Abstract
The mammalian genome encodes thousands of noncoding RNAs. These noncoding transcripts are broadly categorized into short noncoding RNAs, such as micro-RNAs (miRNAs), and long noncoding RNAs (lncRNAs) of greater than 200 nt. While the role of miRNAs in development and cancer biology has been extensively studied, much less is known about the vast majority of noncoding transcripts represented by lncRNAs. LncRNAs are emerging as key regulators of developmental processes and as such, their frequent misregulation in tumorigenesis and disease in not unexpected. The role of lncRNAs in mammary gland development and breast cancer is just beginning to be elucidated. This review will discuss the role of lncRNAs in mammalian and mammary gland development. In addition, we will review the contributions of lncRNAs to the stepwise progression of tumorigenesis, highlighting the role of lncRNAs in breast cancer.
Keywords: Noncoding RNA, Mammary gland development, Alveologenesis, Epigenetics, Breast cancer
Introduction
Although coding regions occupy less than 2% of the mammalian genome, a much larger portion of the genome is transcribed [1–5]. The initial reports of this pervasive transcription generated some controversy regarding their biological relevance. However, several large scale analyses have since provided evidence that many noncoding transcripts are highly regulated and functional [6–9].
In the past decade, there has been a profound shift in our understanding of the role of RNA molecules. They were once regarded as mere templates for generating proteins, but noncoding RNAs are emerging as highly versatile molecules that potentially function at every level of cellular regulation. The expanding catalogue of noncoding RNAs provides evidence that they are becoming as vast and varied as their protein-coding mRNA counterparts. Just as there are ubiquitously expressed housekeeping proteins, there are also housekeeping noncoding RNAs including, ribosomal, transfer, small nuclear and small nucleolar RNAs. On the other hand, regulatory noncoding RNAs generally show restricted patterns of expression during development and are frequently involved in regulating gene expression. Regulatory noncoding RNAs includes short RNAs (<200 nt) such as, small interfering RNAs, Piwi-associated RNAs, and of course, microRNAs. This review will focus on another class of regulatory noncoding RNAs, long noncoding RNAs (>200 nt), which have recently moved to the forefront of noncoding RNA research.
Long noncoding RNAs (lncRNAs), like mRNAs, can be capped, spliced and polyadenylated and fall into a variety of categories based on their genomic context as well as their functions. Some are located sense or antisense to other transcriptional units. Others are bidirectional, being transcribed near another gene from the opposite strand. LncRNAs can also be located within the intron of another transcript. Alternatively, intergenic noncoding RNAs lie between two genes or sets of genes. Functionally, lncRNAs have been shown to affect a wide range of cellular processes, including splicing [10], localization [11], transcription [12], survival [13], cell cycle [14], migration [15], metabolism [16], and organization of cellular compartments [17]. However, for the relatively small fraction of lncRNAs whose functions have been revealed, their predominant role seems to be the regulation of gene expression.
LncRNAs utilize a large arsenal of mechanisms to regulate gene expression. One well-studied mechanism is transcriptional interference, where the act of transcribing a lncRNA interferes with transcription initiation, elongation or termination of another sense or antisense gene [18]. Some lncRNAs act as decoys, containing sequences that mimic transcription factor binding sites [16] or miRNA target sites [19] to titer these factors away from their primary targets thereby inhibiting their function. Other lncRNAs act as co-activators, binding to transcription factors and enhancing their transcriptional activity [12, 20, 21]. LncRNAs can also affect transcription by binding to transcription factors and shuttling them into the cytoplasm to keep them away from their nuclear targets [11]. Recent evidence also suggests that some lncRNAs may have enhancer-like function [22], activating expression of nearby genes by an unknown mechanism.
Many lncRNAs interact with chromatin-modifying complexes to regulate gene expression. The idea that lncRNAs associate with chromatin-modifying complexes initially came from studies of X chromosome inactivation and genomic imprinting, but more recent global analyses have revealed that this may be a more universal characteristic of lncRNAs than originally anticipated. Almost 40% of large intergenic noncoding RNAs (lincRNAs) interact with chromatin-modifying complexes, such as the Polycomb Repressive Complex 2 (PRC2) and the CoREST/REST repressor complex [23]. At least 24% of lincRNAs interact specifically with PRC2 [23] and a recent study has identified thousands of additional lncRNAs that interact with PRC2 [24], many of which are not intergenic RNAs. In addition, several lncRNAs have been shown to interact with the histone methyltransferase associated with the activating trithorax complex, MLL1 [25, 26], and the H3-K9 methyltransferase, G9a [27, 28]. While the functional relevance of these interactions is not yet well understood, several lncRNAs have also been shown to be required for targeting of these complexes to specific loci, either in cis or in trans, to regulate gene expression [27, 29–31]. Precisely how lncRNAs interact with chromatin-remodeling complexes and how they target specific genes is still largely unknown.
The predominant role of lncRNAs in regulating gene expression, combined with their strict spatiotemporal expression patterns, suggests that they may have a significant role in regulating key processes during development. Indeed, mechanistic evidence is accumulating for the role of lncRNAs in regulating differentiation and development, often epigenetically [32–34]. In addition, numerous lncRNAs have been identified that are misregulated in cancer [35–37]. However, the mechanisms of action of lncRNAs in tumor initiation and progression are largely unknown. In this review, we will discuss the role of lncRNAs during development and tumorigenesis, emphasizing what is currently known in the field of mammary gland biology.
LncRNAs and Development
LncRNAs Regulating Differentiation and Development
LncRNAs were originally discovered in association with distinct epigenetic processes, such as X chromosome inactivation in dosage compensation and genomic imprinting, and were therefore regarded by many as anomalies in the field of gene regulation. In mammals, dosage compensation is the process of inactivation of a single X chromosome in female cells to equalize X-linked gene expression with that of male cells. It involves a complex interplay of several lncRNAs including Xist, Jpx, RepA and Tsix. These lncRNAs recruit chromatin-modifying complexes to the targeted X chromosome, leading to histone modifications, heterochromatin formation and silencing of the targeted X chromosome [38, 39]. Genomic imprinting is the process whereby certain genes undergo monoallelic silencing during gametogenesis, and are therefore only expressed from the maternal or paternal allele for the lifetime of the organism. Imprinting, like X-inactivation, is mediated by chromatin-remodeling complexes and has been shown to require lncRNAs, such as Kcnq1ot1 and Airn (formerly known as Air), that are transcribed from the imprinted loci and silence genes in cis [40]. X inactivation and genomic imprinting are essential developmental processes whose mechanisms have been, and continue to be, studied intensely. While these processes were initially considered unique, an increasing number of lncRNAs are being discovered that interact with chromatin-modifying complexes, and it is now apparent that the lncRNAs associated with X-inactivation and imprinting serve as paradigms to understand precisely how lncRNAs can function as epigenetic regulators.
LncRNAs also play a role in regulating the delicate balance between stem cell self-renewal and differentiation during embryogenesis. Large-scale analysis of mouse embryonic stem cells (mESCs) has identified hundreds of lncRNAs that are differentially expressed in the pluripotent state as compared to various stages of differentiation [26]. Of these, two novel lncRNAs, Evx1as and Hoxb5/6as, are transcribed from homeotic loci in differentiated ES cells and interact with the methyltransferase MLL1. In addition, the lncRNA Mira is thought to play a role in germ layer specification by interacting with MLL1 to activate the expression of the homeotic genes Hoxa6 and Hoxa7 during mESC differentiation [25]. These data suggest likely roles for lncRNAs in epigenetically regulating differentiation genes in early embryonic development. In another recent study, functional analysis was carried out on over 200 lincRNAs that are expressed in mESCs [41]. Knockdown of many of these lincRNAs resulted in drastic changes in gene expression in trans. Furthermore, knockdown of dozens of these lincRNAs led to the reduced expression of pluripotency genes as well as the induction of differentiation genes, suggesting that these lincRNAs are indeed functioning in the maintenance of the pluripotent state of mESCs. Finally, these investigators also showed that many of these lincRNAs are themselves regulated by pluripotency-associated transcription factors, such as Oct4, Nanog and Sox2. These same transcription factors were reported previously to regulate the expression of other lncRNAs in mESCs [8, 42]. Together, these studies demonstrate lncRNAs function to regulate cellular homeostasis during embryonic development.
