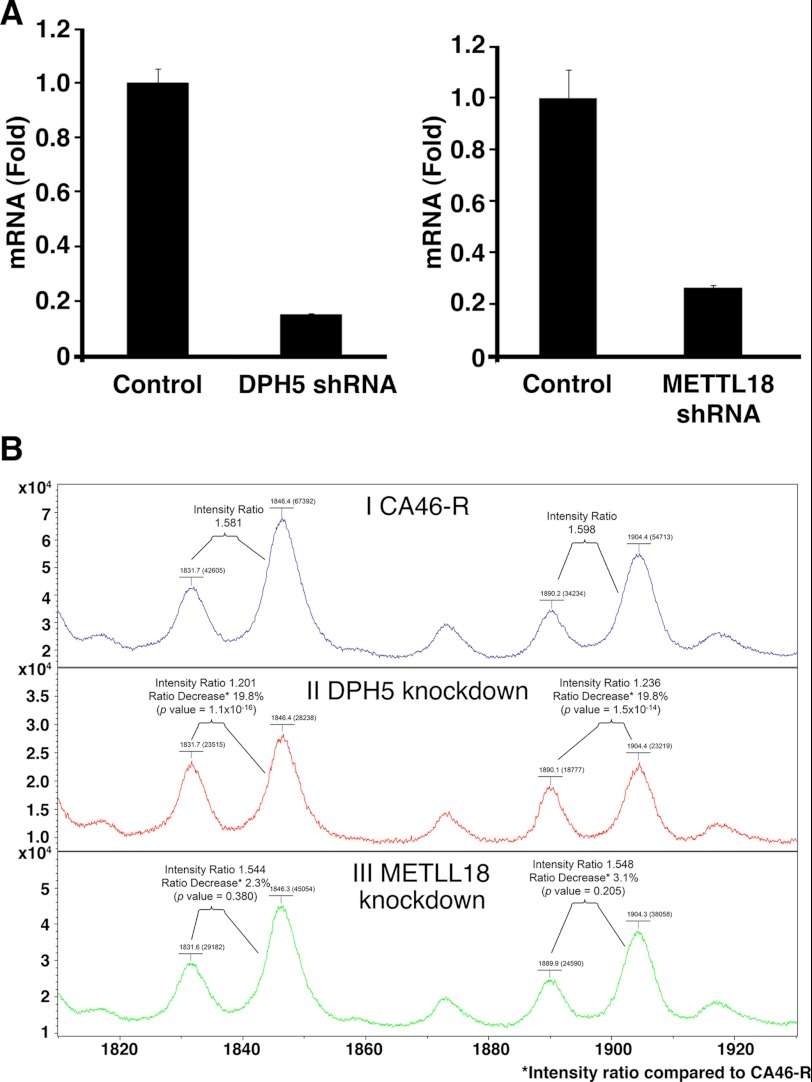
Because DPH5 is a methyltransferase (21), we hypothesized that DPH5 could be responsible for the abnormal methylation of diphthamide observed in the absence of WDR85. To confirm this, we knocked down DPH5 expression in CA46-R cells and found that DPH5 mRNA decreased to 15% of the control level (Fig. 6A) and that the amount of diphthamide with the additional methyl group in EF2 also significantly decreased compared with the control CA46-R cells (Fig. 6B, panels I and II). As a specificity control, we knocked down the expression of another methylation enzyme, METLL18, and did not observe a change in the relative amount of diphthamide with the abnormal methylation (Fig. 6B, panel III). This finding suggests that DPH5 is responsible for the abnormal methylation of diphthamide in cells that lack WDR85 expression.
FIGURE 6.
