Background: α-l-Rhamnosidase hydrolyzes α-linked l-rhamnose from rhamnoglycosides or polysaccharides.
Results: The crystal structure of Streptomyces avermitilis α-l-rhamnosidase belonging to glycoside hydrolase family 78 was determined.
Conclusion: The l-rhamnose complexed structure revealed the catalytic mechanism of the enzyme and a calcium-dependent carbohydrate-binding module.
Significance: Efficient catalysis of an exo-rhamnosidase requires a novel carbohydrate-binding module that binds terminal l-rhamnose sugars.
Keywords: Carbohydrate-binding Protein, Crystal Structure, Enzyme Mechanisms, Enzyme Structure, Glycoside Hydrolases, α-l-Rhamnose Binding Module, α-l-Rhamnosidase, Carbohydrate Binding Module Family xx, Glycoside Hydrolase Family 78
Abstract
α-l-Rhamnosidases hydrolyze α-linked l-rhamnosides from oligosaccharides or polysaccharides. We determined the crystal structure of the glycoside hydrolase family 78 Streptomyces avermitilis α-l-rhamnosidase (SaRha78A) in its free and l-rhamnose complexed forms, which revealed the presence of six domains N, D, E, F, A, and C. In the ligand complex, l-rhamnose was bound in the proposed active site of the catalytic module, revealing the likely catalytic mechanism of SaRha78A. Glu636 is predicted to donate protons to the glycosidic oxygen, and Glu895 is the likely catalytic general base, activating the nucleophilic water, indicating that the enzyme operates through an inverting mechanism. Replacement of Glu636 and Glu895 resulted in significant loss of α-rhamnosidase activity. Domain D also bound l-rhamnose in a calcium-dependent manner, with a KD of 135 μm. Domain D is thus a non-catalytic carbohydrate binding module (designated SaCBM67). Mutagenesis and structural data identified the amino acids in SaCBM67 that target the features of l-rhamnose that distinguishes it from the other major sugars present in plant cell walls. Inactivation of SaCBM67 caused a substantial reduction in the activity of SaRha78A against the polysaccharide composite gum arabic, but not against aryl rhamnosides, indicating that SaCBM67 contributes to enzyme function against insoluble substrates.
Introduction
l-Rhamnose is widely distributed in plants and bacteria as a component of polysaccharides, such as rhamnogalacturonan and arabinogalactan-protein (1, 2), whereas the sugar is also found in some glycol conjugates such as naringin and rutin (3, 4). Naringin, a major flavonoid in grapefruit, contains α-1,2-linked l-rhamnose. Rutin is a citrus flavonoid found in buckwheat, and contains α-1,6-linked l-rhamnose. α-l-Rhamnosidases (EC 3.2.1.40) catalyze the hydrolysis of α-l-rhamnosyl-linkages in l-rhamnose containing compounds. According to the CAZy database (5, 6), currently α-l-rhamnosidases are classified into three glycoside hydrolase families (GHs):5 GH28, GH78, and GH106. To date, only two crystal structures of α-l-rhamnosidases have been determined: α-l-rhamnosidase B (BsRhaB) from Bacillus sp. GL1 (7) and the putative α-l-rhamnosidase BT1001 from Bacteroides thetaiotaomicron VPI-5482 (8). Both enzymes belong to GH78. BsRhaB is composed of five distinct domains and BT1001 contains four domains. The catalytic module of GH78 enzymes is an (α/α)6-barrel, but the detailed catalytic mechanism of GH78 enzymes is unclear, and the role of the surrounding domains are unknown.
In a previous study (9), we characterized a GH78 α-l-rhamnosidase from Streptomyces avermitilis (SaRha78A), showing that the enzyme hydrolyzed aryl rhamnosides and rhamnose-containing polysaccharides. In this article, we provide the crystal structure of SaRha78A in apo form and in complex with l-rhamnose. The data revealed the catalytic mechanism of the enzyme and identified a novel non-catalytic carbohydrate-binding module (CBM), SaCBM67, the founding member of CBM67. Analysis of the ligand specificity of SaCBM67 showed that the protein module bound l-rhamnose through a calcium-dependent mechanism in a short binding cleft. Inactivation of SaCBM67 through mutagenesis or chelation of calcium showed that the CBM made a substantial contribution to the activity of SaRha78A against polysaccharides, but not against aryl rhamnosides.
EXPERIMENTAL PROCEDURES
Crystallization of SaRha78A
Recombinant SaRha78A was expressed in Escherichia coli and purified by histidine tag affinity chromatography as described by Ichinose et al. (9). The purified protein solution in 2 mm Tris-HCl buffer, pH 7.0, containing 20 mm NaCl, was concentrated to 10 mg ml−1 (A1 mm280 = 2.2 units) by ultrafiltration using a YM-30 membrane (Millipore, Billerica, MA), and filtered through a 0.1-μm membrane. The protein was crystallized by the sitting-drop vapor diffusion method using a precipitant solution composed of 10% (w/v) PEG6000, 5% (v/v) MPD, and 0.1 m HEPES pH 6.8. Plate-shaped crystals with 500 × 50 × 10 μm dimensions were consistently obtained using 50 μl of the reservoir solution with a drop consisting of 1 μl of protein solution and 1 μl of reservoir solution. Selenomethionine-labeled SaRha78A was expressed in LeMaster medium using the methionine-auxotrophic strain B834(DE3) (10) and crystallized in the same condition used for the native enzyme.
Data Collection and Structure Determination
Diffraction experiments for native crystals were conducted at the beamline BL41XU of the synchrotron facility SPring-8, Hyogo, Japan. Crystals were soaked into the reservoir solution containing 13% (v/v) glycerol, scooped in a 0.5-mm nylon loop (Hampton Research, Aliso Viejo, CA), and flash-frozen in a nitrogen stream at 95 K. Diffraction data were collected at a wavelength of 0.97915 Å with a Quantum 210 CCD detector (Area Detector Systems Corp., Poway, CA). Diffraction experiments for the selenomethionine-substituted crystals were conducted at beamline BL-5 of the Photon Factory, High Energy Accelerator Research Organization, Tsukuba, Japan. Diffraction data were collected at a wavelength of 0.97909 Å with a Quantum 315 CCD detector (ADSC). For structural analyses of the enzyme in complex with l-rhamnose (Wako Pure Chemical Industry), SaRha78A crystals were soaked into a drop containing 1% (w/v) l-rhamnose in the precipitant solution for 10 min before the diffraction experiment. Diffraction data of the l-rhamnose complex to 1.9-Å resolution were collected at beamline BL-NW12 of the Photon Factory Advanced Ring. Diffraction data were collected at a wavelength of 1.0000 Å with a Quantum 210 CCD detector (ADSC). All data were integrated and scaled using the program DENZO and SCALEPACK in the HKL2000 program suite (11).
The crystal structure was determined by the single-wavelength anomalous dispersion method using selenomethionine-labeled crystals. In total 12 selenium atom positions were determined, and initial phases were calculated using the program SOLVE/RESOLVE (12, 13). The initial model building was conducted by the automodeling program ARP/wARP (14) within the CCP4 program suite (15). Manual model building and molecular refinement were performed using Coot (16) and Refmac5 (17, 18).
For analysis of the l-rhamnose-protein complex, structural determination was conducted using the ligand-free structure as the starting model and the bound l-rhamnose was observed in the difference electron density map. Data collection and refinement statistics are given in Table 1. Stereochemistry of the models was analyzed with the program Rampage (19). Structural drawings were prepared by the PyMol program (DeLano Scientific LLC, Palo Alto, CA).