Many lncRNAs are regulated both spatially and temporally. Thus, it is not surprising that lncRNAs are also involved in regulating proliferation and differentiation of specific stem and progenitor populations during organogenesis. For example, the lncRNAs EGO, HOTAIRM1 and LincRNA-EPS are induced by, and required for, differentiation of distinct lineages during hematopoiesis [43–45]. In addition, global analysis has revealed a large number of lncRNAs that are expressed in the brain and nervous system [7, 46–48] and some lncRNAs have been shown to be necessary for proper neural development. For instance, loss of Evf2 in mice leads to reduced numbers of GABAergic interneurons in early development and disrupts formation of GABA-dependent neuronal circuitry in the adult brain [49]. Other lncRNAs, such as Tug1 and RNCR (also known as MIAT or Gomafu), regulate specification and differentiation of various cell types in the developing retina [50, 51]. LncRNAs also have been found that regulate muscle differentiation by novel mechanisms. For example, Lnc-MD1 acts as a competing endogenous RNA (ceRNA) that binds miRNAs, which normally repress myogenic transcription factors, and inhibits their function, thereby promoting muscle differentiation [19]. Another recent study identified an lncRNA called ANCR, which is downregulated during epidermal differentiation and is required to maintain epidermal progenitors [52]. Since the functions of only a relatively small fraction of lncRNAs currently are known, thousands of lncRNAs still have putative undetermined roles in important developmental processes.
LncRNAs Regulating Mammary Gland Development
The mammary gland begins as the epithelial layer of the skin folds inward to form the mammary bud during embryonic development [53]. At birth, the mammary fat pad contains a simple, branched epithelial structure that quiescently lies beneath the nipple until puberty. During puberty, the mammary epithelial cells of the terminal end buds proliferate and drive ductal elongation toward the ends of the fat pad resulting in a branched tree-like structure that fills the fat pad of the adult mammary gland. The virgin mammary gland then remains mostly quiescent until pregnancy.
In the post-pubertal mammary gland, pregnancies result in dramatic cycles of hormonally-regulated proliferation, differentiation, and apoptosis of mammary epithelial cells making the mammary gland an excellent model to study essential developmental processes such as stem and progenitor cell regulation and cell fate commitment [54–57]. In early to mid-pregnancy, mammary epithelial cells undergo proliferation as they expand to generate lobulo-alveolar structures. During mid to late-pregnancy, proliferation slows as mammary epithelial cells begin to differentiate into alveolar cells. At parturition, alveolar cells undergo terminal differentiation to produce and secrete the high volume of milk required to nurse the young during lactation. Finally, following lactation, most of the differentiated alveolar cells undergo apoptosis in early involution. The mammary gland remodels to a pre-pregnancy-like state and again becomes mostly quiescent until the next pregnancy.
Many coding genes, as well as a few miRNAs (see miRNA reviews in this issue), have well-documented roles in regulating embryonic, post-natal and post-pubertal mammary gland development [58, 59]. However, the role of lncRNAs remains largely unexplored. A few lncRNAs have been mentioned in the literature as having a potential role in mammary development, but without any in depth follow-up. For example, H19 was shown to be hormonally-regulated and expressed highly in alveolar cells during pregnancy and involution, but the specific role of H19 in alveolar cells remains unknown [60]. Additionally, the lncRNA SRA may also play a role in the post-pubertal mammary gland. Overexpression of human SRA in mammary epithelial cells of MMTV-SRA transgenic mice results in excessive side branching of the adult virgin transgenic gland and precocious alveolar development during pregnancy [61]. However, SRA is barely detectable in the virgin mouse mammary gland and subsequent loss-of-function studies to support a role for SRA in mammary development are lacking.
Recently, a large-scale microarray analysis using RNA from mouse mammary glands at day 15 of pregnancy, day 7 of lactation and day 2 of involution, identified 97 lncRNAs that are differentially expressed during post-pubertal development. However, the functions of most of these lncRNAs are not yet understood [62]. Only two lncRNAs, PINC and Zfas1, have been studied more extensively in the post-pubertal mammary gland (Table 1) [62–64]. Interestingly, both of these lncRNAs are highly expressed in alveolar cells during pregnancy. Their expression decreases during lactation and increases again during early involution. PINC and Zfas1 also appear to play similar roles in regulating cell cycle and differentiation of mammary epithelial cells. Each will be discussed in more detail below.
Table 1.
LncRNAs involved in mammary gland development and tumorigenesis
| Mammary Gland Development | ||
| LncRNA | Developmental Expression/Role | Mechanism |
| PINC | Downregulated during lactation and by lactogenic differentiation, knockdown and overexpression studies show that PINC inhibits lactogenic differentiation (unpublished data). Also regulates cell cycle of mammary epithelial cells [63]. | Interacts with PRC2, unknown targets (unpublished data) |
| Zfas1 | Downregulated during lactation and by lactogenic differentiation, negatively regulates proliferation and lactogenic differentiation as shown by knockdown of Zfas1 [62]. | Unknown |
| Breast Cancer | ||
| LncRNA | Clinical Expression/Role | Mechanism |
| HOTAIR | Independent predictor or breast cancer survival. Promotes invasion and metastasis [15]. | Interacts with PRC2 and LSD1/CoREST/REST complexes in gene silencing [29, 117]. |
| H19 | Highly expressed in stromal cells of breast tumors and associated with the presence of ER and PR [85]. Upregulated by E2F1 and promotes cell cycle progression [86]. | Unknown |
| LSINCT5 | Overexpressed in breast and ovarian cancer cell lines and tumors. Enhances cellular proliferation [92]. | Unknown |
| BC200 | Overexpressed in several solid tumors including breast. In DCIS lesions, BC200 expression is associated with high nuclear grade [93, 94]. | Unknown |
| SRA | High expression of noncoding SRA RNA in invasive breast cancer cell lines and PR+ clinical breast cancer samples [112, 113]. | Interacts with and coactivates a large number of nuclear receptors and transcription factors [20, 21, 111]. |
| GAS5 | Low expression in breast cancers associated with poor clinical outcome [116]. | Acts as a decoy that prevents GR from binding to its targets [16]. |
This table contains a summary of lncRNAs that are expressed in the mammary gland and have been shown to regulate mammary development as well as lncRNAs that are misregulated in breast cancer and are thought to contribute to tumor initiation and/or progression
PINC
Pregnancy-Induced NonCoding RNA (PINC) is polyadenylated, alternatively spliced and conserved specifically among mammals [63]. There are two sets of splice forms of mouse PINC (mPINC), mPINC1.0 and mPINC1.6. Knockdown of mPINC1.0 and mPINC1.6 individually in mammary epithelial HC11 cells leads to increased rates of apoptosis and proliferation, respectively. These data suggest mPINC may play a role in regulating survival and cell cycle of mammary epithelial cells.
Recent data suggest that mPINC may play a specific role in regulating alveolar differentiation during post-pubertal mouse development (Shore and Rosen, unpublished data). mPINC splice isoform levels are coordinately regulated, declining between late pregnancy and early lactation in the mammary gland when alveolar cells undergo terminal secretory differentiation. Interestingly, mPINC levels also decrease upon lactogenic differentiation of HC11 cells following hormone treatment. This reduction of mPINC expression may be required for lactation as overexpression of mPINC inhibits lactogenic differentiation of HC11 cells, while conversely knockdown of mPINC enhances differentiation.
PINC and retinoblastoma associated protein 46 (RbAp46) were originally identified as genes that are persistently upregulated following pregnancy in the rat mammary gland and thought to potentially mediate the protective effect of an early pregnancy [64]. PINC and RbAp46 are both expressed in alveolar cells during pregnancy and their expression persists in the regressed lobules of the involuted rat mammary gland. RbAp46 is an important regulator of development and a potential tumor suppressor [65–67]. RbAp46 also associates with the chromatin-modifying PRC2 complex that has been shown to interact with many lncRNAs [68]. Using RNA immunoprecipitation assays, we found that mPINC splice forms interact specifically with RbAp46 and other PRC2 members in HC11 cells and in mammary epithelial cells isolated from day 16 pregnant mammary gland (Shore and Rosen, unpublished data). These data suggest a potential epigenetic mechanism for the regulation of alveolar progenitor cells by mPINC during pregnancy and involution of the mammary gland.
