Abstract
The natural catalytic cycle of cytochrome (cyt) P450 enzymes in human liver microsome (HLM) films was activated electrochemically via the electron transfer sequence electrode→cyt P450 reductase (CPR)→cyt P450. Cyclic voltammograms for HLM films had midpoint potentials of −0.50 V vs. SCE at pH 7.4 characteristic of CPR, not cyt P450s. HLM and CPR microsomes without cyt P450s did not electrocatalytically reduce H2O2, and did not shift midpoint potential when CO was added, also indicating that the peaks do not correspond to iron heme cyt P450 enzymes. Electrochemical activation of the natural cyt P450 cycle for substrate conversion via CPR in HLM films was confirmed by catalytic electrolysis in an electrochemical microfluidic array designed to generate and detect reactive metabolites by measuring their reactivity with DNA.
Keywords: Cytochrome P450, Electrochemical enzyme activation, Human liver microsomes, Protein film voltammetry, Redox enzyme
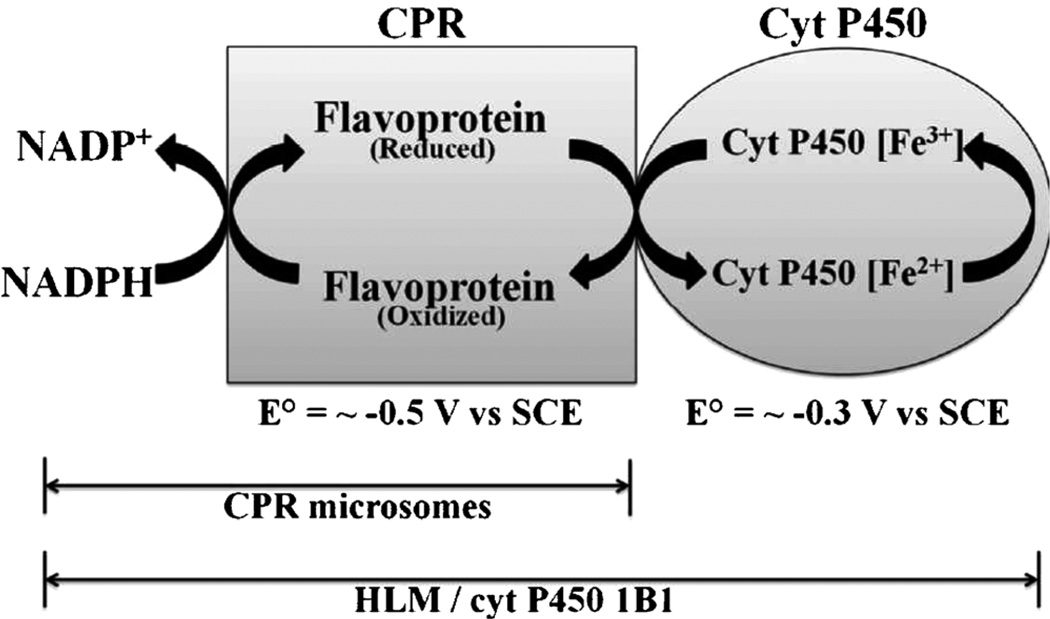
Liver microsomes provide a representative source of enzymes for metabolic toxicity studies that can provide good comparability with in vivo bioassays [1]. Human liver microsomes (HLM) are liver cell membrane fractions containing cytochrome P450s (cyt P450), a superfamily of enzymes responsible for the major fraction of human oxidative metabolism. HLMs contain a collection of cyt P450s accompanied by redox partner cyt P450 reductase (CPR), and activation of oxidative metabolic reactions requires only an electron donor such as NADPH. The proposed pathway involves initial donation of an electron from NADPH via CPR to the cyt P450 followed by dioxygen binding to the heme iron [2–4]. A second electron transfer from CPR leads to the active ferryloxy radical cation of cyt P450 that transfers oxygen to substrates.