Knockdown of DPH5 or METTL18 by shRNA in CA46-R cells. A, quantitative RT-PCR showing efficient suppression of DPH5 or METTL18 mRNA in CA46-R cells by shRNA. B, MALDI-TOF-MS of EF2 tryptic peptides in linear mode. Panel I, peptides from CA46-R cells. Panel II, peptides from DPH5 knockdown cells. Panel III, peptides from METLL18 knockdown cells. Peaks at m/z 1831 correspond to the diphthamide-containing peptide that lost the trimethylamino group. Peaks at m/z 1846 correspond to the diphthamide-containing peptide with an additional methyl group in the imidazole group of His-715 but with a lost trimethylamino group. Peaks at m/z 1890 correspond to the peptide with intact diphthamide. Peaks at m/z 1904 correspond to the peptide with diphthamide with an additional methyl group. Comparison of the intensity ratios of related peaks at m/z 1846/1831 and 1904/1890 in DPH5 knockdown and METLL18 knockdown cells with the ratios of the corresponding peaks in CA46-R cells suggests that knockdown of DPH5 significantly reduced the relative intensity of the methylated diphthamide peptides (m/z 1846 and 1904). Knockdown of METLL18 did not reduced the relative intensity of the methylated diphthamide peptides.
Deletion of the WDR85 Gene Is Responsible for Decreased WDR85 Expression
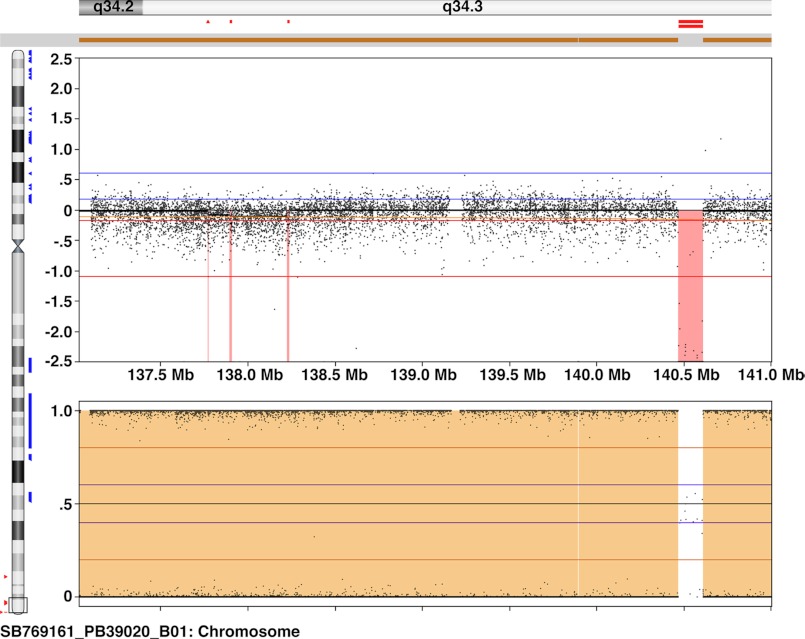
There are multiple mechanisms that have been shown to explain drug resistance (22). We have recently reported that immunotoxin resistance in the HAL-01 ALL cell line is mediated by reversible DNA methylation in the CpG island of the DPH4 promoter region (13). To study the WDR85 promoter in CA46-R cells, we attempted to amplify the promoter region (spanning −1588 to +17 bp) by PCR using three primer pairs and were unable to do so, suggesting that the region was deleted in the resistant cell line. To further evaluate this possibility, we performed high resolution SNP array analysis of CA46 and CA46-R cells, which revealed a homozygous deletion at 9q34.3 from 140,468,104 to 140,610,581 bp (human genome sequence build 19) in the resistant cells (Fig. 7). Thus, deletion of the WDR85 promoter and part of the coding region accounts for the inability of CA46-R cells to make WDR85 protein. There are four additional genes within this deleted region in the resistant cell line (supplemental Table S2). EHMT1 encodes a protein that has been implicated in silencing Myc- and E2F-responsive genes and might play a role in cell cycle transition (23). The functions of the other three genes, ZMYND19, ARRDC1, and C9orf37, are not known.
FIGURE 7.
Copy number variation/SNP array results showing a homozygous deletion at 9q34.3 in CA46-R cells. The upper panel shows the log relative ratio, and the lower panel shows the B-allele frequency for the probes used in CA46-R cells assuming CA46 as the parent cell line. Mb, megabases.