Zfas1
The lncRNA Zfas1 initially was identified by microarray analyses of various stages of post-pubertal mouse mammary gland development [62]. Zfas1 is transcribed in the antisense direction from the promoter region of the Znfx1 gene. Zfas1 and Znfx1 expression is coordinately regulated in several tissues including, brain, kidney, lung and spleen. However, they show distinct patterns of expression during post-pubertal mammary development, suggesting Zfas1 may have a distinct function from Znfx1 in the mammary gland. The primary Zfas1 transcript also contains snoRNAs in three consecutive introns that are differentially regulated in the post-pubertal mammary gland. However, the expression of these snoRNAs is not precisely coordinated with that of Zfas1. While it is likely that transcription from this complex locus is interconnected, the mechanistic implications of this relationship are currently unknown.
Zfas1 is highly expressed in alveolar cells of the pregnant mammary gland, but declines 10-fold during lactation. Zfas1 expression levels also decline upon lactogenic differentiation of HC11 cells. Upon lactogenic hormone treatment, Zfas1 knockdown leads to enhanced dome formation and increased β-casein levels. Knockdown of Zfas1 in HC11 cells also results in increased proliferation. Together these data suggest Zfas1 may be highly expressed in the late pregnant mammary gland to regulate proliferation and repress terminal differentiation of alveolar cells until parturition when Zfas1 levels drastically decline.
Perspectives on LncRNAs and Mammary Gland Development
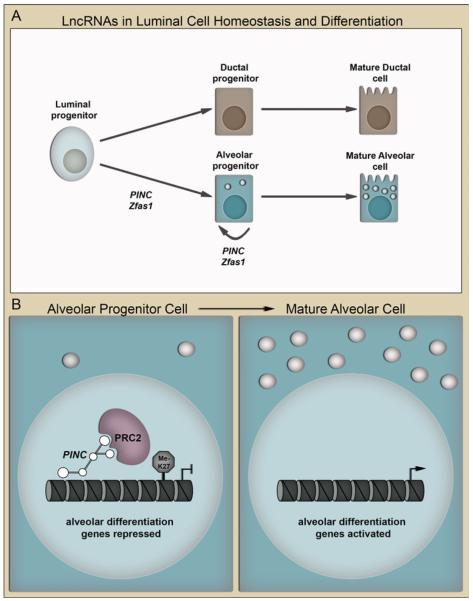
mPINC and Zfas1 are both expressed in alveolar cells of the pregnant and involuting mammary gland and are downregulated during lactation when alveolar cells undergo terminal secretory differentiation. In addition, mPINC and Zfas1 levels decline upon hormonally-induced differentiation of HC11 cells. Furthermore, knockdown studies demonstrate that they both negatively regulate lactogenic differentiation of HC11 cells. Therefore, high expression of mPINC and Zfas1 in the late pregnant gland may be necessary to prevent precocious terminal differentiation and maintain alveolar progenitors until lactation (as illustrated in Fig. 1a). Likewise, increased mPINC and Zfas1 levels during early involution may function to preserve alveolar progenitors in the involuted gland so that they can rapidly differentiate into milk-producing cells in subsequent pregnancies. While the mechanism of action of Zfas1 is unknown, we propose a model where mPINC interacts with PRC2 to target and repress alveolar differentiation genes epigenetically, thereby maintaining alveolar progenitors in the pregnant and involuting mammary gland (see hypothetical model in Fig. 1b).
Figure 1.
LncRNAs regulate mammary epithelial differentiation. a A schematic diagram illustrates a mammary epithelial luminal progenitor and its progeny. PINC and Zfas1 may contribute to alveolar cell fate commitment. PINC and Zfas1 inhibit terminal alveolar differentiation thereby maintaining alveolar progenitor cells. b A hypothetical model shows the role of PINC in alveolar progenitor cells. A PINC-PRC2 complex targets and represses alveolar differentiation genes by methylating histone H3 at lysine 27 (me-K27). In cells lacking PINC, the repressive marks are removed and alveolar differentiation genes are transcribed to induce terminal differentiation
While only a couple of lncRNAs have documented roles in mammary gland development, it is anticipated that the application of RNA-seq in mammary development and breast cancer, through The Cancer Genome Atlas (TCGA) project, will lead to the discovery of many more lncRNAs. The mammary gland is unique to mammals. It has been postulated that the mammary gland evolved from the innate immune system [69], as it provides immunological protection as well as nutrition to offspring during lactation. Previous studies have identified over a thousand lncRNAs that are highly conserved among mammals and, based on functional genomics approaches, a large portion of these have putative roles in regulating innate immunity [8]. Potentially, many of these lncRNAs have a mammalian-specific function in the mammary gland. In addition, milk protein genes and other mammary gland genes are sometimes organized in large genomic clusters that are epigenetically regulated [70, 71]. With the emerging association of lncRNAs and chromatin-modifying complexes, it would not be surprising to identify lncRNAs that govern the coordinated expression of these gene clusters during mammary development. Finally, current studies only reflect a role for lncRNAs in the post-pubertal mammary gland. However, several lncRNAs are induced by signaling pathways that are known to regulate embryonic and post-natal mammary development, such as those regulated by Notch, Wnt, Shh, and BMP [12, 72–75]. Therefore, it is likely that these pathways induce lncRNAs that contribute to early mammary development. Understanding the function of lncRNAs in regulating developmental processes in the mammary gland should help elucidate the potential roles of aberrantly expressed lncRNAs in the initiation and progression of breast cancer.
LncRNAs in Breast Cancer
Transcriptional profiling has revealed highly aberrant lncRNA expression in human cancers [76]. In breast tumors, expression profiling suggests fundamental differences in global RNA regulation between normal and cancer cells with the latter showing a lower percentage of genes expressing antisense transcripts [77]. The expression of highly conserved lncRNAs is altered in human cancers including breast [78], with some recent evidence of their susceptibility to mutation. Using mRNA profiling, breast cancer has reproducibly been divided into a number of clinically relevant subtypes [79, 80]. These subtypes also show differential expression of non coding RNAs including both miRNAs [81] and lncRNAs (Herschkowitz, Perou, and Rosen unpublished results). Likewise, the expression of lncRNAs can be predictive of clinical outcome and they may be useful as biomarkers. One example is HOTAIR (discussed below) which has been shown to be an independent predictor of breast cancer survival [15].
While the study of lncRNA function is still in its infancy, a role for a number of these transcripts has recently been established in cancer, in general, as well as specifically in breast cancer. Like protein-coding genes and miRNAs, lncRNAs play key roles in tumorigenesis. They have been shown to play a functional role in a number of fundamental processes associated with cancer including cell cycle regulation, apoptosis, the DNA damage response, and metastasis.
Oncogenic and Tumor Suppressor LncRNAs
Like protein-coding genes, lncRNAs can function as oncogenes or tumor suppressors. Many lncRNAs shuttle between the nucleus and cytoplasm suggesting they may have dual functions, while others are restricted to the nucleus. In the nucleus, lncRNAs are often part of the nuclear architecture and in some cases are critical for the maintenance of sub-nuclear structures.
MALAT1/NEAT2
The nuclear-retained lncRNA MALAT1, which is highly expressed in many tumor types, regulates alternative splicing by modulating the phosphorylation of SR splicing factors and influencing their distribution to nuclear speckles [10]. While MALAT1 expression is restricted to the nucleus, a small tRNA-like 61-bp ncRNA is generated by RNase P dependent cleavage of the primary transcript and exported into the cytoplasm [82]. However, the function of this processed RNA in normal development and cancer is not known.
Recent evidence also suggests that non coding RNAs are involved in the localization of transcriptional units to specific sub-nuclear architectural structures, thus regulating cellular proliferation. Specifically by binding to unmethlyated Polycomb 2 protein (Pc2)/Cbx4, MALAT1 has been shown to relocate growth control genes to interchromatin granules, which are thought to represent sites of active transcription. In contrast, the lncRNA TUG1 binds to methylated Pc2, relocating genes to the repressive environment of Polycomb bodies [83]. Like MALAT1, many lncRNAs can give rise to a number of distinct processed RNAs with separate functions, any and/or all of which may contribute to their role in tumorigenesis.
