Abstract
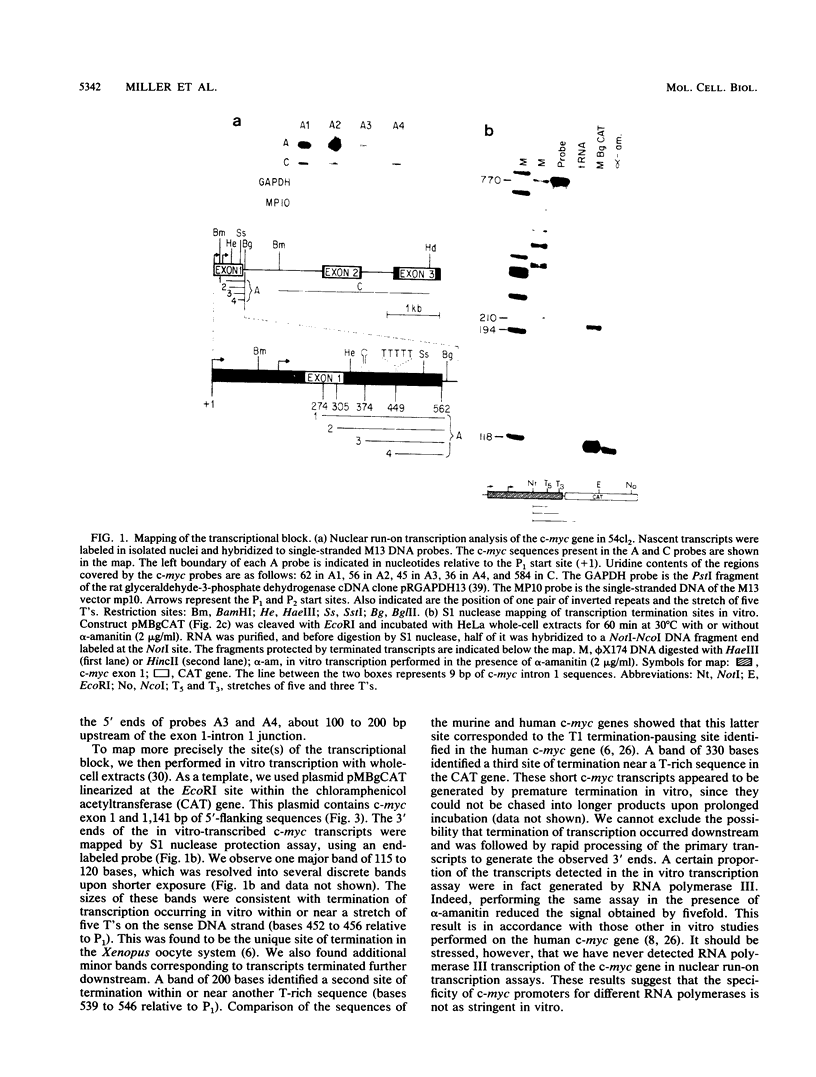
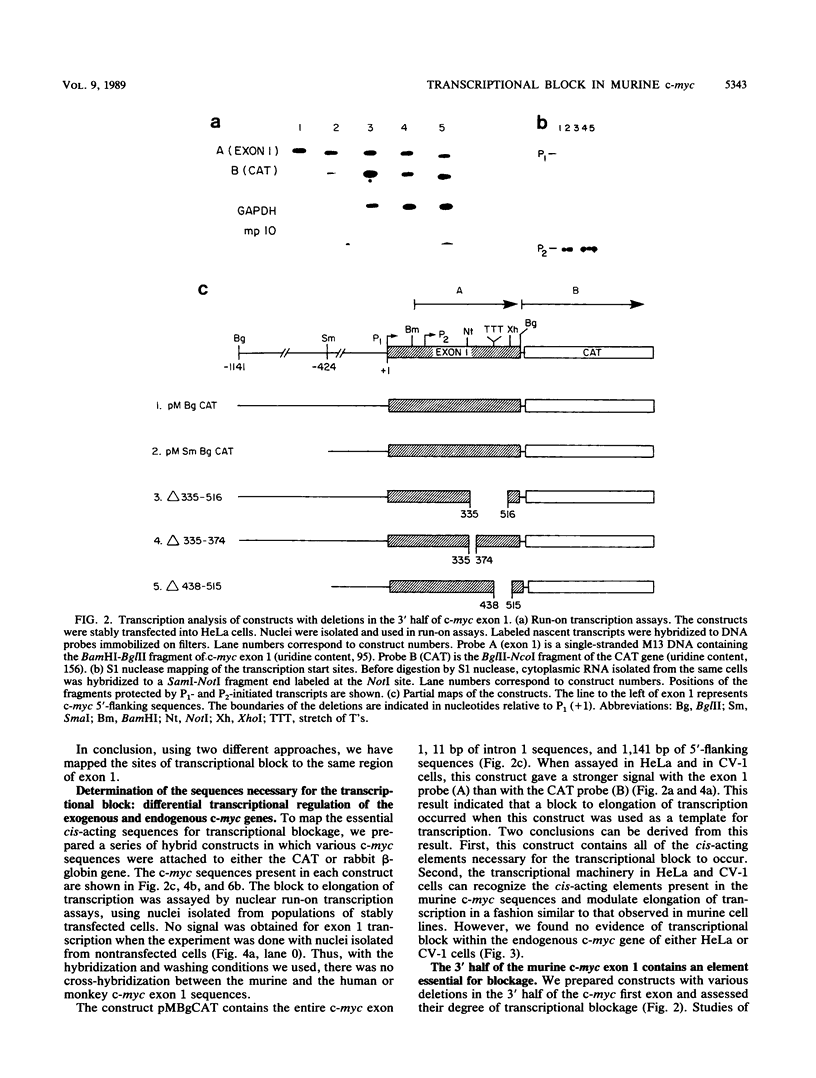
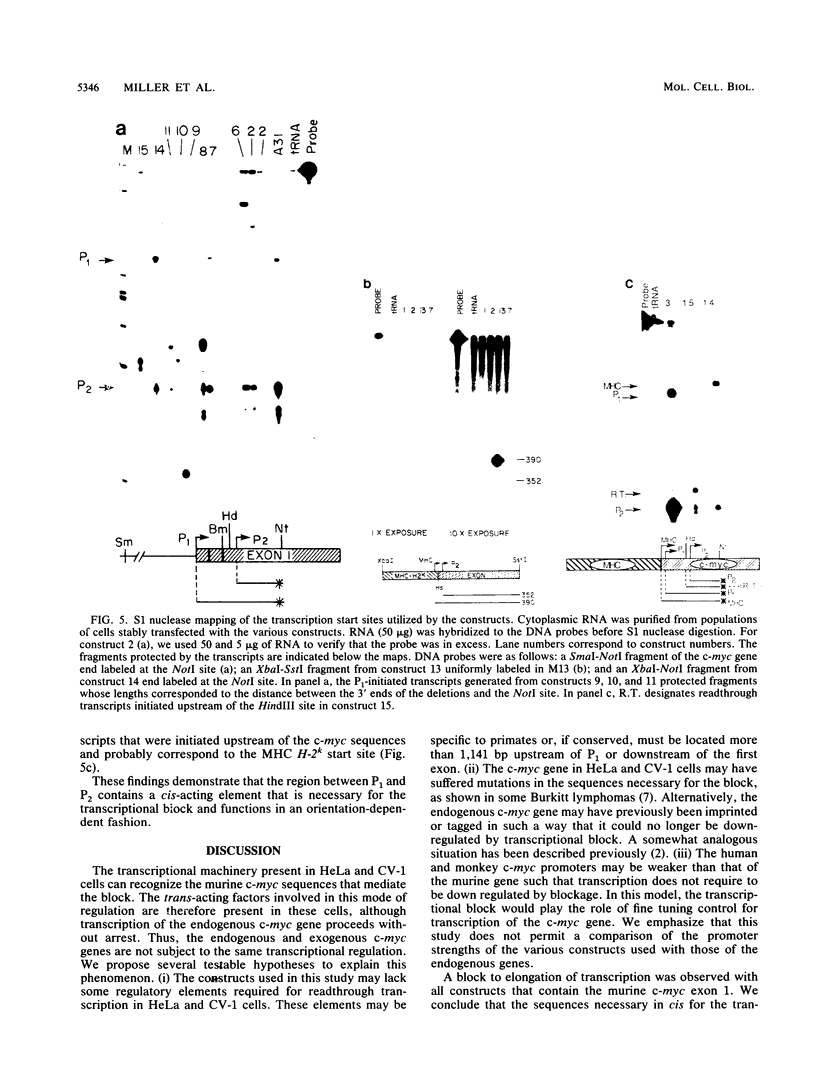
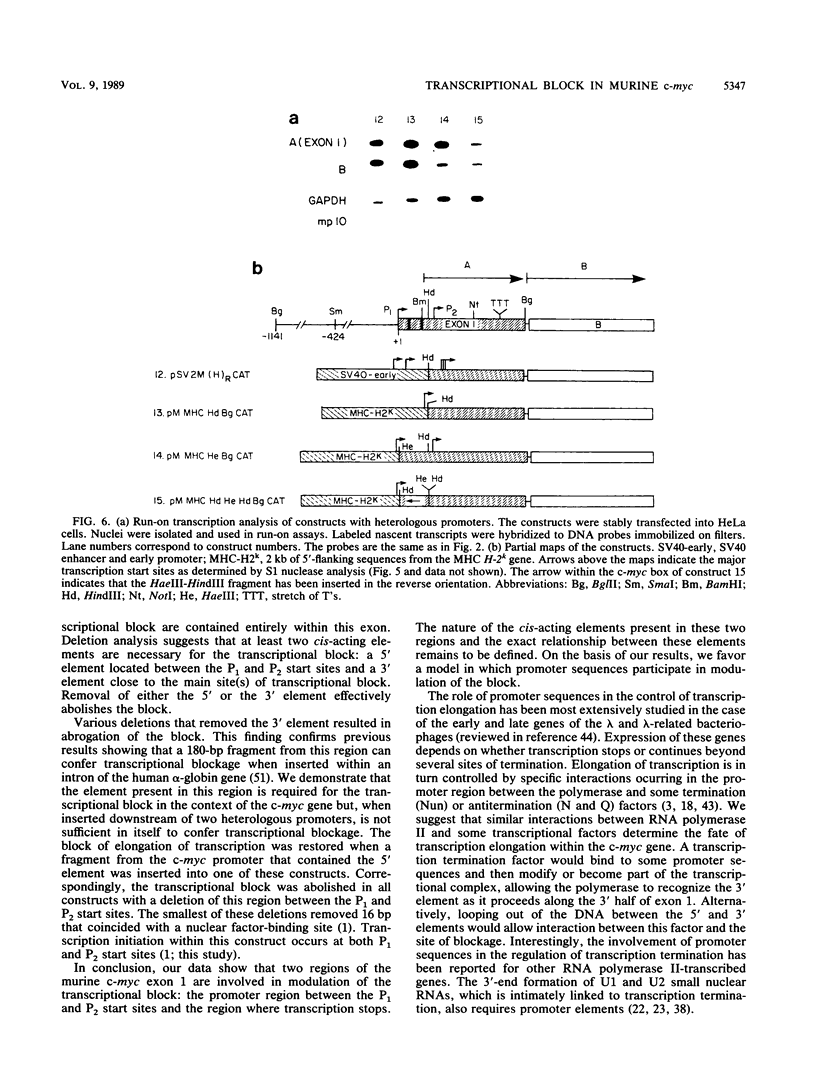
A block to elongation of transcription has been shown to occur within the first exon of the human and murine c-myc genes. The extent of this block was found to vary with the physiological state of cells, indicating that modulation of the transcriptional block can serve to control the expression of this gene. To determine which sequences are required in cis for the transcriptional block, we generated a series of constructs containing various portions of murine c-myc 5'-flanking and exon 1 sequences. We established populations of HeLa and CV-1 cells stably transfected with these constructs. The transcription start sites were determined by S1 nuclease mapping analysis, and the extent of transcriptional block was measured by nuclear run-on transcription assays. Our results demonstrate that at least two cis-acting elements are necessary for the transcriptional block. A 3' element was found to be located in the region where transcription stopped and showed features reminiscent of some termination sites found in procaryotes. A 5' element was positioned between the P1 and P2 (C. Asselin, A. Nepveu, and K. B. Marcu, Oncogene 4:549-558, 1989). Removal of the more 3' binding site abolished the transcriptional block.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asselin C., Nepveu A., Marcu K. B. Molecular requirements for transcriptional initiation of the murine c-myc gene. Oncogene. 1989 May;4(5):549–558. [PubMed] [Google Scholar]
- Atchison M. L., Perry R. P. The role of the kappa enhancer and its binding factor NF-kappa B in the developmental regulation of kappa gene transcription. Cell. 1987 Jan 16;48(1):121–128. doi: 10.1016/0092-8674(87)90362-x. [DOI] [PubMed] [Google Scholar]