TABLE 1.
Data collection and refinement statistics of α-l-rhamnosidase
Values in parentheses refer to the highest resolution shell.
| Data collection | Native Apo, PDB code 3W5M | l-Rhamnose complex, PDB code 3W5N | Selenomethionine peak |
|---|---|---|---|
| Space group | P21 | P21 | P21 |
| Cell parameters | |||
| Å | a = 52.8, b = 128.4, c = 75.2 | a = 53.1, b = 128.6, c = 75.3 | a = 52.9, b = 128.8, c = 75.0 |
| Degree | β = 99.3 | β = 99.8 | β = 99.4 |
| X-ray source | Spring-8 BL41XU | PF-AR BL-NW12 | PF BL-5 |
| Wavelength (Å) | 0.97915 | 1.00000 | 0.97909 |
| Resolution (Å) | 50–1.8 (1.86–1.80) | 50–1.8 (1.86–1.80) | 50–2.0 (2.07–2.00) |
| No. reflections | 640,389 | 694,122 | 499,517 |
| No. unique reflections | 92,680 (9,140) | 91,813 (9,161) | 66,810 (6,533) |
| Completeness (%) | 99.8 (98.5) | 100.0 (99.9) | 99.8 (97.9) |
| Multiplicity | 6.9 (5.7) | 7.6 (7.3) | 7.5 (6.6) |
| R-merge | 0.099 (0.632) | 0.084 (0.558) | 0.085 (0.387) |
| Average I/σ | 36.9 (4.3) | 27.5 (3.9) | 25.8 (4.5) |
| Refinement | |||
| Resolution (Å) | 35.7–1.8 (1.85–1.80) | 40.6–1.8 (1.85–1.80) | |
| R-factor | 0.180 (0.275) | 0.163 (0.230) | |
| R-free | 0.214 (0.378) | 0.194 (0.287) | |
| No. waters | 758 | 1099 | |
| Average B-value (Å2) | 30.3 | 19.6 | |
| Root mean square deviations from ideals | |||
| Lengths (Å) | 0.010 | 0.009 | |
| Angles (°) | 1.40 | 1.26 | |
| Ramachandran plot (%) (favored/allowed/disallowed) | 98.1/1.9/0.0 | 97.6/2.4/0.0 | |
Sugar Binding Analysis by Isothermal Titration Calorimetry
To produce SaCBM67, the appropriate region of the SaRha78A gene (encoding residues Pro133-Ala296) was amplified using the following primers: forward, 5′-CATATGCCTTCCCTGGAGGGTAGTTCGTGG-3′ and reverse, 5′-AAGCTTTCACGCGACCCGGCCCCACGGTCCTGC-3′. The amplified DNA was cloned into pET28 vector (Novagen) at NdeI and HindIII restriction enzyme sites (underlined). The protein was expressed by the same method as SaRha78A, and purified by immobilized metal ion affinity chromatography using the N-terminal histidine tag. The D179A and N180A mutants of SaCBM67 were constructed using the PCR-based QuikChangeTM site-directed mutagenesis kit (Stratagene) of the SaRhaCBM plasmid using the following primers: mutant D179A, 5′-CTGGCCATCAGCGCGGCGAACGTCTACGCCGTC-3′ and 5′-GACGGCGTAGACGTTCGCCGCGCTGATGGCCAG-3′; N180A, 5′-GCCATCAGCGCGGACGCGGTCTACGCCGTCTCC-3′, and 5′-GGAGACGGCGTAGACCGCGTCCGCGCTGATGGC-3′ (mutated bases underlined).
Binding of SaCBM67 to its target ligands was determined by isothermal titration calorimetry using a MicroCal VP-ITC. Titrations were carried out at 25 °C in 20 mm Na-HEPES buffer, pH 7.5, containing 2 mm CaCl2, unless otherwise stated. The reaction cell contained protein at 700–800 μm (depending on the ligand), whereas the syringe contained 20 mm of the potential ligands (l-fucose, l-mannose, and l-rhamnose purchased from Sigma). The data were analyzed using MicroCal Origin version 7.0, which yielded the change in enthalpy (ΔH), association constants (Ka) and stoichiometry of binding (n), enabling changes in entropy (ΔS) and Gibbs free energy (ΔG) to be calculated using the equation −RTInKa = ΔG = ΔH–TΔS.
Mutant Generation and Enzyme Assays
Site-directed mutagenesis was used to generate the SaRha78A mutants of catalytically important residues. The pET30-SaRha78A plasmid, encoding full-length SaRha78A (9), was used as a template and PCR with KOD -plus- neo polymerase (Toyobo, Osaka, Japan) was performed using the following sets of primers: E636D-F, 5′-CGGCCCGTGATGATCGGCTCGGCTGGA-3′, and E636D-R, 5′-CAGCCGAGCCGATCATCACGGGCCGGAGT-3′; E636Q-F, 5′-CGGCCCGTGATCAACGGCTCGGCTGGA-3′, and E636Q-R, 5′-CAGCCGAGCCGTTGATCACGGGCCGGAGT-3′; E895Q-F, 5′-ACCACCATGTGGCAGCGCTGGGACTCCAT-3′, and E895Q-R, 5′-AGTCCCAGCGCTGCCACATGGTGGTGGA-3′; E895D-F, 5′-ACCACCATGTGGGATCGCTGGGACTCCAT-3′, and E895D-R, 5′-AGTCCCAGCGATCCCACATGGTGGTGGA-3′. The underlined sequences represent mutation sites. Recombinant SaRha78A mutants were expressed in E. coli Tuner (DE3) (Novagen, Merck KGaA, Darmstadt, Germany) and purified, and the kinetic parameters of the wild type and mutants of SaRha78A for p-nitrophenyl-α-l-rhamnopyranoside (PNP-α-l-Rha) were determined as described previously (9). The activity of the wild type and mutants of SaRha78A for gum arabic were determined as follows. Gum arabic was treated with 0.05 n H2SO4 at 100 °C for 1 h for acid hydrolysis, and subjected to enzyme digestion by α-l-arabinofuranosidase (20) and β-l-arabinopyranosidase (21), to increase enzyme accessibility to the polysaccharide. After removal of released l-arabinose by dialysis, the partially digested gum arabic was freeze dried and then used as the substrate. The reactions were performed in McIlvaine buffer, pH 5.0, containing 1–5% (w/v) substrates, 0.1% (w/v) bovine serum albumin, 2 mm CaCl2, and 0.9 nm to 10.0 mm enzyme at 40 °C for up to 360 min. The amount of l-rhamnose released was quantified by high-performance anion-exchange chromatography with pulsed amperometric detection. The assay was performed in duplicate. The kinetic parameters kcat and Km were determined by Lineweaver-Burk plot from three independent experiments and at five substrate concentrations.
RESULTS
Overall Structure of SaRha78A
The crystal structure of SaRha78A was determined by the single-wavelength anomalous dispersion method using selenomethionine crystal data. The structure of the apo form of SaRha78A and the protein in complex with l-rhamnose were determined. Structure refinement statistics are summarized in Table 1. The quality and accuracy of the final structures are indicated by the observation that no residues fell within the disallowed region of the Ramachandran stereochemistry plot. The recombinant SaRha78A molecule is composed of a single polypeptide chain of 1043 amino acids, where the C-terminal His tag, 1031KLAAALEHHHHHH1043, were derived from the expression vector and purification tag, respectively. The two residues N-terminal Met1 and Ser2 and the 11 C-terminal residues, Ala1033 to His1043, were not identified due to lack of electron density. The final model consisted of one SaRha78A molecule in the asymmetric unit bound to a single calcium ion. The ligand free structure contained three Tris molecules, and the l-rhamnose complex structure contained three sodium ions.