H19
H19 is an imprinted gene expressed exclusively from the maternal allele that maintains the silencing of IGF2. Aberrant expression of H19 is observed in a number of solid tumors. The majority of breast cancers express high levels of H19 as compared to normal tissues. H19 is generally overexpressed in stromal cells, rarely in tumor epithelial cells in tumors and was found to be associated with the presence of ER and PR [84]. This is not surprising given that H19 has been shown to be hormonally regulated in the mammary gland as discussed earlier. There are data indicating both oncogenic and tumor suppressive roles for H19 in different cancers [85]. In breast cancer cell lines, H19 RNA expression is directly regulated by E2F1 and promotes cell cycle progression [86]. This supports a putative oncogenic function for H19 in breast tumorigenesis. Additionally, H19 has been shown to be upregulated by hypoxia through HIF1-alpha and suppressed by p53 [87].
ANRIL/p15AS
The INK4/ARF (CDKN2a/b) locus, located at chromosome 9p21 contains three well-characterized tumor suppressors, p16INK4a, p15INK4b, and p14ARF. The lncRNA ANRIL (Antisense Noncoding RNA in the INK4 Locus) was initially identified by examining the deletion of this region in hereditary neural tumors [88]. ANRIL is a polyadenylated lncRNA antisense to CDKN2A (p16INK4a) and CDKN2B (p15INK4b). ANRIL has been shown to repress p16INK4a and p15INK4b, but not p14ARF, in cis through direct binding to CBX7 [89], a PRC1 complex member, and SUZ12 [31], part of the PRC2 complex. Upon recruitment, these complexes apply repressive histone modifications resulting in heterochromatin formation and gene silencing at this locus. Interestingly, genome-wide association studies (GWAS) have revealed that ANRIL is associated with increased susceptibility to several diseases, including breast cancer [90].
ncRNACCND1
A low-copy number lncRNA, ncRNACCND1, transcribed 2 kb upstream of CCND1 is induced by ionizing radiation and regulates transcription of CCND1 in cis by forming a ribonucleoprotein repressor complex [91]. This lncRNA, tethered to the 5′ regulatory region of CCND1, acts as a ligand and recruits the RNA-binding protein TLS (translated in liposarcoma). TLS, allosterically modulated by ncRNACCND1, inhibits histone acetyltransferases resulting in inhibition of CCND1 transcription.
There is extensive evidence of inactivation of the cyclinD/p16/Rb tumor suppressor pathway in breast cancer, which is often characterized by cyclin D1 overexpression and amplification, p16INK4a silencing, and/or Rb loss. It is not yet known whether ANRIL or ncRNACCND1 play a role in inactivating this pathway in breast cancer.
LSINCT5
LSINCT5 (long stress-induced non-coding transcript 5) is a 2.6 Kb polyadenylated lncRNA expressed from the negative strand, that is localized in the nucleus, and overexpressed in breast and ovarian cancer cell lines and tumors [92]. Knockdown experiments revealed that LSINCT5 enhances cellular proliferation. Interestingly, following knockdown, another nuclear lncRNA, NEAT-1 as well as PSPC1 (paraspeckle component 1) are downregulated. These results are suggestive of a potential role of LSINCT5 in paraspeckle formation, but this needs to be explored further.
BC200
BC200, also known as BCYRN1 (brain cytoplasmic RNA 1), is a 200 nt long ncRNA selectively expressed in the nervous system and usually not detected in somatic cells other than neurons. It is, however, overexpressed in several solid cancers including breast cancer [93]. BC200 RNA is expressed at high levels in invasive breast carcinomas, but is barely detectable in normal tissue or in benign tumors [94]. Interestingly, in ductal carcinomas in situ (DCIS), significant BC200 expression is associated with high nuclear grade. This suggests that BC200 may be a useful marker for early detection of breast cancer and that the presence of BC200 RNA in early lesions might have utility as a prognostic indicator of tumor progression.
LncRNAs and p53
The p53 tumor suppressor is the most commonly altered gene in human breast cancer. It is mutated in about 30–40% of all human breast tumors with a much higher frequency associated with poor clinical outcome and basal-like breast cancers. P53 is an example of an increasing number of genes that encode both RNAs and proteins with distinct functions. Thus, while p53 is best known for its function as a transcription factor, p53 mRNA also has evolved the additional function of regulating the MDM2 protein [95].
As a transcription factor, p53 also regulates a number of ncRNAs, including both miRNAs and lncRNAs. For example, p53 regulates both miRNA processing [96] as well as the transcription of miRNAs, such as miR-34 family members [97] and miR-200c [98, 99]. Furthermore, p53 regulates the expression of specific lncRNAs (discussed in detail below), such as those transcribed from the regulatory sequence upstream of CDKN1A/p21, a well-known p53 transcriptional target.
LincRNA-p21
A lincRNA located 15 kb upstream of CDKN1A, named lincRNA-p21, is induced following the activation of p53 in murine tumors [100]. LincRNA-p21 represses p53 target genes by binding to hnRNP-K, a repressor protein that binds the promoters of genes involved in p53 signaling. LincRNA-p21 is further required for the induction of apoptosis. It has not been reported whether there is a conserved and functional human ortholog corresponding to lincRNA-21.
PANDA
DNA damage induces five lncRNAs from the CDKN1A locus [13]. One of these, located approximately 5 kb upstream of the CDKN1A transcriptional start site, is PANDA (P21 associated ncRNA DNA damage activated). PANDA is a 1.5 kb 5′-capped and polyadenylated non-spliced lncRNA that is transcribed antisense to CDKN1A following p53-dependent induction. Functionally, PANDA interacts with the NF-YA transcription factor, preventing it from occupying target gene promoters, and thus limiting the expression of pro-apoptotic genes including FAS, PUMA, and NOXA. PANDA knockdown significantly sensitized human fibroblasts to apoptosis following treatment with doxorubicin. This suggests that in parallel to p53-mediated induction of CDKN1A to regulate cell cycle arrest, PANDA is a p53 effector that acts independently of p21 to promote cell survival by impeding apoptosis. Interestingly, the gain-of-function p53 (p.Arg273His) mutant, observed in Li-Fraumeni syndrome lacks the ability to induce CDKN1A, while maintaining the ability to induce PANDA. Similar observations were reported in metastatic ductal carcinomas. This may be important because in many cancers including breast cancer, gain-of-function p53 missense mutations rather than just inactivating mutations are frequently observed. This has led to the speculation that these mutations may have been selected for at least in part as a function of increased cell survival due to their ability to induce PANDA.
MEG3
Maternally expressed gene 3 (MEG3) is an imprinted lncRNA located on chromosome 14q32 and, as its name implies, is expressed exclusively from the maternal allele. MEG3 has been shown to activate p53 and facilitate p53 signaling, including the enhancement of p53 binding to target genes [101]. MEG3 overexpression in meningioma, hepatocellular carcinoma, and breast cancer cell lines leads to suppression of cell proliferation [102–104]. MEG3 is downregulated in human tumors and hypermethylated in leukemias and pituitary tumors [105, 106], but its role in breast cancer has not been established
Steroid Receptors and Co-activators
Steroid hormones and their receptor play major roles in the etiology, progression, and treatment of breast cancer. Greater than 70% of breast cancers are estrogen receptor (ER) positive and/or progesterone receptor (PR) positive. LncRNAs have been recently shown to be important as regulators, targets, and co-activators of steroid receptor signaling.
Estrogen Receptor
Global nuclear run-on and sequencing (GRO-seq) has been used to explore the rapid effects of estrogen treatment on the entire transcriptome of MCF-7 human breast cancer cells [107]. Estrogen signaling directly regulates a large fraction of the transcriptome including not only protein-encoding genes, but also noncoding transcripts. These transcripts include divergent (transcribed in the opposite direction from transcriptional start sites, also produced at enhancers), antisense, and intergenic transcripts. A large number of previously undetected estrogen-regulated intergenic transcripts were identified, many of which are located proximal to ER binding sites.
Progesterone Receptor
Antisense RNA transcripts have also been identified overlapping the PR promoter [108]. Synthetic antigene RNAs (agRNAs) complementary to the PR promoter, rather than binding DNA, form a complex with and recruit AGO2 to the antisense transcript that overlaps the promoter. These agRNAs can modulate the expression of PR. miRNA mimics predicted to target the PR promoter and inhibit PR expression have also been identified [109]. Interestingly, the antisense RNA transcript serves as the target for these mimics in inhibiting PR transcription.
These findings clearly have a much broader significance than just the regulation of PR expression, which is important in mammary gland development and breast cancer. Non-coding transcripts may overlap with many other gene promoters. Thus, the recognition and interplay of miRNAs and lncRNAs at promoters may provide a general mechanism for regulating gene transcription.