- Barik S., Ghosh B., Whalen W., Lazinski D., Das A. An antitermination protein engages the elongating transcription apparatus at a promoter-proximal recognition site. Cell. 1987 Sep 11;50(6):885–899. doi: 10.1016/0092-8674(87)90515-0. [DOI] [PubMed] [Google Scholar]
- Bender T. P., Thompson C. B., Kuehl W. M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987 Sep 18;237(4821):1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Cesarman E., Dalla-Favera R., Bentley D., Groudine M. Mutations in the first exon are associated with altered transcription of c-myc in Burkitt lymphoma. Science. 1987 Nov 27;238(4831):1272–1275. doi: 10.1126/science.3685977. [DOI] [PubMed] [Google Scholar]
- Chung J., Sussman D. J., Zeller R., Leder P. The c-myc gene encodes superimposed RNA polymerase II and III promoters. Cell. 1987 Dec 24;51(6):1001–1008. doi: 10.1016/0092-8674(87)90586-1. [DOI] [PubMed] [Google Scholar]
- Eick D., Berger R., Polack A., Bornkamm G. W. Transcription of c-myc in human mononuclear cells is regulated by an elongation block. Oncogene. 1987;2(1):61–65. [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R., Weber J., Ziff E., Darnell J. E. Premature termination during adenovirus transcription. Nature. 1979 Mar 22;278(5702):367–370. doi: 10.1038/278367a0. [DOI] [PubMed] [Google Scholar]
- Feramisco J. R., Smart J. E., Burridge K., Helfman D. M., Thomas G. P. Co-existence of vinculin and a vinculin-like protein of higher molecular weight in smooth muscle. J Biol Chem. 1982 Sep 25;257(18):11024–11031. [PubMed] [Google Scholar]
- Fort P., Rech J., Vie A., Piechaczyk M., Bonnieu A., Jeanteur P., Blanchard J. M. Regulation of c-fos gene expression in hamster fibroblasts: initiation and elongation of transcription and mRNA degradation. Nucleic Acids Res. 1987 Jul 24;15(14):5657–5667. doi: 10.1093/nar/15.14.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G., Guise J. W., McDevitt M. A., Tucker P. W., Nevins J. R. Relative position and strengths of poly(A) sites as well as transcription termination are critical to membrane versus secreted mu-chain expression during B-cell development. Genes Dev. 1987 Jul;1(5):471–481. doi: 10.1101/gad.1.5.471. [DOI] [PubMed] [Google Scholar]