The protein forms a multidomain structure comprised of six distinct domains, one α-domain and five β-domains (Fig. 1). They were designated as domain N, domain D (designated SaCBM67 as it fulfills a carbohydrate binding function, see below), domain E, domain F, domain A (designated SaRha78CM as it comprises the catalytic module, see below), and domain C from the N terminus to the C terminus. Domain N (Ala3-Gly114) consists of 10 β-strands folding into 4- and 3-stranded β-sheets and adopt a fibronectin type 3 fold. Domain D (SaCBM67 Pro133-Pro297) and domain E (Ala115-Ala132, Val298-Gly426, and Glu444-Gly478) display a β-jellyroll fold consisting of 11 and 13 β-strands, respectively (SaCBM67 is described in detail below). Domain E provided two β-strands (Pro427-Lys443) that were inserted into one of the β-sheets of Domain F (Pro479-Ala596 and Pro427-Lys443), which also displayed a β-jellyroll fold. Domain F consisted of 16 β-strands organized into two parallel β-sheets.
FIGURE 1.
Structure of SaRha78A. A, stereoview of the ribbon model of the SaRha78A·l-rhamnose complex structure. Each domain is drawn in a different color: domains N, D, E, F, A, and C are in blue, cyan, green, yellow, orange, and red colors, respectively. Calcium ion as a pink sphere, and the bound l-rhamnose molecules in gray stick models. B, topology diagram of SaRha78A. α-Helices, 310-helices, and β-strands are shown in filled cylinders, shaded cylinders, and filled arrows, respectively. Domains are colored in accordance with A. β-Strands are numbered in domain D.
SaRha78CM (Val606-Ala931) comprised an (α/α)6-barrel, which displays structural similarity to the catalytic module of BsRhaB, the prototypic GH78 rhamnosidase from Bacillus sp. (7), and is thus likely to fulfill the same function in SaRha78A. SaRha78CM contains 13 α-helices and lacks any β-structures. Domain C (Gly932-Val1030 and Pro597-Asn605) adopts a β-sandwich structure in which the two β-sheets consist of 5- and 6-anti-parallel β-strands, respectively.
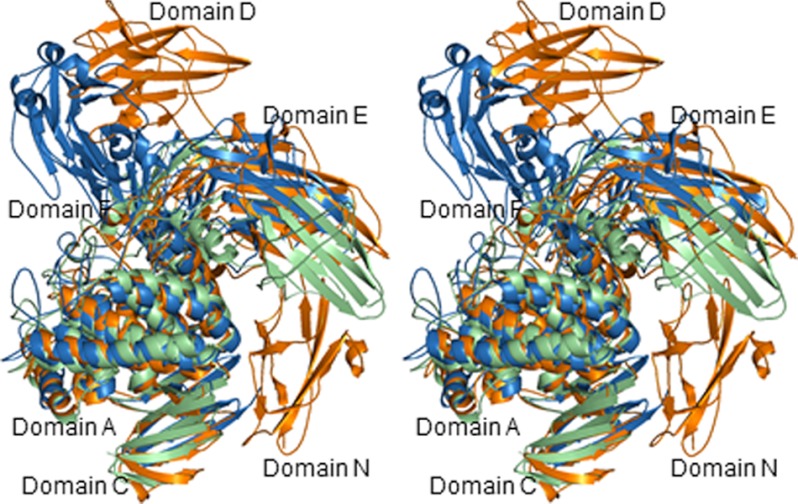
Domains N, E, F, and C are located around SaRha78CM. Domains F and C interact with SaRha78CM through large interfaces, whereas domains N and E also abut onto the catalytic module, but through a small contact area. SaCBM67 protrudes from domain E through two linker peptides, and makes no contact with the other domains. The domain arrangement of SaRha78A is partially different to the other two GH78 proteins, BsRhaB and BT1001, for which crystal structures are available. When the three proteins were superimposed it is evident that a domain equivalent to domain N is not present in BT1001 or BsRhaB, whereas SaRha78CM domains F and C are common to all three proteins and superimposed with a root mean square deviation of 2.3 (SaRha78CM), 2.1 (domain F), and 1.5 Å (domain C) between SaRha78A and BT1001 and 2.0 (SaRha78CM), 2.0 (domain F), and 1.3 Å (domain C) between SaRha78A and BsRhaB (Fig. 2). Domain E also superimposed with an equivalent domain in BsRhaB with a root mean square deviation of 1.8 Å, but the equivalent domain is displaced in BT1001. SaCBM67 were essentially in a different position in SaRha78A and BsRhaB, but was absent in BT1001 (Fig. 2).
FIGURE 2.
Stereoview of the superimposed ribbon-models of GH78 proteins. SaRha78A, BsRhaB (PDB code 2OKX) (7), BT1001 (PDB code 3CIH) are drawn in orange, sky blue, and light green, respectively.
Crystal Structure of SaRha78A in Complex with l-Rhamnose
To explore the mechanism of enzyme action the structure of SaRha78A in complex with l-rhamnose, its reaction product, was determined. The bound l-rhamnose molecules were observed in two biologically significant regions of the enzyme (Fig. 1A); in the active site of the catalytic module SaRha78CM (Rha1501) and bound to SaCBM67 (Rha1511). The average B-factors of the cyclic six atoms of the l-rhamnose rings were 20.9 and 17.9 Å2 for Rha1501 and Rha1511, respectively. The overall structure of the SaRha78A·l-rhamnose complex was almost identical to that of ligand-free SaRha78A with a root mean square deviation of 0.21 Å, implying little effect of ligand binding upon the overall structure.