SRA
The SRA gene encodes both noncoding as well as protein-generating RNA isoforms. SRA RNA is highly expressed in a number of solid tumor types including breast, ovarian, and uterine tumors [61, 110]. SRA is a spliced, polyadenylated transcript that has been shown to enhance ligand-activated steroid receptor transcription on luciferase reporters and is found in SRC-1/SRC-2 coactivator complexes [20, 111]. SRA RNA is capable of interacting with and coactivating a large number of nuclear receptors (steroid and non-steroid) and other transcription factors. Thus, it may act as a scaffold in assembling coregulator complexes. Interestingly, SRA RNAs that retain intron-1 are noncoding, and are expressed at higher levels in invasive breast cancer cell lines (MDA-MB-231 and MDA-MB-468) as compared to non-invasive cell lines (MCF-7, T47D, and BT20). Non-transformed MCF10A cells express the lowest levels [112]. Higher expression of SRA RNAs that retain intron-1 is observed in clinical breast tumor samples with higher PR levels [113]. siRNA knockdown has been used to deplete SRA RNA in two human cancer cell lines (HeLa and MCF7 cells) followed by genome-wide expression analysis to determine target genes [114]. The majority of significantly changed genes following knockdown were downregulated, suggesting that SRA RNAs play a larger role in co-activation than co-repression. Similar knockdown experiments were performed in E2-treated and -untreated MCF7 cells. Surprisingly, despite SRAs putative role as an ER coactivator, only a small number of direct ER target genes were affected by SRA depletion in estradiol-treated MCF-7 cells indicating that SRA serves as a coactivator for only a minor subset of ER targets. Overall, these target gene expression data is suggestive of roles for SRA in glucose uptake, cellular signaling, T3 hormone generation, and invasion/metastasis. SRA depletion in MDA-MB-231 cells reduced invasiveness and expression of genes consistent with the invasive phenotype. In agreement with the knockdown results and consistent with its role as a coactivator, overexpression of SRA was shown to co-activate certain target promoters and enhance the activity of some co-regulatory proteins. As mentioned previously, transgenic overexpression of human SRA in the murine mammary gland results in increased epithelial hyperplasia [61]. These mice fail to develop palpable tumors, however, likely due to enhanced apoptosis in the preneoplastic lesions. Together, while these data suggest that SRA RNA is playing an oncogenic role in mammary tumorigenesis, the precise mechanisms involved remain to be elucidated.
GAS5
GAS5 is an lncRNA existing as multiple splice isoforms containing up to 12 exons. Following overexpression in HeLa cells GAS5 modulates cell survival and metabolism [16]. It functions by serving as a RNA transcript decoy that prevents the glucocorticoid receptor (GR) from binding to target genes. Mechanistically, the 3′ end of GAS5 contains a stem-loop structure resembling the GR DNA-binding element and this interacts with the GR DNA-binding domain. Upon stimulation with dexamethasone, a GR agonist, GAS5 is sufficient to repress GR-induced genes, such as cIAP2. By the same means, GAS5 also seems to regulate other receptors including the androgen, mineralcorticoid, and progesterone receptors.
In breast tumors, GAS5 transcript levels were significantly reduced relative to adjacent unaffected normal breast epithelial tissues [115]. Decreased expression of GAS5 and RNU44, an intronic snoRNA contained within the GAS5 transcript, were associated with a poor prognosis in breast cancers [116]. When overexpressed in breast cancer cell lines GAS5 induced apoptosis and suppressed proliferation [115]. These data are suggestive of a tumor suppressor role for GAS5 in breast cancer.
Breast Cancer Metastasis
Metastatic disease is the major cause of mortality from breast cancer. Metastasis is an inefficient multi-step process which involves local invasion of the primary tumor cells and entry into the vasculature, survival in the circulation, invasion into other tissues, establishment of micrometastases, and finally the growth of secondary tumors. While miRNAs have been shown to play major roles in both the promotion and suppression of metastasis (reviewed elsewhere in this issue), the role of lncRNAs is only just starting to be explored. However early indications suggest that lncRNAs will also be of major importance.
HOTAIR
One lncRNA that recently has been shown to play a key role in breast cancer metastasis is HOTAIR (Hox transcript antisense intergenic RNA). HOTAIR is expressed from the HOXC locus and was the first lncRNA shown to be acting in trans. HOTAIR binds to and targets the PRC2 complex to the HOXD locus [29]. Furthermore, HOTAIR functions as a RNA scaffold containing two main functional domains. The 5′ domain of HOTAIR binds PRC2, whereas a 3′ domain binds the LSD1/CoREST/REST H3K4 demethylase complex [117], thus bridging two repressive complexes in order to coordinate their functions in gene silencing. Enforced HOTAIR expression in epithelial cancer cells induces genome-wide retargeting of PRC2 leading to widespread changes in repressive (H3K27me3) and active (H3K4me3) chromatin marks resembling those found in embryonic fibroblasts. This results in more invasive and metastatic cells. Thus, it is not surprising that HOTAIR expression is predictive of breast cancer survival [15]. Finally, Chromatin Isolation by RNA Purification (ChIRP) followed by DNA sequencing has been employed recently to map the genome-wide occupancy of three lncRNAs including HOTAIR [118]. These results suggest that HOTAIR binds to focal target sites in the genome, and is responsible for recruiting PRC2 to these sites. PRC2 then spreads from these focal target sites along the chromosome in a HOTAIR-independent manner. The mechanisms, however, by which HOTAIR recognizes these sites remain to be established.
Long Meets Short
As mentioned above with reference to the role of linc-MD1 in muscle differentiation [19], lncRNAs can regulate gene expression through their interactions with miRNAs by acting as decoys for target mRNAs. Another example of this mechanism is the lncRNA HULC (highly upregulated liver cancer transcript), which binds to and inhibits miR-372 in liver cancer [119].
PTENP1
miRNAs have been shown to be major regulators of the PTEN tumor suppressor and thus contribute to cell transformation through aberrant PI3K/AKT pathway activation. PTENP1, a pseudogene of PTEN is able to compete for miRNA binding sites with PTEN [120]. Accordingly, PTENP1 displays a tumor suppressive role in in vitro assays and is genomically lost in cancer. A similar relationship is also found between KRAS and its pseudogene, KRAS1P. Overexpression of the KRAS1P 3′ UTR results in increased KRAS mRNA abundance and an increase in cellular proliferation. These findings show that transcribed pseudogenes can serve as decoys for miRNAs that target protein-encoding mRNA transcripts of their cognate genes. Not only pseudogenes and long non coding transcripts can play this role, but protein-encoding RNA transcripts also can act as ceRNAs (competing endogenous RNAs). These can communicate and co-regulate each other by competing for miRNAs that target both transcripts [121, 122]. Several putative PTEN ceRNAs have been identified and validated. This includes the ZEB2 transcript, which can modulate PTEN protein levels and thus PI3K/AKT signaling in a miRNA-dependent, protein-coding-independent manner [121].
Not surprisingly, miRNAs can also target lncRNAs just as they do mRNAs [123]. miR-155 has been shown to target transcribed ultraconserved regions (UCRs), regions of the genome that are completely conserved between orthologous regions in the human, rat, and mouse genomes [124]. We are just beginning to understand these complex networks containing all combinations of RNA:RNA interactions and their ultimate impact on gene expression. Thus, it is likely that similar mechanisms may be operable and play an important role in breast cancer.
Genetic Association
Genetic association studies have recently uncovered a number of single nucleotide polymorphisms (SNPs) associated with susceptibility to common diseases including breast cancer. While most of these studies have initially focused on coding genes many of these polymorphisms may be found in regulatory regions and regions containing non-coding RNAs. With this in mind recently discovered SNPs in lncRNAs are associated with prostate cancer risk [125]. In addition, among six SNPs that are located within UCRs, two were significantly associated with familial breast cancer risk [126]. At present the significance of variations in the sequence of lncRNAs as compared with protein-coding genes will be difficult to verify experimentally.
Clinical Significance, Prognostic Implications, and Therapeutics
LncRNAs that show dramatic differences in expression between tumors and normal tissues may also be detected in body fluids. Thus, they potentially represent promising prognostic biomarkers for the diagnosis and treatment of breast cancer. As an example, the lncRNA PCA3/DD3 is being tested as a potential biomarker for early detection in prostate cancer [127, 128]. PCA3 originally was discovered in a differential display analysis comparing normal prostate and tumor samples [129]. As mentioned previously, lncRNAs like HOTAIR and BC200 may represent promising prognostic and/or predictive biomarkers for breast cancer.