Cyt P450s in thin films have enormous potential in selective biosynthesis and biosensors. Thin-film voltammetry of a number of cyt P450s has been studied [5–9]. Three general pathways to activate cyt P450s on electrodes have been elaborated: (a) direct electron transfer to pure cyt P450FeIII in films in aerobic solutions results in catalytic reduction of oxygen to hydrogen peroxide (H2O2), which transfers oxygen to the heme iron [5]; (b) fusion proteins with an electron acceptor protein linked to a cyt P450 feature electron flow from electrode, to electron acceptor protein, to the cyt P450 [7]; and (c) microsomal CPR/cyt P450 films where electrons flow from electrode to CPR to the cyt P450 as in the natural catalytic pathway [8]. All these approaches substitute the electrode for NADPH as an electron donor. We have reported successful electrochemical activation of cyt P450 via CPR in films that combined CPR microsomes with excess cyt P450 1A2 and 2E1 [8, 10], as well as in films of rat liver microsomes [11, 12]. Such a pathway may also occur in genetically enriched cyt P450 1A2 and cyt P450 3A4 supersomes [13] in polyion films on electrodes. However, it has not yet been confirmed that cyt P450s in HLMs can be electrochemically activated in the same fashion.
In this communication, we report successful electrochemical activation of the natural catalytic pathway of cyt P450s in films of HLM and polyions on pyrolytic graphite (PG) electrodes. Cyt P450 1B1 supersomes (cyt P450 1B1), which contain cyt P450 1B1 and CPR, and CPR microsomes which contains only CPR as a negative control were also used in this study. We demonstrate direct conversion of a substrate using thin films of the HLM to generate reactive metabolites from 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), which are shown to react with DNA in a toxicity screening assay.
Quartz crystal microbalance (QCM) studies [14] confirmed stable, reproducible formation of the films containing enzymes and polyions on PG electrodes used in this work (Table 1). Cyclic voltammograms (CVs) of (PDDA/HLM)3 and (PDDA/cyt P450 1B1)3 showed the characteristics of non-ideal quasireversible protein film voltammetry [15]. Reduction–oxidation peak pairs had midpoint potentials (E°) −0.50 V vs. saturated calomel reference electrode (SCE) for HLM and −0.51 V for cyt P450 1B1 and CPR microsomes. Peak current was linear vs. scan rate up to 1000 mV s−1, reduction peak currents shifted negative with increasing scan rate, and oxidation-reduction peak current ratios were ~1.
Table 1.
Characteristics of films of polyions and enzyme sources from QCM results.
| Film assembly [a] | Nominal thickness (nm) | Mass of enzyme source (µg cm−2) | Mass of polyion (µg cm−2) |
|---|---|---|---|
| (PDDA/HLM)3 | 30 | 8.6±0.7 | 1.5±0.4 |
| (PDDA/CPR microsomes)3 | 14 | 3.6±0.5 | 0.9±0.2 |
| (PDDA/cyt P450 1B1)3 | 18 | 5.1±0.5 | 0.9±0.2 |
| (Mb/PSS)3 | 19 | 5.6±0.4 | 0.8±0.1 |
Abbreviations: PDDA: poly(diallyldimethylammonium chloride), PSS: poly(sodium 4-styrenesulfonate), Mb: Myoglobin.
Subscripts refer to number of bilayers
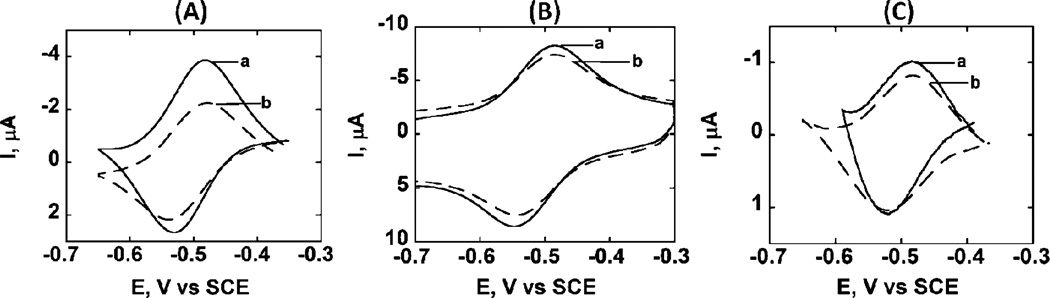
For FeIII heme protein films in solutions containing CO, protein FeII-CO complexes form upon reduction that result in 35–50 mV positive shift of midpoint potentials due to the follow-up chemical reaction of the protein FeII form with CO [8, 10, 16]. CV peaks of CPR microsome, HLM or cyt P450 1B1 supersome films did not shift with CO in solution (Figure 1). This lack of midpoint potential shifts for CPR microsomes, HLM and cyt P450 1B1 films suggests that CV peaks originated not from cyt P450s but from CPR [8, 11, 13, 17]. CPR contains FAD and FMN cofactors, and CPR voltammetry has been interpreted previously [11, 13, 17]. Further, midpoint potentials near −0.5 V vs. SCE are characteristic of CPR and not cyt P450s, which have CV midpoint potentials in similar films of about −0.30 to −0.35 V vs. SCE [6, 8].