Catalytic Module
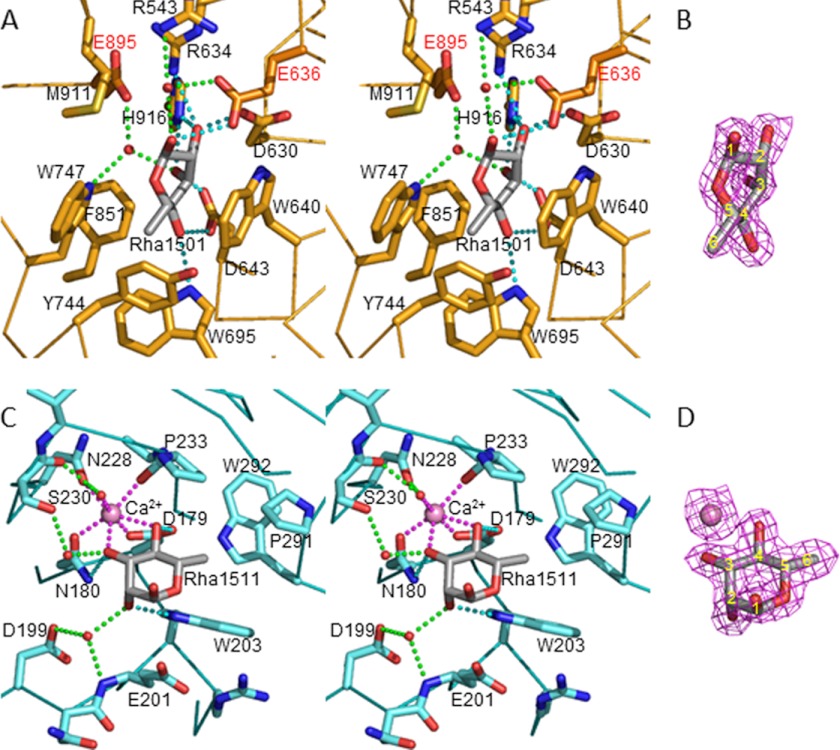
Rha1501, located in the active site of SaRha78CM, adopted an intermediate structure between skewboat 5S1 and boat 2,5B conformations (Figs. 1 and 3, A and B), and the bound l-rhamnose was observed in mainly an α-anomeric configuration. The bound l-rhamnose was sandwiched between two aromatic residues, Trp747 and Trp640, which thus make extensive hydrophobic interactions with the pyranose ring of the sugar. The pocket topology is completed by additional hydrophobic residues, Trp695, Tyr744, and Phe851. The C-6 methyl group, which comprises the signature feature of l-rhamnose, distinguishing the sugar from l-mannose, was buried in a hydrophobic hollow comprising Trp695, Tyr744, and Trp747. The likely polar contacts between Rha1501 and the protein are as follows: O1 is within hydrogen bonding distance with Glu636 Oϵ2, and makes water-mediated polar contacts with Glu636 Oϵ1 and Arg543 Nη2. O2 hydrogen bonded with Asp630 Oδ2, His916 Nϵ2, and Arg634 Nη2, where the closest contact was with Asp630 (2.4 Å). O3 makes polar contacts with Asp643 Oδ2 and His916 Nϵ2 and, through water-mediated interactions, with Trp747 Nϵ1 and Glu895 Oϵ2. This water molecule was situated in close proximity to the C1 atom of Rha1501 with a distance of 2.9 Å. O4 hydrogen bonded with Asp643 Oδ1 and Trp695 Nϵ1. Thus, Rha1501 made eight direct and four solvent-mediated hydrogen bonds with SaRha78A, and all these amino acids are conserved in BsRhaB. The O3 and O4 atoms of Rha1501 were located at the bottom of the pocket, and the O1 atom was at the entrance and was solvent exposed, explaining why SaRha78A is an exo-acting enzyme that removes l-rhamnose residues from the non-reducing end of oligosaccharide or polysaccharide substrates.
FIGURE 3.
The bound l-rhamnose structures in the SaRha78A·l-rhamnose complex. Bound l-rhamnose molecules are shown in gray stick models. Estimated hydrogen bonds are shown in cyan, water-mediating hydrogen bonds in green, ion coordination in magenta break lines. A, stereo view of the catalytic pocket. Two catalytic residues are in pale-red color. B, 2Fobs − Fcalc electron density map of the bound l-rhamnose in the catalytic pocket. Contour level is 1 σ. C, stereo view of the sugar-binding pocket in domain D (SaCBM67). Calcium ion as pink sphere. D, electron density map of the bound l-rhamnose and calcium ion shown with contour level of 1 σ.
SaCBM67
SaCBM67 displays a β-jellyroll fold in which the convex (β-sheet 1) and concave (β-sheet 2) β-sheets comprise six and five anti-parallel strands, respectively. The order of the β-strands is as follows: β-sheet 1: β-6, β-3, β-8, β-9/β-1 (forms an interrupted β-strand), β-11; β-sheet 2: β-10, β-2, β-7, β-4, β-5 (Fig. 1B). Although this fold is typical of the vast majority of CBMs (for review see Ref. 22), unusually, the two sheets are not fully solvent exposed; β-sheet 2 is partially occluded by a loop extending from Thr260 to Lys282, whereas the two loops that link SaCBM67 with the rest of the enzyme lie over β-sheet 1. Rha1511 was positioned in a blind canyon interacting primarily with loops connecting β-strands β-3 and β-4, β-5 and β-6, β-7 and β-8. A central feature of the l-rhamnose binding site is a calcium ion that makes coordinate bonds with O3 and O4 of the sugar, whereas the metal interacts with the protein through Asp179 Oδ1, Asn180 Oδ1, Asn228 Oδ1, Pro233 main chain O, and a water-mediated contact with Ser230 main chain O (Fig. 3, C and D). The bound l-rhamnose also makes direct hydrogen bonds with Trp203 Nϵ1, Asn180 Nδ2, and Asp179 Oδ2 through O2, O3, and O4 atoms, respectively. It is noteworthy that the peptide linkage between two calcium coordinating residues Asp179 Oδ1 and Asn180 Oδ1 was in cis-configuration. Two water molecules also mediated interactions between l-rhamnose and the protein. The C-6 methyl group pointed toward a small hydrophobic pocket comprising Trp203, Pro233, Pro291, and Trp292. The bound l-rhamnose adopted a relaxed 1C4 chair conformation with the O1 atom in the α-anomeric configuration. Although O2, O3, and O4 pointed at the protein surface, O1 was solvent exposed, indicating that SaCBM67 binds to l-rhamnose residues at the non-reducing termini of complex carbohydrates.
Catalytic Residues of SaRha78A and the Specificity and Function of SaCBM67
To verify the proposed identity of the catalytic residues of SaRha78A, aspartic acid and glutamine mutants of Glu636 and Glu895 were constructed. As expected, the activities of all mutants were at least 40 times lower than those of the wild-type enzyme (Table 2). The kcat/Km values of E636D, E636Q, E895D, and E895Q mutants were 4.66, 11.3, 24.5, and 24.9 mm−1 s−1, respectively. These data support a structural suggestion that Glu636 and Glu895 are involved in the enzyme catalysis.
TABLE 2.
Activities of SaRha78A wild type and mutants for PNP-α-l-Rha
| Enzyme | Substrate | CaCl2a | Km | kcat | kcat/Km |
|---|---|---|---|---|---|
| mm | s−1 | mm−1 s−1 | |||
| WT | PNP-α-l-Rha | − | 0.026 ± 0.003 | 26.4 ± 1.7 | 1014 |
| WT | PNP-α-l-Rha | + | 0.024 ± 0.002 | 27.2 ± 1.2 | 1115 |
| WT | Gum arabic | + | 238 ± 61b | 0.003 ± 0.001 | 1.4 × 10−5 ± 7 × 10−6c |
| E636D | PNP-α-l-Rha | − | 0.015 ± 0.002 | 0.07 ± 0.004 | 4.66 |
| E636Q | PNP-α-l-Rha | − | 0.22 ± 0.03 | 2.49 ± 0.18 | 11.3 |
| E895D | PNP-α-l-Rha | − | 0.022 ± 0.002 | 0.54 ± 0.20 | 24.5 |
| E895Q | PNP-α-l-Rha | − | 0.012 ± 0.003 | 0.29 ± 0.13 | 24.9 |
| D179A | PNP-α-l-Rha | + | 0.028 ± 0.002 | 27.5 ± 1.3 | 975 |
| D179A | Gum arabic | + | NDd | ND | 2.7 × 10−7 ± 4 × 10−9c |
| N180A | PNP-α-l-Rha | + | 0.026 ± 0.004 | 24.4 ± 0.78 | 939 |
| N180A | Gum arabic | + | ND | ND | 2.7 × 10−7 ± 2 × 10−8c |
a Reaction mixture contained final 2 mm CaCl2 indicated as +.
b The number is given in mg/ml.
c The numbers are given in mg/ml−1s−1.
d Not determined as Km was too high to measure.