LncRNAs also have the potential to be useful as therapeutic agents or targets. Currently epigenetic cancer therapy uses HDAC and DNMT inhibitors to induce genome-wide modifications of histone acetylation and DNA methylation, respectively, but these are relatively non-specific approaches. An alternative approach of targeting RNA-protein interfaces with a focus on cancer specific lncRNAs may allow for site-specific epigenetic modulation and the development of cancer-specific therapeutics [130].
LncRNAs can be targeted with synthetic siRNAs or miRNAs. However, because of their more extensive secondary structures as compared to mRNAs, it is likely that many siRNAs will need to be screened in order to achieve successful knockdown [131]. Another way to target lncRNAs is to utilize oligonucleotides that bind and disrupt RNA-protein interactions. These “antagolincs” potentially can be selected for using a method such as systematic evolution of ligands by exponential enrichment (SELEX) [131]. Finally small molecules can be screened for their interaction with lncRNAs. For successful application of these approaches, more insight into lncRNA structure and function will be required.
Substantial technical hurdles, including the development of specific delivery systems, must be overcome before it will be feasible to rationally design and utilize synthetic RNA molecules as novel strategies to target tumor cells. However, one might envision that the same way that GAS5 serves as a decoy for the GR, the activity of transcription factors might be regulated using RNA molecules designed to form hairpin structures mimicking DNA transcription factor-binding sites. Likewise, synthetic RNA sponges can be designed that contain miRNA-binding sites to sequester and reduce the expression level of oncogenic miRNAs. With analogy to lncRNAs like HOTAIR it should also be possible to rationally design chimeric RNA molecules that possess the RNA structural motifs to recruit chromatin modifying complexes in order to regulate specific gene expression. At present all of these approaches represent wishful thinking, but they provide a future direction for lncRNA research.
Perspectives on LncRNAs and Breast Cancer
In summary, lncRNAs function in many important oncogenic and tumor suppressor pathways. This includes playing important roles in the regulation of two important tumor suppressor pathways often perturbed in cancers, the RB pathway and the p53 pathway. The extensive association of lncRNAs with chromatin modifying complexes is of particular interest with regards to cancer. Mutations in proteins involved in chromatin modification have been a recurrent theme in recent cancer sequencing studies, e.g. mutations have been identified in components of chromatin modifying complexes including Ezh2 in B-cell lymphoma [132], MLL2 and MLL3 in medulloblastomas [133], and MEN1 in pancreatic neuroendocrine tumors [134]. When cancer sequencing studies turn their attention to noncoding portions of the genome, it is highly likely that mutations will be found in lncRNAs that interact with these protein complexes. Targeting the interaction between lncRNAs and chromatin modifying complexes, rather than these ubiquitous proteins themselves, therefore, may represent a promising new approach for cancer specific therapeutics.
LncRNAs have been shown to extensively bind and regulate protein complexes. Their interactions with chromatin modifying complexes and transcription factors are at present best understood. This has led some to hypothesize that lncRNA functions may extend well beyond their roles in the nucleus associated with gene regulation. They may also modulate signal transduction pathways by linking signaling proteins to specific downstream target effectors thus regulating how the signals are propagated in a context specific manner [135]. If true, this could open up a new paradigm for looking at how signal transduction pathways are wired in cancer and also provide novel therapeutic approaches. Finally, genes that encode bi-functional RNAs like SRA and TP53, as well as mRNAs like ZEB2 that can act as ceRNAs, challenge the existing paradigm classifying RNAs as either coding or noncoding. It is becoming increasingly clear that coding-independent functions possibly can be bestowed on any RNA.
Acknowledgements
ANS was supported by a Department of Defense Pre-doctoral Fellowship Award (W81XWH-06-1-0717). This work was supported by National Cancer Institute (NCI) Grants CA-16303 and CA148761.
Abbreviations
- miRNA
microRNA
- lncRNA
long/large noncoding RNA
- lincRNA
long/large intergenic noncoding RNA
- PRC2
polycomb repressive complex-2
- CoREST
co-repressor for REST
- REST
repressor element-1 (RE1) silencing transcription factor
- MLL1
mixed lineage leukemia 1
- Xist
X-inactive specific transcript
- RepA
Repeat A
- Kcnq1ot1
Kcnq1 overlapping transcript 1
- Airn
antisense Igf2r RNA noncoding
- mESC
mouse embryonic stem cell
- Oct4
octamer-binding transcription factor 4
- Sox2
SRY (sex determining region Y)-box 2
- EGO
eosinophil granule ontogeny
- HOTAIRM1
HOX antisense intergenic RNA myeloid 1
- LincRNA-EPS
lincRNA erythroid prosurvival 1
- Evf2
embryonic ventral forebrain 1
- GABA
gamma-aminobutryic acid
- Tug1
taurine-upregulated gene 1
- RNCR
retinal noncoding RNA
- MIAT
myocardial infarction associated transcript
- ceRNA
competing endogenous RNA
- ANCR
anti-differentiation noncoding RNA
- SRA
steroid receptor RNA activator
- MMTV
mouse mammary tumor virus
- PINC
pregnancy induced noncoding RNA
- Zfas1
Znfx1 antisense RNA 1
- mPINC1.0
mouse pregnancy induced noncoding RNA 1.0 Kb
- mPINC1.6
mouse pregnancy induced noncoding RNA 1.6 Kb
- RbAp46
retinoblastoma associated protein 46
- Znfx1
zinc finger, NFX1-type containing 1
- snoRNA
small nucleolar RNA
- TCGA
the cancer genome atlas
- Shh
sonic hedgehog
- BMP
bone morphogenetic protein
- MALAT1
metastasis associated lung adenocarcinoma transcript 1
- ER
estrogen receptor
- PR
progesterone receptor
- AGO2
argonaute 2
- UCR
ultraconserved region
- GWAS
genome-wide association study
References
- 1.Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. doi:10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309(5740):1564–6. doi: 10.1126/science.1112009. doi:10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 3.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–63. doi: 10.1126/science.1112014. doi:10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 4.Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306(5705):2242–6. doi: 10.1126/science.1103388. doi:10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 5.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308(5725):1149–54. doi: 10.1126/science.1108625. doi:10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 6.Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16(1):11–9. doi: 10.1101/gr.4200206. doi:10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A. 2008;105(2):716–21. doi: 10.1073/pnas.0706729105. doi:10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. doi:10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic non-coding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–27. doi: 10.1101/gad.17446611. doi:10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–38. doi: 10.1016/j.molcel.2010.08.011. doi:10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, et al. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309(5740):1570–3. doi: 10.1126/science.1115901. doi:10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 12.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20(11):1470–84. doi: 10.1101/gad.1416106. doi:10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–9. doi: 10.1038/ng.848. doi:10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meola N, Pizzo M, Alfano G, Surace EM, Banfi S. The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA. 2012;18(1):111–23. doi: 10.1261/rna.029454.111. doi:10.1261/rna.029454.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–6. doi: 10.1038/nature08975. doi:10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. doi:10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–26. doi: 10.1016/j.molcel.2009.01.026. doi:10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazo A, Hodgson JW, Petruk S, Sedkov Y, Brock HW. Transcriptional interference: an unexpected layer of complexity in gene regulation. J Cell Sci. 2007;120(Pt 16):2755–61. doi: 10.1242/jcs.007633. doi:10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- 19.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147(2):358–69. doi: 10.1016/j.cell.2011.09.028. doi:10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97(1):17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 21.Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, et al. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell. 2006;11(4):547–60. doi: 10.1016/j.devcel.2006.08.003. doi:10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. doi:10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–72. doi: 10.1073/pnas.0904715106. doi:10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–53. doi: 10.1016/j.molcel.2010.12.011. doi:10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43(6):1040–6. doi: 10.1016/j.molcel.2011.08.019. doi:10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18(9):1433–45. doi: 10.1101/gr.078378.108. doi:10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32(2):232–46. doi: 10.1016/j.molcel.2008.08.022. doi:10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 28.