Fig. 1.
Background subtracted cyclic voltammograms of the films on PG electrodes at 1 Vs−1 in 50 mM phosphate buffer pH 7.4+0.1 M NaCl: (A) (PDDA/HLM)3, (B) (PDDA/CPR microsomes)3, (C) (PDDA/cyt P450 1B1 supersomes)3. Curve labels: (a) buffer saturated with N2 (b) buffer saturated with CO.
Catalytic reduction of H2O2 can be observed by voltammetrically in films of iron heme proteins including cyt P450s. [11, 13, 15]. However, films of HLM or CPR microsomes did not show catalytic behavior in 2 mM H2O2 (see Figure S1). In contrast, films of positive control heme protein myoglobin (Mb) gave a large peak for catalytic reduction of H2O2 (Figure S1). These findings further confirm that the redox peaks of HLM and CPR microsomes (Figure 1, Scheme 1) originate from CPR and not cyt P450s. Species responsible for the CV peaks of HLM films did not catalytically reduce H2O2, nor did peaks shift under the influence of CO. All these results suggest peak assignment to CPR, which does not catalytically reduce H2O2 or react with CO in its reduced form [11, 13, 17].
Scheme 1.
Electrochemical processes occurring in the microsomal films.
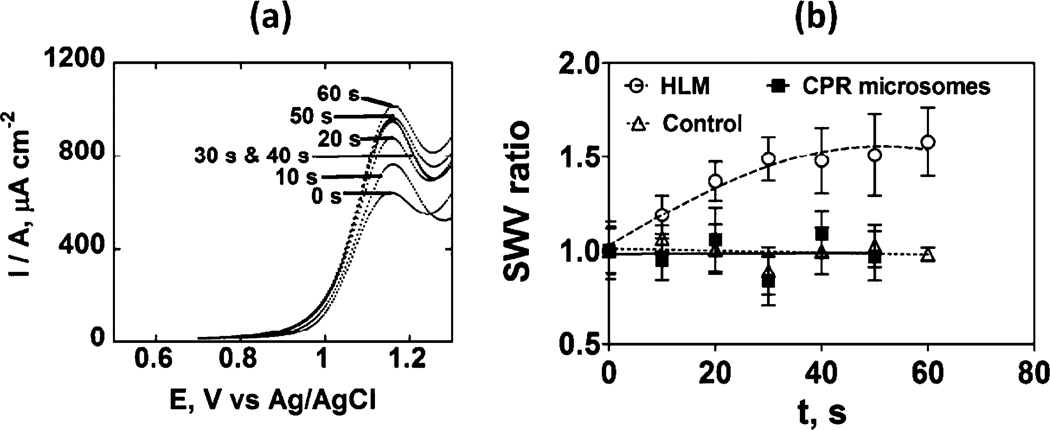
To demonstrate that electrons injected into CPR can be used to activate the cyt P450 enzymes in the HLM film, we used a microfluidic electrochemical toxicity screening device reported previously using rat liver enzymes [12]. This device is designed to produce and detect reactive metabolites formed by cyt P450 enzymes by monitoring their reactions with DNA in DNA/RuPVP ([Ru(bpy)2(PVP)10](ClO4); bpy=2,2-bipyridyl; PVP=poly(4-vinylpyridine))/enzyme films. Electrolysis was done at a potential negative of the film’s reduction peak (−0.7 V vs. SCE, see Figure 1) to deliver electrons to CPR and activate the natural cyt P450 catalytic cycle. The enzyme reactions were done using constant flow of oxygenated 50 µM 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). After the enzyme reaction, square wave voltammetry (SWV) detects DNA damage caused by reactive metabolites. Cyt P450-catalyzed NNK metabolism involves the formation of an N-methyl hydroxylated metabolite, which reacts with DNA to produce a covalently attached guanine adduct [18]. The difference SWV peak at ~1.1 V vs. SCE originates from oxidation of RuII sites in the polymer to RuIII, which catalytically oxidizes guanines in the DNA. If DNA forms covalent adducts with the metabolites, the double stranded structure is disrupted and RuIII sites gain better access to guanines. This results in larger catalytic oxidation rates and larger peak currents in SWV [19, 20]. The ratio of the SWV difference peak current after the enzyme reaction (Ip,f) to that of controls with no enzyme reaction (Ip,i)) is the measured quantity indicative of DNA damage [5, 12].