To explore the biological function of SaCBM67, a recombinant form of the protein module was expressed in E. coli and, after purifying by immobilized metal ion affinity chromatography, its binding properties were explored using isothermal titration calorimetry. The results are summarized in Table 3. SaCBM67 bound l-rhamnose with a Ka of 7.2 × 103 m−1 and free energy of binding ΔG of −5.3 kcal/mol. SaCBM67 did not bind to l-galactose or l-fucose, demonstrating that stereochemistry of the sugar at C4 and/or C2 are important specificity determinants. SaCBM67 bound to l-mannose, albeit with a 2-fold reduction in affinity. The C-6 methyl group of the bound l-rhamnose was partially buried in a hydrophobic pocket, however, there was sufficient solvation in the region of C-6 to accommodate a hydroxyl group. Because l-mannose seldom exists in natural polysaccharides, SaCBM67 binds primarily to l-rhamnose in biological systems.
TABLE 3.
Binding parameters for the recognition of sugars by SaCBM67 and mutants
The concentration of SaCBM67 in the cell was 700 μm for l-rhamnose and 800 μm for l-mannose. The concentration of both ligands in the syringe was 20 mm.
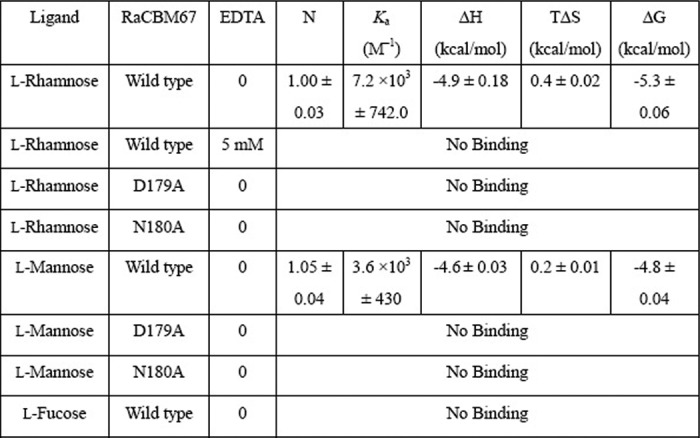
SaCBM67 did not bind to l-rhamnose in the presence of EDTA, which chelates calcium, demonstrating that the metal ion plays a key role in ligand recognition. Similarly the D179A and N180A mutations, which remove calcium-mediated and direct hydrogen bonds with l-rhamnose, abrogate ligand binding, confirming the importance of calcium in the binding of SaCBM67 to its ligand. To summarize, these data are entirely consistent with the structural data in showing that domain D (hence defined as SaCBM67) in SaRha78A comprises a novel CBM with a biologically relevant specificity for l-rhamnose residues.
To investigate the role of SaCBM67 in the function of SaRha78A, the activities of the wild-type enzyme, in the presence of EDTA, and mutants D179A and N180A were explored. The data (Table 2) showed that inactivation of SaCBM67, through either calcium chelation with EDTA or mutation of key residues, did not influence activity against aryl-rhamnosides, but caused a substantial reduction (∼50-fold) in activity against the rhamnose-containing polysaccharide composite, gum arabic. These results demonstrate that SaCBM67 plays a central role in the action of SaRha78A against polysaccharides; the mechanism for this catalytic potentiation is discussed below.
DISCUSSION
In this study, we determined the crystal structure of SaRha78A, which was the third example of a GH78 protein. The structure in conjunction with biochemical studies show that the three proteins contain four highly conserved domains. Domain N of SaRha78A comprising a fibronectin type 3 fold is not evident in the other GH78 proteins, whereas the other five domains of the Sterptomyces enzyme are present in BsRhaB or in both BsRhaB and BT1001. Despite the structural similarities of the proteins, the total amino acid identity was less than 15% compared with the other two proteins. These differences in the domain arrangement in the three α-l-rhamnosidases points to unusual evolutionary pathways within GH78.
Catalytic Mechanism of GH78 Enzymes
Previous studies have established that GH78 enzymes hydrolyze glycosidic bonds through an acid base-assisted single displacement or inverting mechanism (23). Analysis of the crystal structure of the GH78 enzyme BsRhaB soaked with rhamnose revealed electron density in a deep pocket which, by analogy with related inverting (α/α)6-barrel glycoside hydrolases, is likely to comprise the active site. Although the electron density was sufficiently large to comprise a sugar, rhamnose, in its relaxed 1C4 conformation, could not be modeled into the observed density (7). In the structure of the SaRha78A·l-rhamnose complex, an l-rhamnose molecule (Rha1501) was observed in the proposed active site pocket. The modeled l-rhamnose adopted an intermediate conformation between a skewboat 5S1 and boat 2,5B. Previously, in some inverting glycoside hydrolases, including a Clostridium thermocellum GH8 endoglucanase (CtCel8A) that also displays a (α/α)6 fold, the sugar bound at the active site (−1 subsite) adopted distorted 2SO/2,5B ring conformations (24–26). Further molecular dynamics studies indicated that the sugar ring in its 2SO conformation represented the Michaelis complex, which would then adopt a 2,5B conformation at the transition state, and, finally, the reaction product would display a 5S1 conformation (27, 28). By analogy, Rha1501 bound in the active site of SaRha78A appears to be migrating between the transition state and product conformations adopted during the reaction trajectory. Thus, it would appear that a boat 2,5B is the conformation adopted by the transition state of glycans hydrolyzed by GH78 rhamnosidases. It is interesting to note that although Cui et al. (7) were unable to model an l-rhamnose in its relaxed confirmation into the active site electron density, the authors suggested that this may be because the bound sugar adopted a distorted conformation, as observed here.