DISCUSSION
We have described the isolation and characterization of a Burkitt lymphoma cell line that is resistant to the cytotoxic effect of the immunotoxin HA22. The resistant cell line has a deletion of the WDR85 gene. In the absence of WDR85, diphthamide in EF2 of resistant cells has a novel modification revealed by mass spectrometry (Fig. 3), which results in a form of EF2 that cannot be ADP-ribosylated and inactivated by an immunotoxin containing the catalytic portion of PE.
The CA46 cells used to select the HA22-resistant mutant were originally isolated by Magrath et al. (24) at NCI and were given to us 15 years ago. Because the CA46 cells were not cloned prior to selection with HA22, we assume that the cells with a deletion of WDR85 and surrounding genes were present in a small proportion of the parental cells used for the selection of HA22 resistance and that the deletion was not induced by the immunotoxin, which is not a mutagen.
The human WDR85 gene is an ortholog of the yeast YBR246W gene and encodes a 50-kDa protein. It belongs to the WD repeat protein family; members of this family have important biological roles in signal transduction, transcription regulation, cell cycle control, and apoptosis. The underlying common function of WD repeat proteins is to coordinate multiprotein complex assemblies, where the WD repeat domain functions as a scaffold for protein-protein interactions (25). Carette et al. (10) have reported that His-715 in EF2 is not modified in cells with insertional inactivation of the WDR85 gene and suggested that WDR85 is required for the first step of diphthamide synthesis. Subsequently, Su et al. (11) investigated yeast cells with a deletion of the YBR246W gene in which they overexpressed EF2 and did not detect diphthamide but did detect some accumulation of diphthamide precursor. On the basis of this finding, they suggested that the YBR246W gene in yeast is required for the last step of diphthamide biosynthesis: amidation of diphthine.
In this study, we investigated an immunotoxin-resistant cell line in which the WDR85 gene is deleted. Using EF2 from resistant cells, we analyzed the peptide of EF2 containing the diphthamide residue by mass spectrometry. Consistent with the report by Carette et al. (10), we found that the amount of diphthamide-containing peptide was drastically reduced, but we did not detect any unmodified His-715 in EF2, although we did detect unmodified EF2 from HAL-01-R cells with reduced DPH4 expression, which we analyzed as a control. Instead, we found that diphthamide from CA46-R cells has an additional modification, which is a methyl group attached to N1 (Nτ; or chemically almost equivalent N3 (Nπ)) of the imidazole ring of His-715 (Fig. 4). A methyl group at either of these positions would prevent ADP-ribosylation of diphthamide because this additional methyl group occupies the nitrogen atom required for ADP-ribosylation. We also found that DPH5 is probably responsible for adding this additional methyl group. The mechanism by which WDR85 protein prevents this methylation is not yet established, but we have noted that the WDR85 protein contains a WD40 repeat domain, and these domains are involved in protein-protein interactions. Carette et al. (10) reported that in the absence of WDR85, DPH5 interacts more strongly with EF2, indicating that WDR85 modulates the interaction of EF2 and DPH5. It is possible that WDR85 acts as an adaptor molecule in an enzymatic complex that regulates the methylation step of the diphthamide biosynthesis pathway. We suggest that previous investigators did not find an abnormally methylated form of diphthamide in EF2 because of a difference in the experimental approach. The experimental difference that may account for the divergent findings is that we analyzed endogenous EF2 from WDR85 mutant cells, whereas overexpressed EF2 from transfected cells was analyzed in the other studies. It is possible that EF2 overexpression and perturbation of diphthamide biosynthesis in cells lacking WDR85 could have led to accumulation of unmodified His-715 (10) and diphthine (11). It also seems possible that a modified diphthamide peptide with a larger than expected m/z could have been overlooked during the analysis.
The deleted region in resistant CA46 cells is located at 9q34 in chromosome 9 from 140,468,104 to 140,610,581 bp (based on build 19 of the human genome sequence). Recurrent deletion of 9q34 has been found in adult normal karyotype precursor B-cell ALL (26), although it is not a common finding, and the reported deleted region does not contain the WDR85 gene. DPH1, also named OVCA1, is ubiquitously expressed in normal human tissues (27). It was reported that DPH1 is deleted in ovarian cancer (28, 29). Overexpression of DPH1 suppresses colony formation in cultured human ovarian cancer cells (30). In gene knock-out mice, loss of Dph1 decreases the growth rate of embryonic fibroblasts and leads to tumorigenesis (31). These studies led to the suggestion that DPH1 is a tumor suppressor gene. Until now, there has been no report about the possible role of the other diphthamide genes in relation to cancer. In this study, we found that WDR85 is deleted in immunotoxin-resistant cells derived from the Burkitt lymphoma cell line CA46. We believe that the WDR85 deletion existed in some cells before selection because we did not use mutagens.