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–20. doi: 10.1126/science.1163802. doi:10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 29.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–23. doi: 10.1016/j.cell.2007.05.022. doi:10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–6. doi: 10.1126/science.1163045. doi:10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–62. doi: 10.1038/onc.2010.568. doi:10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12(2):136–49. doi: 10.1038/nrg2904. doi:10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19(7–8):454–92. doi: 10.1007/s00335-008-9136-7. doi:10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 34.Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: an answer to the `genome complexity' conundrum. Genes Dev. 2007;21(1):11–42. doi: 10.1101/gad.1484207. doi:10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- 35.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–61. doi: 10.1016/j.tcb.2011.04.001. doi:10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–61. doi: 10.1093/hmg/ddq353. doi:10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalil AM, Rinn JL. RNA-protein interactions in human health and disease. Semin Cell Dev Biol. 2011;22(4):359–65. doi: 10.1016/j.semcdb.2011.02.016. doi:10.1016/j.semcdb.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brockdorff N. Chromosome silencing mechanisms in X-chromosome inactivation: unknown unknowns. Development. 2011;138(23):5057–65. doi: 10.1242/dev.065276. doi:10.1242/dev.065276. [DOI] [PubMed] [Google Scholar]
- 39.Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol. 2011;12(12):815–26. doi: 10.1038/nrm3231. doi:10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- 40.Bartolomei MS, Ferguson-Smith AC. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol. 2011;3:7. doi: 10.1101/cshperspect.a002592. doi:10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. doi:10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16(2):324–37. doi: 10.1261/rna.1441510. doi:10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner LA, Christensen CJ, Dunn DM, Spangrude GJ, Georgelas A, Kelley L, et al. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood. 2007;109(12):5191–8. doi: 10.1182/blood-2006-06-027987. doi:10.1182/blood-2006-06-027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, et al. A myelopoiesis-associated regulatory intergenic non-coding RNA transcript within the human HOXA cluster. Blood. 2009;113(11):2526–34. doi: 10.1182/blood-2008-06-162164. doi:10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011;25(24):2573–8. doi: 10.1101/gad.178780.111. doi:10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5(8):e1000617. doi: 10.1371/journal.pgen.1000617. doi:10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31(3):522–33. doi: 10.1038/emboj.2011.459. doi:10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. doi:10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12(8):1020–7. doi: 10.1038/nn.2371. doi:10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15(6):501–12. doi: 10.1016/j.cub.2005.02.027. doi:10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 51.Rapicavoli NA, Poth EM, Blackshaw S. The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol. 2010;10:49. doi: 10.1186/1471-213X-10-49. doi:10.1186/1471-213X-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez-Pajares V, et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev. 2012 doi: 10.1101/gad.182121.111. doi:10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watson CJ, Khaled WT. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008;135(6):995–1003. doi: 10.1242/dev.005439. doi:10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- 54.Anderson SM, Rudolph MC, McManaman JL, Neville MC. Key stages in mammary gland development. Secretory activation in the mammary gland: it's not just about milk protein synthesis! Breast Cancer Res. 2007;9(1):204. doi: 10.1186/bcr1653. doi:10.1186/bcr1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brisken C, Rajaram RD. Alveolar and lactogenic differentiation. J Mammary Gland Biol Neoplasia. 2006;11(3–4):239–48. doi: 10.1007/s10911-006-9026-0. doi:10.1007/s10911-006-9026-0. [DOI] [PubMed] [Google Scholar]
- 56.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23(22):2563–77. doi: 10.1101/gad.1849509. doi:10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oakes SR, Hilton HN, Ormandy CJ. The alveolar switch: coordinating the proliferative cues and cell fate decisions that drive the formation of lobuloalveoli from ductal epithelium. Breast Cancer Res. 2006;8(2):207. doi: 10.1186/bcr1411. doi:10.1186/bcr1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6(9):715–25. doi: 10.1038/nrm1714. doi:10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- 59.Siegel PM, Muller WJ. Transcription factor regulatory networks in mammary epithelial development and tumorigenesis. Oncogene. 2010;29(19):2753–9. doi: 10.1038/onc.2010.43. doi:10.1038/onc.2010.43. [DOI] [PubMed] [Google Scholar]
- 60.Adriaenssens E, Lottin S, Duqimont T, Fauquette W, Coll J, Dupouy JP, Boilly B, Curgy JJ. Steroid hormones modulate H19 gene expression in both mammary gland and uterus. Oncogene. 1999;18(31):4460–73. doi: 10.1038/sj.onc.1202819. [DOI] [PubMed] [Google Scholar]
- 61.Lanz RB, Chua SS, Barron N, Soder BM, DeMayo F, O'Malley BW. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol Cell Biol. 2003;23(20):7163–76. doi: 10.1128/MCB.23.20.7163-7176.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, et al. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA. 2011;17(5):878–91. doi: 10.1261/rna.2528811. doi:10.1261/rna.2528811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, et al. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci U S A. 2006;103(15):5781–6. doi: 10.1073/pnas.0600745103. doi:10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ginger MR, Gonzalez-Rimbau MF, Gay JP, Rosen JM. Persistent changes in gene expression induced by estrogen and progesterone in the rat mammary gland. Mol Endocrinol. 2001;15(11):1993–2009. doi: 10.1210/mend.15.11.0724. [DOI] [PubMed] [Google Scholar]
- 65.Guan LS, Rauchman M, Wang ZY. Induction of Rb-associated protein (RbAp46) by Wilms' tumor suppressor WT1 mediates growth inhibition. J Biol Chem. 1998;273(42):27047–50. doi: 10.1074/jbc.273.42.27047. [DOI] [PubMed] [Google Scholar]
- 66.Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331(6015):304–8. doi: 10.1126/science.1199082. doi:10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson AE, Karandikar UC, Pepple KL, Chen Z, Bergmann A, Mardon G. The enhancer of trithorax and polycomb gene Caf1/p55 is essential for cell survival and patterning in Drosophila development. Development. 2011;138(10):1957–66. doi: 10.1242/dev.058461. doi:10.1242/dev.058461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–905. doi: 10.1101/gad.1035902. doi:10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vorbach C, Capecchi MR, Penninger JM. Evolution of the mammary gland from the innate immune system? Bioessays. 2006;28(6):606–16. doi: 10.1002/bies.20423. doi:10.1002/bies.20423. [DOI] [PubMed] [Google Scholar]
- 70.Rijnkels M, Kabotyanski E, Montazer-Torbati MB, Hue Beauvais C, Vassetzky Y, Rosen JM, et al. The epigenetic landscape of mammary gland development and functional differentiation. J Mammary Gland Biol Neoplasia. 2010;15(1):85–100. doi: 10.1007/s10911-010-9170-4. doi:10.1007/s10911-010-9170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lemay DG, Lynn DJ, Martin WF, Neville MC, Casey TM, Rincon G, et al. The bovine lactation genome: insights into the evolution of mammalian milk. Genome Biol. 2009;10(4):R43. doi: 10.1186/gb-2009-10-4-r43. doi:10.1186/gb-2009-10-4-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeda K, Ichijo H, Fujii M, Mochida Y, Saitoh M, Nishitoh H, et al. Identification of a novel bone morphogenetic protein-responsive gene that may function as a noncoding RNA. J Biol Chem. 1998;273(27):17079–85. doi: 10.1074/jbc.273.27.17079. [DOI] [PubMed] [Google Scholar]
- 73.Yochum GS, Cleland R, McWeeney S, Goodman RH. An antisense transcript induced by Wnt/beta-catenin signaling decreases E2F4. J Biol Chem. 2007;282(2):871–8. doi: 10.1074/jbc.M609391200. doi:10.1074/jbc.M609391200. [DOI] [PubMed] [Google Scholar]