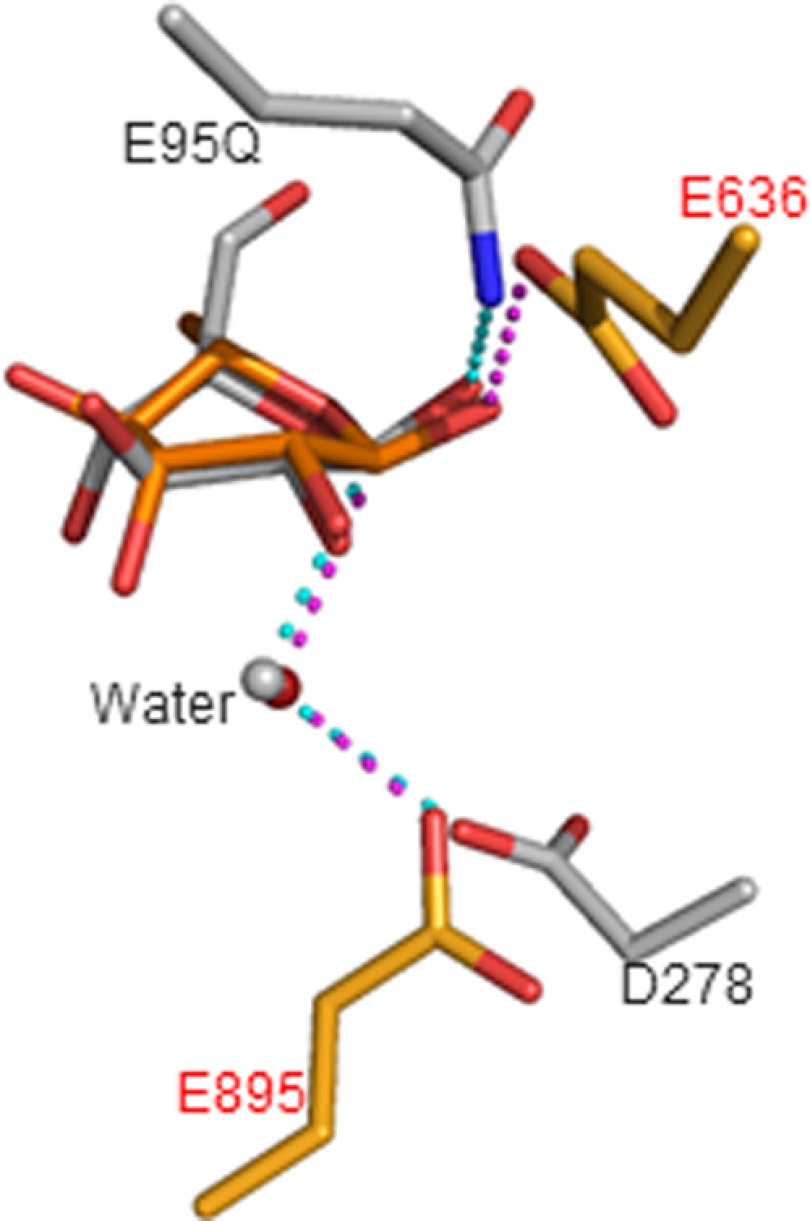
Generally, two acidic amino acids, either aspartate or glutamate, are employed by most inverting GHs to catalyze hydrolysis (23, 29). One carboxylate protonates the scissile glycosidic oxygen atom and the other coordinates the nucleophilic water molecule. In the SaRha78A·l-rhamnose complex, O1 of Rha1501 was within hydrogen bonding distance with Glu636 Oϵ2 atom. The C-1 hydroxyl group was observed in an α-anomeric configuration, and the O1 atom position could be considered as the scissile bond position of the substrate. Therefore, Glu636 appeared to comprise the catalytic proton donor (acid) of the enzyme. A water molecule makes a strong hydrogen bond with O1 of Rha1501, occupying the β-anomeric position, and thus is likely to comprise the solvent nucleophile utilized by the “inverting” enzyme. This water molecule makes strong hydrogen bonds with Trp747 Nϵ1 and Glu895 Oϵ2, and thus Glu895 appears to be the catalytic general base. These catalytic residues and nucleophilic water are conserved in CtCel8A (24) (Fig. 4). Thus, based on the criteria of conservation of fold, catalytic apparatus, and catalytic mechanism, we propose that GH78 be included in clan GH-M that currently contains families GH8 and GH48.
FIGURE 4.
Superimposed model of the catalytic site. SaRha78A (orange) on that of C. thermocellum endoglucanase CelA E95Q mutant complexed with cellopentaose (gray) (24). Two catalytic residues, the nucleophilic waters in the inverting reaction mechanism, bound sugars at subsite −1 are shown in stick models.
Consistent with the proposed catalytic role of Glu895, the glutamate is conserved in the other two GH78 enzymes for which structures are available. Although, Glu636, the catalytic acid, corresponds to Glu572 in BsRhaB, in BT1001 the equivalent residue is Asp337. When Glu572 and Glu841 in BsRhaB (corresponding to Glu636 and Glu895 in SaRha78A) were mutated to glutamine, catalytic efficiency decreased by 4–5 orders of magnitude, supporting their proposed catalytic function. Furthermore, the observation that BT1001 is catalytically inactive (data not shown) is entirely consistent with the replacement of Glu636 in SaRha78A (catalytic acid) with an aspartate, which would be too distant from the nucleophilic water to activate the solvent molecule. Reflecting a pH optimum of 6.0 the catalytic acid of SaRha78A requires a pKa modulator which, typically, is a carboxylate residue. The only candidate pKa modulator in SaRha78A is Asp630, which is within hydrogen bonding distance with Glu636. Consistent with the role of this aspartate is the observation that mutation of the equivalent residue in BsRhaB, Asp567, causes a 105-fold reduction in catalytic activity (7).
l-Rhamnose Binding SaCBM67
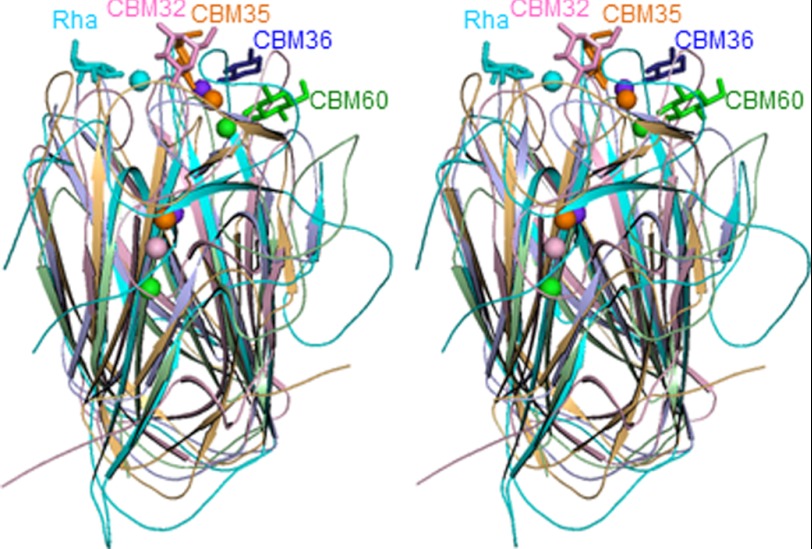
A DALI search indicated that BsRhaB (PDB code 2OKX) (7), but not BT1001, contains a domain that is structurally equivalent to SaCBM67 with a Z-score of 20.6. SaCBM67 also displays weak structural homology with a CBM32 (Z-score of 5.0, PDB code 2VCA) (30), CBM60 (Z-score of 4.4, PDB 2XFE) (31), CBM36 (Z-score of 4.1, PDB 2DCJ) (32), and CBM35 (Z-score of 3.9, PDB 2W87) (33), which recognize their different ligands through either an exo- (CBM32 and CBM35) or endo-mode (CBM36 and CBM60) of binding. These CBMs all comprise a β-jellyroll structure and, in addition to a structural calcium (which is absent in CBM67), contain a second calcium atom that is integral to ligand recognition. In SaCBM67 calcium also plays a key role in l-rhamnose recognition, although the metal binding site is displaced compared with the other CBMs (Fig. 5).
FIGURE 5.
Stereoview of the superimposed ribbon models of SaCBM67 and the different CBM structures. Calcium ions are drawn as spheres, and the bound sugars in stick models. SaCBM67, cyan; CBM32 in sialidase toxin from Clostridium perfringens (PDB code 2V72) (42), pink; CBM35 in xylanase from Cellvibrio japonicus (PDB code 2W87) (33), light orange; CBM36 in xylanase from Paenibacillus polymyxa ATCC 842 (PDB code 1uX7) (43), blue; CBM60 from uncultured bacterium (PDB code 2XFD) (31), light green.