In the region deleted in CA46-R cells, four other genes are removed. These are EHMT1, ZMYND19, ARRDC1, and C9orf37. EHMT1 was reported to be involved in the silencing of Myc- and E2F-responsive genes and could play a role in the G0/G1 cell cycle transition (23). Defects in the EHMT1 gene are the cause of the chromosome 9q subtelomeric deletion syndrome. However, the 9q subtelomeric deletion syndrome is not known to increase susceptibility to cancer (32). Because the abnormal methylation of diphthamide occurs in cells with a deletion of EHMT1, it is unlikely that this protein has a role in the abnormal methylation we observed. ZMYND19 is a MYND zinc finger domain-containing protein that binds to the C terminus of melanin-concentrating hormone receptor-1 and to the N termini of α-tubulin and β-tubulin (33, 34). The function of ZMYND19 is not clear. There is minimal reported information about the ARRDC1 and C9orf37 genes.
In summary, we isolated an immunotoxin-resistant cell line and found that resistance is associated with loss of the diphthamide synthesis gene WDR85 and formation of a novel form of diphthamide in EF2 that cannot be ADP-ribosylated or inactivated by HA22. Reduced expression of WDR85 is due to a deletion of the WDR85 gene. This represents a novel mechanism of immunotoxin resistance that will be assessed in patient samples when they are available.
Acknowledgments
We thank Javed Khan, Young Song, Jun Wei, and Rajesh Patidar for assistance with the SNP array; Matthew J. Fivash for statistical analysis of mass spectrometry data; and Sergei Dikler and Paul Kowalski (Bruker Daltonics) for help with MALDI-TOF-MS data analysis.
This work was supported, in whole or in part, by the Intramural Research Program of the Center for Cancer Research, NCI, National Institutes of Health, in part with federal funds from NCI, National Institutes of Health, under Contract HHSN261200800001E and under a Cooperative Research and Development Agreement with MedImmune, LLC. Dr. Pastan is an inventor of patents on immunotoxins that have all been assigned to the National Institutes of Health.

This article contains supplemental “Experimental Procedures,” Fig. S1, Tables S1 and S2, and additional references.
- ALL
- acute lymphoblastic leukemia
- EF2
- elongation factor-2
- PE
- Pseudomonas exotoxin A
- WST
- water-soluble tetrazolium salt.
REFERENCES
- 1. Howlader N., Noone A. M., Krapcho M., Neyman N., Aminou R., Altekruse S. F., Kosary C. L., Ruhl J., Tatalovich Z., Cho H., Mariotto A., Eisner M. P., Lewis D. R., Chen H. S., Feuer E. J., Cronin K. A. (eds) (2009) SEER Cancer Statistics Review, 1975–2009 (Vintage 2009 Populations), National Cancer Institute, Bethesda, MD, http://seer.cancer. gov/csr/1975_2009_pops09/ [Google Scholar]
- 2. Ries L. A. G., Percy C. L., Bunin G. R. (1999) Introduction. in Cancer Incidence and Survival among Children and Adolescents: United States SEER Program 1975–1995 (Reis L. A. G., Smith M. A., Gurney J. G., Linet M., Tamra T., Young J. L., Bunin G. R., eds) NIH Publication No. 99-4649, pp. 1–16, National Cancer Institute, SEER Program, Bethesda, MD, http://seer.cancer.gov/publications/childhood/ [Google Scholar]
- 3. Oeffinger K. C., Mertens A. C., Sklar C. A., Kawashima T., Hudson M. M., Meadows A. T., Friedman D. L., Marina N., Hobbie W., Kadan-Lottick N. S., Schwartz C. L., Leisenring W., Robison L. L., and Childhood Cancer Survivor Study (2006) Chronic health conditions in adult survivors of childhood cancer. N. Engl. J. Med. 355, 1572–1582 [DOI] [PubMed] [Google Scholar]