- 74.Kohtz JD, Fishell G. Developmental regulation of EVF-1, a novel non-coding RNA transcribed upstream of the mouse Dlx6 gene. Gene Expr Patterns. 2004;4(4):407–12. doi: 10.1016/j.modgep.2004.01.007. doi:10.1016/j.modgep.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 75.Tsutsumi M, Itoh M. Novel transcript nort is a downstream target gene of the Notch signaling pathway in zebrafish. Gene Expr Patterns. 2007;7(3):227–32. doi: 10.1016/j.modgep.2006.10.002. doi:10.1016/j.modgep.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 76.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6(10):e25915. doi: 10.1371/journal.pone.0025915. doi:10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maruyama R, Shipitsin M, Choudhury S, Wu Z, Protopopov A, Yao J, et al. Altered antisense-to-sense transcript ratios in breast cancer. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1010559107. doi:10.1073/pnas.1010559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perez DS, Hoage TR, Pritchett JR, Ducharme-Smith AL, Halling ML, Ganapathiraju SC, et al. Long, abundantly expressed non-coding transcripts are altered in cancer. Hum Mol Genet. 2008;17(5):642–55. doi: 10.1093/hmg/ddm336. doi:10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- 79.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–74. doi: 10.1073/pnas.191367098. doi:10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100(14):8418–23. doi: 10.1073/pnas.0932692100. doi:10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8(10):R214. doi: 10.1186/gb-2007-8-10-r214. doi:10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135(5):919–32. doi: 10.1016/j.cell.2008.10.012. doi:10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147(4):773–88. doi: 10.1016/j.cell.2011.08.054. doi:10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Adriaenssens E, Dumont L, Lottin S, Bolle D, Lepretre A, Delobelle A, et al. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am J Pathol. 1998;153(5):1597–607. doi: 10.1016/S0002-9440(10)65748-3. doi:10.1016/S0002-9440(10)65748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32(6):473–80. doi: 10.1002/bies.200900170. doi:10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- 86.Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, et al. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem. 2005;280(33):29625–36. doi: 10.1074/jbc.M504033200. doi:10.1074/jbc.M504033200. [DOI] [PubMed] [Google Scholar]
- 87.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803(4):443–51. doi: 10.1016/j.bbamcr.2010.01.010. doi:10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 88.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67(8):3963–9. doi: 10.1158/0008-5472.CAN-06-2004. doi:10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 89.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38(5):662–74. doi: 10.1016/j.molcel.2010.03.021. doi:10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pasmant E, Sabbagh A, Vidaud M, Bieche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J. 2011;25(2):444–8. doi: 10.1096/fj.10-172452. doi:10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 91.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454(7200):126–30. doi: 10.1038/nature06992. doi:10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Silva JM, Boczek NJ, Berres MW, Ma X, Smith DI. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011;8(3):496–505. doi: 10.4161/rna.8.3.14800. [DOI] [PubMed] [Google Scholar]
- 93.Chen W, Bocker W, Brosius J, Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol. 1997;183(3):345–51. doi: 10.1002/(SICI)1096-9896(199711)183:3<345::AID-PATH930>3.0.CO;2-8. doi:10.1002/(SICI)1096-9896(199711)183:3<345∷AIDPATH930>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 94.Iacoangeli A, Lin Y, Morley EJ, Muslimov IA, Bianchi R, Reilly J, et al. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 2004;25(11):2125–33. doi: 10.1093/carcin/bgh228. doi:10.1093/carcin/bgh228. [DOI] [PubMed] [Google Scholar]
- 95.Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10(9):1098–105. doi: 10.1038/ncb1770. doi:10.1038/ncb1770. [DOI] [PubMed] [Google Scholar]
- 96.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460(7254):529–33. doi: 10.1038/nature08199. doi:10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 97.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939. doi:10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13(3):317–23. doi: 10.1038/ncb2173. doi:10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208(5):875–83. doi: 10.1084/jem.20110235. doi:10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–19. doi: 10.1016/j.cell.2010.06.040. doi:10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem. 2007;282(34):24731–42. doi: 10.1074/jbc.M702029200. doi:10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 102.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30(47):4750–6. doi: 10.1038/onc.2011.193. doi:10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70(6):2350–8. doi: 10.1158/0008-5472.CAN-09-3885. doi:10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, et al. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88(11):5119–26. doi: 10.1210/jc.2003-030222. [DOI] [PubMed] [Google Scholar]
- 105.Benetatos L, Hatzimichael E, Dasoula A, Dranitsaris G, Tsiara S, Syrrou M, et al. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res. 2010;34(2):148–53. doi: 10.1016/j.leukres.2009.06.019. doi:10.1016/j.leukres.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 106.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. doi:10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–34. doi: 10.1016/j.cell.2011.03.042. doi:10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15(8):842–8. doi: 10.1038/nsmb.1444. doi:10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Younger ST, Corey DR. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011;39(13):5682–91. doi: 10.1093/nar/gkr155. doi:10.1093/nar/gkr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Leygue E, Dotzlaw H, Watson PH, Murphy LC. Expression of the steroid receptor RNA activator in human breast tumors. Cancer Res. 1999;59(17):4190–3. [PubMed] [Google Scholar]
- 111.Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 2001;20(6):1341–52. doi: 10.1093/emboj/20.6.1341. doi:10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 112.Hube F, Guo J, Chooniedass-Kothari S, Cooper C, Hamedani MK, Dibrov AA, et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25(7):418–28. doi: 10.1089/dna.2006.25.418. doi:10.1089/dna.2006.25.418. [DOI] [PubMed] [Google Scholar]
- 113.Cooper C, Guo J, Yan Y, Chooniedass-Kothari S, Hube F, Hamedani MK, et al. Increasing the relative expression of endogenous non-coding Steroid Receptor RNA Activator (SRA) in human breast cancer cells using modified oligonucleotides. Nucleic Acids Res. 2009;37(13):4518–31. doi: 10.1093/nar/gkp441. doi:10.1093/nar/gkp441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Foulds CE, Tsimelzon A, Long W, Le A, Tsai SY, Tsai MJ, et al. Research resource: expression profiling reveals unexpected targets and functions of the human steroid receptor RNA activator (SRA) gene. Mol Endocrinol. 2010;24(5):1090–105. doi: 10.1210/me.2009-0427. doi:10.1210/me.2009-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28(2):195–208. doi: 10.1038/onc.2008.373. doi:10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 116.Gee HE, Buffa FM, Camps C, Ramachandran A, Leek R, Taylor M, et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 2011;104(7):1168–77. doi: 10.1038/sj.bjc.6606076. doi:10.1038/sj.bjc.6606076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–93. doi: 10.1126/science.1192002. doi:10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–78. doi: 10.1016/j.molcel.2011.08.027. doi:10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38(16):5366–83. doi: 10.1093/nar/gkq285. doi:10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–8. doi: 10.1038/nature09144. doi:10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell. 2011;147(2):382–95. doi: 10.1016/j.cell.2011.09.032. doi:10.1016/j.cell.2011.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147(2):344–57. doi: 10.1016/j.cell.2011.09.029. doi:10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30(21):4414–22. doi: 10.1038/emboj.2011.359. doi:10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–29. doi: 10.1016/j.ccr.2007.07.027. doi:10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 125.Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, et al. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis. 2011;32(11):1655–9. doi: 10.1093/carcin/bgr187. doi:10.1093/carcin/bgr187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yang R, Frank B, Hemminki K, Bartram CR, Wappenschmidt B, Sutter C, et al. SNPs in ultraconserved elements and familial breast cancer risk. Carcinogenesis. 2008;29(2):351–5. doi: 10.1093/carcin/bgm290. doi:10.1093/carcin/bgm290. [DOI] [PubMed] [Google Scholar]
- 127.Day JR, Jost M, Reynolds MA, Groskopf J, Rittenhouse H. PCA3: from basic molecular science to the clinical lab. Cancer Lett. 2011;301(1):1–6. doi: 10.1016/j.canlet.2010.10.019. doi:10.1016/j.canlet.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 128.Lee GL, Dobi A, Srivastava S. Prostate cancer: diagnostic performance of the PCA3 urine test. Nat Rev Urol. 2011;8(3):123–4. doi: 10.1038/nrurol.2011.10. doi:10.1038/nrurol.2011.10. [DOI] [PubMed] [Google Scholar]
- 129.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA, et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res. 1999;59(23):5975–9. [PubMed] [Google Scholar]
- 130.Moskalev EA, Schubert M, Hoheisel JD. RNA-directed epigenomic reprogramming: an emerging principle of a more targeted cancer therapy? Genes Chromosomes Cancer. 2012;51(2):105–10. doi: 10.1002/gcc.20943. doi:10.1002/gcc.20943. [DOI] [PubMed] [Google Scholar]
- 131.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7. doi: 10.1158/0008-5472.CAN-10-2483. doi:10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–5. doi: 10.1038/ng.518. doi:10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–9. doi: 10.1126/science.1198056. doi:10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199–203. doi: 10.1126/science.1200609. doi:10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rotblat B, Leprivier G, Sorensen PH. A possible role for long non-coding RNA in modulating signaling pathways. Med Hypotheses. 2011;77(6):962–5. doi: 10.1016/j.mehy.2011.08.020. doi:10.1016/j.mehy.2011.08.020. [DOI] [PubMed] [Google Scholar]