CBM67 members are distributed not only in many bacterial GH78 α-l-rhamnosidases, but also in some Basidiomycete lectins, family 1 pectate lyases, peptidases, and proteins of unknown functions. Protein alignment of candidate members of CBM67 (Fig. 6) identified five subfamilies within the constructed phylogenetic tree (supplemental Fig. S1). In addition to SaCBM67, the only member of CBM67 for which a function is known is the lectin from Pleurotus cornucopiae. The lectin, which shows highest affinity against N-acetyl-d-galactosamine (34, 35), contains two CBM67-like sequences in tandem with sequence identities against SaRhaCBM67 of 25 and 35% for the N- and C-terminal modules, respectively. Hemagglutinating activity of this lectin is inhibited by EDTA and restored by calcium chloride, indicating that calcium binding is necessary for sugar binding. The calcium coordinating residues Asp179 and Asn228 of SaCBM67 are conserved in the two lectin modules, whereas Asn180, which is conserved in the C-terminal module, is replaced by an aspartate in the N-terminal module. Thus, the calcium binding site is conserved in the CBM67 ligand recognition site in the lectin, a putative polysaccharide lyase family 1 pectate lyase from Sorangium cellulosum (GenBankTM accession number CAN95071) and α-l-rhamnosidase from Paenibacillus sp. Y412MC10 (ACX62649), and both modules in a putative peptidase S8 and S53 from Pseudoalteromonas atlantica T6c (ABG39167). Although the l-rhamnose binding residues in SaCBM67 are conserved in CBM67 from the Paenibacillus α-l-rhamnosidase, suggesting that both proteins bind to l-rhamnose, these amino acids are not retaining in other CBM67 members. In particular, the loop extending from Glu197-Gly202 in SaCBM67, which contains Asp199 that makes a water-mediated interaction with the bound l-rhamnose, is missing in the Basidiomycete lectin, pectate lyase, and peptidase, which might cause different sugar specificity. Indeed, whereas SaCBM67 binds to l-rhamnose, the protein module does not recognize GlcNAc (data not shown), the ligand recognized by the P. cornucopiae lectin.
FIGURE 6.
Amino acid sequence alignment of SaCBM67 with selected CBM67 modules from related proteins. PcLec_N, P. cornucopiae lectin N-terminal domain (Genbank accession number BAD16583); PcLec_C, P. cornucopiae lectin C-terminal domain; ScPL1, S. cellulosum pectate lyase (CAN95071); PsRha78, Paenibacillus sp. Y412MC10 α-l-rhamnosidase (ACX62649); PaPep_N, P. atlantica T6c N-terminal domain (ABG39167); PaPep_C, P. atlantica T6c C-terminal domain. Residues conserved in at least five sequences are shaded. Residues coordinating calcium ion are indicated by ©, other residues in contact with the bound l-rhamnose are labeled with ®. The secondary structure elements of SaCBM67 are indicated by arrows for β-strands and coils for 310-helices.
Prior to the discovery of the CBM67 family, the only other, non-enzymatic, l-rhamnose-binding proteins were animal lectins for which NMR and crystal structures are available (36, 37). These proteins display an α/β-fold and ligand binding is not metal dependent. Thus, the mechanism of ligand recognition in SaCBM67 is distinct from other proteins that bind to the hexose sugar.
SaRha78A is an example of an enzyme that displays an exo-mode of action in which both the active site of the catalytic module, and the appended CBM, bind to the same terminal sugar. Indeed, the ∼50-fold, increase in catalytic activity afforded by the CBM is substantially greater than the 2–4-fold enhancement mediated by typical endo-binding CBMs (for review see Refs. 2, 38, and 39). Gum arabic is a highly complex mixture of polysaccharides and glycoproteins, in which the predominant structure is an arabionogalactan in which the β-1,3-galactan backbone contains branches that are capped with l-rhamnose or l-arabinofuranose residues (40). This presents a structure in which both the GH78 catalytic module and CBM67 of SaRha78 can bind to different terminal l-rhamnose residues of the same polysaccharide molecule. The ensuing avidity effect will result in much tighter binding of SaRha78 to gum arabic, compared with either the CBM or the catalytic module as discrete entities, leading to the observed, CBM-mediated, enhanced catalytic efficiency. This emerging model for how exo-acting CBMs potentiate the activity of exo-acting glycosidases is supported by a recent study showing that a CBM, which binds to terminal fructose residues, mediates a substantial increase in the activity of an exo-acting β-fructosidase against levan, a highly branched fructose-containing polysaccharide (41).
Acknowledgments
We thank the beamline researchers and staffs at Photon Factory and SPring-8. We thank Dr. Hitomi Ichinose for technical support for SaCBM67 construction and sample preparation for the crystallization.
This work was supported, in part, by JSPS KAKENHI Grant 22580110.

This article contains supplemental Fig. S1.
The atomic coordinates and structure factors (codes 3W5M and 3W5N) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GH
- glycoside hydrolase family
- BsRhaB
- Bacillus sp. GL1 α-l-rhamnosidase B
- BT1001
- Bacteroides thetaiotaomicron VPI-5482 α-l-rhamnosidase
- CBM
- carbohydrate-binding module family
- SaCBM67
- carbohydrate-binding module family 67 of S. avermitilis α-l-rhamnosidase
- SaRha78A
- S. avermitilis α-l-rhamnosidase
- SaRha78CM
- S. avermitilis α-l-rhamnosidase catalytic module
- PDB
- Protein Data Bank.
REFERENCES
- 1. Ochsner U. A., Fiechter A., Reiser J. (1994) Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 269, 19787–19795 [PubMed] [Google Scholar]
- 2. Mutter M., Renard C. M., Beldman G., Schols H. A., Voragen A. G. (1998) Mode of action of RG-hydrolase and RG-lyase toward rhamnogalacturonan oligomers. Characterization of degradation products using RG-rhamnohydrolase and RG-galacturonohydrolase. Carbohydr. Res. 311, 155–164 [DOI] [PubMed] [Google Scholar]
- 3. Drewnowski A., Henderson S. A., Shore A. B. (1997) Taste responses to naringin, a flavonoid, and the acceptance of grapefruit juice are related to genetic sensitivity to 6-n-propylthiouracil. Am. J. Clin. Nutr. 66, 391–397 [DOI] [PubMed] [Google Scholar]
- 4. Vila-Real H., Alfaia A. J., Bronze M. R., Calado A. R., Ribeiro M. H. (2011) Enzymatic synthesis of the flavone glucosides, prunin and isoquercetin, and the aglycones, naringenin and quercetin, with selective α-l-rhamnosidase and β-d-glucosidase activities of naringinase. Enzyme Res. 2011, 692618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) The Carbohydrate-Active EnZymes database (CAZy). An expert resource for glycogenomics. Nucleic Acids Res. 37, D233-D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henrissat B., Davies G. (1997) Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 [DOI] [PubMed] [Google Scholar]
- 7. Cui Z., Maruyama Y., Mikami B., Hashimoto W., Murata K. (2007) Crystal structure of glycoside hydrolase family 78 α-l-rhamnosidase from Bacillus sp. GL1. J. Mol. Biol. 374, 384–398 [DOI] [PubMed] [Google Scholar]
- 8. Bonanno J. B., Almo S. C., Bresnick A., Chance M. R., Fiser A., Swaminathan S., Jiang J., Studier F. W., Shapiro L., Lima C. D., Gaasterland T. M., Sali A., Bain K., Feil I., Gao X., Lorimer D., Ramos A., Sauder J. M., Wasserman S. R., Emtage S., D'Amico K. L., Burley S. K. (2005) New York-Structural GenomiX Research Consortium (NYSGXRC). A large scale center for the protein structure initiative. J. Struct. Funct. Genomics 6, 225–232 [DOI] [PubMed] [Google Scholar]