- 4. Wayne A. S., Reaman G. H., Helman L. J. (2008) Progress in the curative treatment of childhood hematologic malignancies. J. Natl. Cancer Inst. 100, 1271–1273 [DOI] [PubMed] [Google Scholar]
- 5. Wayne A. S., Kreitman R. J., Findley H. W., Lew G., Delbrook C., Steinberg S. M., Stetler-Stevenson M., FitzGerald D. J., Pastan I. (2010) Anti-CD22 immunotoxin RFB4(dsFv)-PE38 (BL22) for CD22 positive hematologic malignancies of childhood: pre-clinical studies and phase I clinical trial. Clin. Cancer Res. 16, 1894–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kreitman R. J., Tallman M. S., Robak T., Coutre S., Wilson W. H., Stetler-Stevenson M., Fitzgerald D. J., Lechleider R., Pastan I. (2012) Phase I trial of anti-CD22 recombinant immunotoxin moxetumomab pasudotox (CAT-8015 or HA22) in patients with hairy cell leukemia. J. Clin. Oncol. 30, 1822–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Salvatore G., Beers R., Margulies I., Kreitman R. J., Pastan I. (2002) Improved cytotoxic activity toward cell lines and fresh leukemia cells of a mutant anti-CD22 immunotoxin obtained by antibody phage display. Clin. Cancer Res. 8, 995–1002 [PubMed] [Google Scholar]
- 8. FitzGerald D. J., Wayne A. S., Kreitman R. J., Pastan I. (2011) Treatment of hematologic malignancies with immunotoxins and antibody-drug conjugates. Cancer Res. 71, 6300–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Greganova E., Altmann M., Bütikofer P. (2011) Unique modifications of translation elongation factors. FEBS J. 278, 2613–2624 [DOI] [PubMed] [Google Scholar]
- 10. Carette J. E., Guimaraes C. P., Varadarajan M., Park A. S., Wuethrich I., Godarova A., Kotecki M., Cochran B. H., Spooner E., Ploegh H. L., Brummelkamp T. R. (2009) Haploid genetic screens in human cells identify host factors used by pathogens. Science 326, 1231–1235 [DOI] [PubMed] [Google Scholar]
- 11. Su X., Chen W., Lee W., Jiang H., Zhang S., Lin H. (2012) YBR246W is required for the third step of diphthamide biosynthesis. J. Am. Chem. Soc. 134, 773–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wayne A. S., Bhojwani D., Silverman L. B., Richards K., Stetler-Stevenson M., Shah N., Jeha S., Pui C.-H., Buzoianu M., Fitzgerald D. J., Kreitman R. J., Ibrahim R., Pastan I. (2011) A novel anti-CD22 immunotoxin, moxetumomab pasudotox: phase I study in pediatric acute lymphoblastic leukemia. Blood 118, Abstr. 248, 1317 [Google Scholar]
- 13. Wei H., Xiang L., Wayne A. S., Chertov O., FitzGerald D. J., Bera T. K., Pastan I. (2012) Immunotoxin resistance via reversible methylation of the DPH4 promoter is a unique survival strategy. Proc. Natl. Acad. Sci. U.S.A. 109, 6898–6903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pastan I., Beers R., Bera T. K. (2004) Recombinant immunotoxins in the treatment of cancer. Methods Mol. Biol. 248, 503–518 [DOI] [PubMed] [Google Scholar]
- 15. Du X., Beers R., Fitzgerald D. J., Pastan I. (2008) Differential cellular internalization of anti-CD19 and -CD22 immunotoxins results in different cytotoxic activity. Cancer Res. 68, 6300–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y., Hansen J. K., Xiang L., Kawa S., Onda M., Ho M., Hassan R., Pastan I. (2010) A flow cytometry method to quantitate internalized immunotoxins shows that Taxol synergistically increases cellular immunotoxins uptake. Cancer Res. 70, 1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chaudhary V. K., Queen C., Junghans R. P., Waldmann T. A., FitzGerald D. J., Pastan I. (1989) A recombinant immunotoxin consisting of two antibody variable domains fused to Pseudomonas exotoxin. Nature 339, 394–397 [DOI] [PubMed] [Google Scholar]