Increase in SWV peak current densities were observed with time of enzyme reaction in the HLM/DNA film indicating reactive metabolites of NNK form and damage DNA (Figure 2 a). Figure 2 b suggests progressive DNA damage by metabolites of NNK until saturation is reached. In contrast, CPR microsomes did not show an increase in SWV ratio, indicating no metabolism of NNK due to the lack of cyt P450s.
Fig. 2.
SWV detection of DNA damage caused by reactive metabolites of 50 µM NNK in anaerobic pH 7.4 phosphate buffer in a microfluidic metabolic toxicity screening assay: (a) Background-subtracted averaged (n=8) difference SWV peak current densities for HLM/DNA/RuPVP films; (b) influence of enzyme reaction time on SWV peak current ratio (Ip,f/Ip,i). Control lacked substrate or included substrate without the enzyme reaction, which gave equivalent results. Error bars represent SD, n=8.
Results from microfluidic genotoxicity screening arrays indicate that electrochemical activation via CPR causes the electrons to be subsequently transferred to cyt P450 enzymes to convert the NNK substrate to reactive metabolites that react with DNA (Figure 2). The turnover rate of NNK oxidation by HLM is 176±22 ({µg protein−1} s−1 mM−1 for 50 mM NNK under these conditions. Reactive metabolites were formed from HLM, but not from CPR microsomes that lack cyt P450s. This result indicates further that the natural catalytic cycle of cyt P450 can be submitted by electrochemical activation in the HLM films. The pathway involves reduction of the cyt P450 to the FeII form, then subsequent reactions with oxygen and a second electron from CPR to yield the active oxidant, ferryloxy cyt P450 [4, 8].
In summary, voltammograms and metabolite generation presented above strongly suggest that electrons are transferred from electrode to CPR to cyt P450s in HLM films. Results document the successful electrode-driven activation of HLM in the sensing arrays used here. This approach was concurrently used to validate a new, higher throughput microfluidic array for genotoxicity screening combining HLMs with representative metabolic bioconjugation enzymes [21].
Experimental
Chemicals and Materials
Ruthenium metallopolymer [Ru(bpy)2(PVP)10](ClO4) (RuPVP (bpy=2,2-bipyridyl; PVP=poly(4-vinylpyridine)) was synthesized as described [22]. Sources of other chemicals are in Supporting Information (SI).
Film Fabrication, Characterization and Voltammetry
Films of (PDDA/microsomes or cyt P450 1B1)3 and (Mb/PSS)3 were constructed layer-by-layer (LbL) on roughened ordinary basal plane PG electrodes (Advanced Ceramics) as reported previously [5], and film formation was monitored by using QCM. (see SI file). CVs were done with a CHI 660A analyzer, using a SCE and Pt counter electrode in an anaerobic 50 mM phosphate + 0.1 M NaCl buffer at pH 7.4.