- Gilmour D. S., Lis J. T. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Mol Cell Biol. 1986 Nov;6(11):3984–3989. doi: 10.1128/mcb.6.11.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayhack E. J., Yang X. J., Lau L. F., Roberts J. W. Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell. 1985 Aug;42(1):259–269. doi: 10.1016/s0092-8674(85)80121-5. [DOI] [PubMed] [Google Scholar]
- Hay N., Aloni Y. Attenuation in SV40 as a mechanism of transcription-termination by RNA polymerase B. Nucleic Acids Res. 1984 Feb 10;12(3):1401–1414. doi: 10.1093/nar/12.3.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Aloni Y. Attenuation of late simian virus 40 mRNA synthesis is enhanced by the agnoprotein and is temporally regulated in isolated nuclear systems. Mol Cell Biol. 1985 Jun;5(6):1327–1334. doi: 10.1128/mcb.5.6.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Skolnik-David H., Aloni Y. Attenuation in the control of SV40 gene expression. Cell. 1982 May;29(1):183–193. doi: 10.1016/0092-8674(82)90102-7. [DOI] [PubMed] [Google Scholar]
- Hernandez N., Lucito R. Elements required for transcription initiation of the human U2 snRNA gene coincide with elements required for snRNA 3' end formation. EMBO J. 1988 Oct;7(10):3125–3134. doi: 10.1002/j.1460-2075.1988.tb03179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Weiner A. M. Formation of the 3' end of U1 snRNA requires compatible snRNA promoter elements. Cell. 1986 Oct 24;47(2):249–258. doi: 10.1016/0092-8674(86)90447-2. [DOI] [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kelly K., Cochran B. H., Stiles C. D., Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983 Dec;35(3 Pt 2):603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988 