- 18. Fei Z., Bera T. K., Liu X., Xiang L., Pastan I. (2011) Ankrd26 gene disruption enhances adipogenesis of mouse embryonic fibroblasts. J. Biol. Chem. 286, 27761–27768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Du X., Youle R. J., FitzGerald D. J., Pastan I. (2010) Pseudomonas exotoxin A-mediated apoptosis is Bak dependent and preceded by the degradation of Mcl-1. Mol. Cell. Biol. 30, 3444–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y., Zhu X., Torelli A. T., Lee M., Dzikovski B., Koralewski R. M., Wang E., Freed J., Krebs C., Ealick S. E., Lin H. (2010) Diphthamide biosynthesis requires an organic radical generated by an iron-sulphur enzyme. Nature 465, 891–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mattheakis L. C., Shen W. H., Collier R. J. (1992) DPH5, a methyltransferase gene required for diphthamide biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 4026–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baylin S. B. (2011) Resistance, epigenetics and the cancer ecosystem. Nat. Med. 17, 288–289 [DOI] [PubMed] [Google Scholar]
- 23. Ogawa H., Ishiguro K., Gaubatz S., Livingston D. M., Nakatani Y. (2002) A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296, 1132–1136 [DOI] [PubMed] [Google Scholar]
- 24. Magrath I. T., Freeman C. B., Pizzo P., Gadek J., Jaffe E., Santaella M., Hammer C., Frank M., Reaman G., Novikovs L. (1980) Characterization of lymphoma-derived cell lines: comparison of cell lines positive and negative for Epstein-Barr virus nuclear antigen. II. Surface markers. J. Natl. Cancer Inst. 64, 477–483 [PubMed] [Google Scholar]
- 25. Li D., Roberts R. (2001) WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58, 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nowak N. J., Sait S. N., Zeidan A., Deeb G., Gaile D., Liu S., Ford L., Wallace P. K., Wang E. S., Wetzler M. (2010) Recurrent deletion of 9q34 in adult normal karyotype precursor B-cell acute lymphoblastic leukemia. Cancer Genet. Cytogenet. 199, 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jensen M. R., Helin K. (2004) OVCA1: emerging as a bona fide tumor suppressor. Genes Dev. 18, 245–248 [DOI] [PubMed] [Google Scholar]
- 28. Phillips N., Ziegler M., Saha B., Xynos F. (1993) Allelic loss on chromosome 17 in human ovarian cancer. Int. J. Cancer 54, 85–91 [DOI] [PubMed] [Google Scholar]
- 29. Phillips N. J., Zeigler M. R., Deaven L. L. (1996) A cDNA from the ovarian cancer critical region of deletion on chromosome 17p13.3. Cancer Lett. 102, 85–90 [DOI] [PubMed] [Google Scholar]
- 30. Bruening W., Prowse A. H., Schultz D. C., Holgado-Madruga M., Wong A., Godwin A. K. (1999) Expression of OVCA1, a candidate tumor suppressor, is reduced in tumors and inhibits growth of ovarian cancer cells. Cancer Res. 59, 4973–4983 [PubMed] [Google Scholar]
- 31. Chen C. M., Behringer R. R. (2004) Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev. 18, 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stewart D. R., Kleefstra T. (2007) The chromosome 9q subtelomere deletion syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 145C, 383–392 [DOI] [PubMed] [Google Scholar]
- 33. Bächner D., Kreienkamp H. J., Richter D. (2002) MIZIP, a highly conserved, vertebrate specific melanin-concentrating hormone receptor 1 interacting zinc-finger protein. FEBS Lett. 526, 124–128 [DOI] [PubMed] [Google Scholar]
- 34. Francke F., Buck F., Bächner D. (2005) MYND domain specific interaction of the melanin-concentrating hormone receptor 1 interacting zinc-finger protein with α- and β-tubulin. Biochem. Biophys. Res. Commun. 334, 1292–1298 [DOI] [PubMed] [Google Scholar]