- 9. Ichinose H., Fujimoto Z., Kaneko S. (2013) Characterization of an α-l-rhamnosidase from Streptomyces avermitilis. Biosci. Biotechnol. Biochem. 77, 213–216 [DOI] [PubMed] [Google Scholar]
- 10. LeMaster D. M., Richards F. M. (1985) 1H-15N heteronuclear NMR studies of Escherichia coli thioredoxin in samples isotopically labeled by residue type. Biochemistry 24, 7263–7268 [DOI] [PubMed] [Google Scholar]
- 11. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 12. Terwilliger T. C. (2003) SOLVE and RESOLVE. Automated structure solution and density modification. Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 13. Terwilliger T. C. (2003) Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perrakis A., Morris R., Lamzin V. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 15. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 17. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 19. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry. ϕ, ψ, and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 20. Yang H., Ichinose H., Nakajima M., Kobayashi H., Kaneko S. (2006) Synergy between an α-l-arabinofuranosidase from Aspergillus oryzae and an endo-arabinanase from Streptomyces coelicolor for degradation of arabinan. Food Sci. Technol. Res. 12, 43–49 [Google Scholar]
- 21. Ichinose H., Fujimoto Z., Honda M., Harazono K., Nishimoto Y., Uzura A., Kaneko S. (2009) A β-l-arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27. J. Biol. Chem. 284, 25097–25106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boraston A. B., Bolam D. N., Gilbert H. J., Davies G. J. (2004) Carbohydrate-binding modules. Fine-tuning polysaccharide recognition. Biochem. J. 382, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zverlov V. V., Hertel C., Bronnenmeier K., Hroch A., Kellermann J., Schwarz W. H. (2000) The thermostable α-l-rhamnosidase RamA of Clostridium stercorarium. Biochemical characterization and primary structure of a bacterial α-l-rhamnoside hydrolase, a new type of inverting glycoside hydrolase. Mol. Microbiol. 35, 173–179 [DOI] [PubMed] [Google Scholar]
- 24. Guérin D. M., Lascombe M. B., Costabel M., Souchon H., Lamzin V., Béguin P., Alzari P. M. (2002) Atomic (0.94 Å) resolution structure of an inverting glycosidase in complex with substrate. J. Mol. Biol. 316, 1061–1069 [DOI] [PubMed] [Google Scholar]
- 25. Brüx C., Ben-David A., Shallom-Shezifi D., Leon M., Niefind K., Shoham G., Shoham Y., Schomburg D. (2006) The structure of an inverting GH43 β-xylosidase from Geobacillus stearothermophilus with its substrate reveals the role of the three catalytic residues. J. Mol. Biol. 359, 97–109 [DOI] [PubMed] [Google Scholar]
- 26. Sidhu G., Withers S. G., Nguyen N. T., McIntosh L. P., Ziser L., Brayer G. D. (1999) Sugar ring distortion in the glycosyl-enzyme intermediate of a family G/11 xylanase. Biochemistry 38, 5346–5354 [DOI] [PubMed] [Google Scholar]
- 27. Petersen L., Ardèvol A., Rovira C., Reilly P. J. (2009) Mechanism of cellulose hydrolysis by inverting GH8 endoglucanases. A QM/MM metadynamics study. J. Phys. Chem. B 113, 7331–7339 [DOI] [PubMed] [Google Scholar]
- 28. Barker I. J., Petersen L., Reilly P. J. (2010) Mechanism of xylobiose hydrolysis by GH43 β-xylosidase. J. Phys. Chem. B 114, 15389–15393 [DOI] [PubMed] [Google Scholar]
- 29. Ly H. D., Withers S. G. (1999) Mutagenesis of glycosidases. Annu. Rev. Biochem. 68, 487–522 [DOI] [PubMed] [Google Scholar]
- 30. Ficko-Blean E., Stubbs K. A., Nemirovsky O., Vocadlo D. J., Boraston A. B. (2008) Structural and mechanistic insight into the basis of mucopolysaccharidosis IIIB. Proc. Natl. Acad. Sci. U.S.A. 105, 6560–6565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Montanier C., Flint J. E., Bolam D. N., Xie H., Liu Z., Rogowski A., Weiner D. P., Ratnaparkhe S., Nurizzo D., Roberts S. M., Turkenburg J. P., Davies G. J., Gilbert H. J. (2010) Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. J. Biol. Chem. 285, 31742–31754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yazawa R., Takakura J., Sakata T., Ihsanawati, Yatsunami R., Fukui T., Kumasaka T., Tanaka N., Nakamura S. (2011) A calcium-dependent xylan-binding domain of alkaline xylanase from alkaliphilic Bacillus sp. strain 41M-1. Biosci. Biotechnol. Biochem. 75, 379–381 [DOI] [PubMed] [Google Scholar]
- 33. Montanier C., van Bueren A. L., Dumon C., Flint J. E., Correia M. A., Prates J. A., Firbank S. J., Lewis R. J., Grondin G. G., Ghinet M. G., Gloster T. M., Herve C., Knox J. P., Talbot B. G., Turkenburg J. P., Kerovuo J., Brzezinski R., Fontes C. M., Davies G. J., Boraston A. B., Gilbert H. J. (2009) Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc. Natl. Acad. Sci. U.S.A. 106, 3065–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oguri S., Ando A., Nagata Y. (1996) A novel developmental stage-specific lectin of the basidiomycete Pleurotus cornucopiae. J. Bacteriol. 178, 5692–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sumisa F., Ichijo N., Yamaguchi H., Nakatsumi H., Ando A., Iijima N., Oguri S., Uehara K., Nagata Y. (2004) Molecular properties of mycelial aggregate-specific lectin of Pleurotus cornucopiae. J. Biosci. Bioeng. 98, 257–262 [DOI] [PubMed] [Google Scholar]
- 36. Vakonakis I., Langenhan T., Prömel S., Russ A., Campbell I. D. (2008) Solution structure and sugar-binding mechanism of mouse latrophilin-1 RBL. A 7TM receptor-attached lectin-like domain. Structure 16, 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shirai T., Watanabe Y., Lee M. S., Ogawa T., Muramoto K. (2009) Structure of rhamnose-binding lectin CSL3. Unique pseudo-tetrameric architecture of a pattern recognition protein. J. Mol. Biol. 391, 390–403 [DOI] [PubMed] [Google Scholar]
- 38. Hall J., Black G. W., Ferreira L. M., Millward-Sadler S. J., Ali B. R., Hazlewood G. P., Gilbert H. J. (1995) The non-catalytic cellulose-binding domain of a novel cellulase from Pseudomonas fluorescens subsp. cellulosa is important for the efficient hydrolysis of Avicel. Biochem. J. 309, 749–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hervé C., Rogowski A., Blake A. W., Marcus S. E., Gilbert H. J., Knox J. P. (2010) Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc. Natl. Acad. Sci. U.S.A. 107, 15293–15298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Defaye J., Wong E. (1986) Structural studies of gum arabic, the exudate polysaccharide from Acacia senegal. Carbohydr. Res. 150, 221–231 [Google Scholar]
- 41. Cuskin F., Flint J. E., Gloster T. M., Morland C., Baslé A., Henrissat B., Coutinho P. M., Strazzulli A., Solovyova A. S., Davies G. J., Gilbert H. J. (2012) How nature can exploit nonspecific catalytic and carbohydrate binding modules to create enzymatic specificity. Proc. Natl. Acad. Sci. U.S.A. 109, 20889–20894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boraston A. B., Ficko-Blean E., Healey M. (2007) Carbohydrate recognition by a large sialidase toxin from Clostridium perfringens. Biochemistry 46, 11352–11360 [DOI] [PubMed] [Google Scholar]
- 43. Jamal-Talabani S., Boraston A. B., Turkenburg J. P., Tarbouriech N., Ducros V. M., Davies G. J. (2004) Ab initio structure determination and functional characterization of CBM36. A new family of calcium-dependent carbohydrate binding modules. Structure 12, 1177–1187 [DOI] [PubMed] [Google Scholar]