Microfluidic Array Fabrication and Metabolite Detection
The microfluidic system features an 8-electrode array and symmetrically placed Pt counter and Ag/AgCl reference electrodes, as reported previously [12]. Full description is in the SI file.
Supplementary Material
Acknowledgement
This work was supported financially by US PHS Grant No. ES03154 from National Institute of Environmental Health Sciences (NIEHS), NIH, USA.
Footnotes
Supporting Information for this article is available on the WWW under http://dx.doi.org/10.1002/elan.201200373
References
- 1.Wienkers LC, Heath TG. Nature Rev. Drug Discov. 2005;4:825. doi: 10.1038/nrd1851. [DOI] [PubMed] [Google Scholar]
- 2.Schenkman JB, Greim H. Cytochrome P450. Berlin: Springer; 1993. [Google Scholar]
- 3.Ortiz de Montellano PR. Cytochrome P450. 3rd ed. New York: Kluwer/Plenum; 2005. [Google Scholar]
- 4.Guengerich FP. Chem. Res. Toxicol. 2001;14:611. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 5.Rusling JF, Hvastkovs EG, Hull DO, Schenkman JB. Chem. Commun. 2008;141:141. doi: 10.1039/b709121b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bistolas N, Wollenberger U, Jung C, Scheller FW. Biosens. Bioelectron. 2005;20:2408. doi: 10.1016/j.bios.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 7.Dodhia VR, Gilardi G. In: Engineering the Bioelectronic Interface: Applications to Analyte Biosensing and Protein Detection. Davis J, editor. RSC Publications; 2009. pp. 153–189. [Google Scholar]
- 8.Krishnan S, Schenkman JB, Rusling JF. J. Phys. Chem. B. 2011;115:8371. doi: 10.1021/jp201235m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadeghi SJ, Fantuzzi A, Gilardi G. Biochim. Biophys. Acta, Proteins Proteomics. 2011;1814:237. doi: 10.1016/j.bbapap.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Krishnan S, Wasalathanthri D, Zhao L, Schenkman JB, Rusling JF. J. Am. Chem. Soc. 2011;133:1459. doi: 10.1021/ja108637s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnan S, Rusling JF. Electrochem. Commun. 2007;9:2359. doi: 10.1016/j.elecom.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wasalathanthri DP, Mani V, Tang CK, Rusling JF. Anal. Chem. 2011;83:9499. doi: 10.1021/ac202269t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sultana N, Schenkman JB, Rusling JF. J. Am. Chem. Soc. 2005;127:13460. doi: 10.1021/ja0538334. [DOI] [PubMed] [Google Scholar]
- 14.Lvov Y. In: Handbook of Surfaces and Interfaces of Materials. Nalwa RW, editor. California: Academic Press; 2001. pp. 170–189. [Google Scholar]
- 15.Rusling JF, Wang B, Yun S. In: Bioelectrochemistry. Bartlett PN, editor. New York: Wiley; 2008. pp. 39–86. [Google Scholar]
- 16.Zhang Z, Nassar A-EF, Lu Z, Schenkman JB, Rusling JF. J. Chem. Soc. Faraday Trans. 1997;93:1769. [Google Scholar]
- 17.Sultana N, Schenkman JB, Rusling JF. Electroanalysis. 2007;24:2499. [Google Scholar]
- 18.Hecht SS. Chem. Res. Toxicol. 1998;11:560. doi: 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- 19.Zhou L, Yang J, Estavillo C, Stuart JD, Schenkman JB, Rusling JF. J. Am. Chem. Soc. 2003;125:1431. doi: 10.1021/ja0290274. [DOI] [PubMed] [Google Scholar]
- 20.Rusling JF, Hvastkovs EG, Schenkman JB. In: Drug Metabolism Handbook. Nassar A, Hollenburg PF, Scatina J, editors. New Jersey: Wiley; 2009. pp. 307–340. [Google Scholar]
- 21.Wasalathanthri DP, Faria RC, Malla S, Joshi AA, Schenkman JB, Rusling JF. Analyst. doi: 10.1039/c2an35993f. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennany L, Forster RJ, Rusling JF. J. Am. Chem. Soc. 2003;125:5213. doi: 10.1021/ja0296529. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.