Oct;8(10):4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Israël A., Le Bail O., Kourilsky P. Detailed analysis of the mouse H-2Kb promoter: enhancer-like sequences and their role in the regulation of class I gene expression. Cell. 1986 Jan 31;44(2):261–272. doi: 10.1016/0092-8674(86)90760-9. [DOI] [PubMed] [Google Scholar]
- Krystal G., Birrer M., Way J., Nau M., Sausville E., Thompson C., Minna J., Battey J. Multiple mechanisms for transcriptional regulation of the myc gene family in small-cell lung cancer. Mol Cell Biol. 1988 Aug;8(8):3373–3381. doi: 10.1128/mcb.8.8.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maderious A., Chen-Kiang S. Pausing and premature termination of human RNA polymerase II during transcription of adenovirus in vivo and in vitro. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5931–5935. doi: 10.1073/pnas.81.19.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Samuels M., Sharp P. A. In vitro transcription: whole-cell extract. Methods Enzymol. 1983;101:568–582. doi: 10.1016/0076-6879(83)01038-1. [DOI] [PubMed] [Google Scholar]
- Mather E. L., Nelson K. J., Haimovich J., Perry R. P. Mode of regulation of immunoglobulin mu- and delta-chain expression varies during B-lymphocyte maturation. Cell. 1984 Feb;36(2):329–338. doi: 10.1016/0092-8674(84)90226-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCachren S. S., Jr, Salehi Z., Weinberg J. B., Niedel J. E. Transcription interruption may be a common mechanism of c-myc regulation during HL-60 differentiation. Biochem Biophys Res Commun. 1988 Feb 29;151(1):574–582. doi: 10.1016/0006-291x(88)90633-x. [DOI] [PubMed] [Google Scholar]
- Mulligan R. C., Howard B. H., Berg P. Synthesis of rabbit beta-globin in cultured monkey kidney cells following infection with a SV40 beta-globin recombinant genome. Nature. 1979 Jan 11;277(5692):108–114. doi: 10.1038/277108a0. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Fahrlander P. D., Yang J. Q., Marcu K. B. Amplification and altered expression of the c-myc oncogene in A-MuLV-transformed fibroblasts. Nature. 1985 Oct 3;317(6036):440–443. doi: 10.1038/317440a0. [DOI] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986 Nov;5(11):2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B., Skoultchi A. I., Lachman H. M. Contributions of transcriptional and post-transcriptional mechanisms to the regulation of c-myc expression in mouse erythroleukemia cells. Genes Dev. 1987 Nov;1(9):938–945. doi: 10.1101/gad.1.9.938. [DOI] [PubMed] [Google Scholar]
- Piechaczyk M., Blanchard J. M., Marty L., Dani C., Panabieres F., El Sabouty S., Fort P., Jeanteur P. Post-transcriptional regulation of glyceraldehyde-3-phosphate-dehydrogenase gene expression in rat tissues. Nucleic Acids Res. 1984 Sep 25;12(18):6951–6963. doi: 10.1093/nar/12.18.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Reines D., Wells D., Chamberlin M. J., Kane C. M. Identification of intrinsic termination sites in vitro for RNA polymerase II within eukaryotic gene sequences. J Mol Biol. 1987 Jul 20;196(2):299–312. doi: 10.1016/0022-2836(87)90691-7. [DOI] [PubMed] [Google Scholar]
- Resnekov O., Aloni Y. RNA polymerase II is capable of pausing and prematurely terminating transcription at a precise location in vivo and in vitro. Proc Natl Acad Sci U S A. 1989 Jan;86(1):12–16. doi: 10.1073/pnas.86.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert J., Sloan S. B., Weisberg R. A., Gottesman M. E., Robledo R., Harbrecht D. The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell. 1987 Nov 6;51(3):483–492. doi: 10.1016/0092-8674(87)90644-1. [DOI] [PubMed] [Google Scholar]
- Roberts J. W. Phage lambda and the regulation of transcription termination. Cell. 1988 Jan 15;52(1):5–6. doi: 10.1016/0092-8674(88)90523-5. [DOI] [PubMed] [Google Scholar]
- Salditt-Georgieff M., Sheffery M., Krauter K., Darnell J. E., Jr, Rifkind R., Marks P. A. Induced transcription of the mouse beta-globin transcription unit in erythroleukemia cells. Time-course of induction and of changes in chromatin structure. J Mol Biol. 1984 Feb 5;172(4):437–450. doi: 10.1016/s0022-2836(84)80016-9. [DOI] [PubMed] [Google Scholar]
- Schibler U., Hagenbüchle O., Wellauer P. K., Pittet A. C. Two promoters of different strengths control the transcription of the mouse alpha-amylase gene Amy-1a in the parotid gland and the liver. Cell. 1983 Jun;33(2):501–508. doi: 10.1016/0092-8674(83)90431-2. [DOI] [PubMed] [Google Scholar]
- Selby M. J., Bain E. S., Luciw P. A., Peterlin B. M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989 Apr;3(4):547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Watson R. J. A transcriptional arrest mechanism involved in controlling constitutive levels of mouse c-myb mRNA. Oncogene. 1988 Mar;2(3):267–272. [PubMed] [Google Scholar]
- Watson R. J. Expression of the c-myb and c-myc genes is regulated independently in differentiating mouse erythroleukemia cells by common processes of premature transcription arrest and increased mRNA turnover. Mol Cell Biol. 1988 Sep;8(9):3938–3942. doi: 10.1128/mcb.8.9.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R., Urlaub G., Chasin L. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., Bishop J. M. DNA sequences that mediate attenuation of transcription from the mouse protooncogene myc. Proc Natl Acad Sci U S A. 1989 Jan;86(2):505–509. doi: 10.1073/pnas.86.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Q., Remmers E. F., Marcu K. B. The first exon of the c-myc proto-oncogene contains a novel positive control element. EMBO J. 1986 Dec 20;5(13):3553–3562. doi: 10.1002/j.1460-2075.1986.tb04682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vegvar H. E., Lund E., Dahlberg J. E. 3' end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986 Oct 24;47(2):259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]