Abstract
Protein interactions are important for understanding many molecular mechanisms underlying cellular processes. So far, interfaces between interacting proteins have been characterized by NMR spectroscopy mostly by using chemical shift perturbations and cross-saturation via intermolecular cross-relaxation. Although powerful, these techniques cannot provide unambiguous estimates of intermolecular distances between interacting proteins. Here, we present an alternative approach, called REDSPRINT (REDduced/Standard PRoton density INTerface identification), to map protein interfaces with greater accuracy by using multiple NMR probes. Our approach is based on monitoring the cross-relaxation from a source protein (or from an arbitrary ligand that need not be a protein) with high proton density to a target protein (or other biomolecule) with low proton density using isotope-filtered nuclear Overhauser spectroscopy (NOESY). This methodology uses different isotropic labeling for the source and target proteins to identify the source-target interface and also determine the proton density of the source protein at the interface for protein-protein or protein-ligand docking. The utility of this technique, including a method for direct determination of the protein surface, is demonstrated for two different protein-protein complexes.
Many biological processes rely on cascades of protein interactions.(Uetz, Giot et al. 2000; Rain, Selig et al. 2001) The structural characterization of protein-protein interfaces is a precondition for understanding biological processes at an atomic level. Indeed, nuclear magnetic resonance (NMR) spectroscopy has been very successfully used to study protein-protein interfaces. So far, three approaches have been developed to study the association of biological macromolecules, (i) the complete structure determination of protein-protein complexes by using intermolecular distance restraints, (ii) the identification of the interfaces on each molecule and (iii) the characterization of the relative orientation (docking) of two binding partners. The first method relies on asymmetric isotopic labeling of the two partners. The combination of isotope filters (Otting and Wuthrich 1990; Ikura and Bax 1992; Breeze 2000) with nuclear Overhauser effect spectroscopy (Kumar, Ernst et al. 1980; Neuhaus and Williamson 2000) (NOESY) allows one to focus on intermolecular distances. However, despite recent progress in studying large complexes,(Williams, Cai et al. 2005; Xu, Zheng et al. 2006) this approach often suffers from unfavorable relaxation properties and ambiguities in the assignment from degenerate proton chemical shifts. Alternatively, the chemical shift perturbations observed upon complex formation between the source and target molecules can be used to identify the interface. Although this approach is easy to implement, provides valuable information, is suitable for systems with high molecular weight (Fiaux, Bertelsen et al. 2002), and even for in-cell experiments, (Burz, Dutta et al. 2006; Burz, Dutta et al. 2006) it may still suffer from ambiguities, since chemical shift perturbations are difficult to interpret in terms of structural effects.(Foster, Wuttke et al. 1998) Therefore any conclusion regarding the complex structures from chemical shift perturbation studies is restricted to semi-quantitative approaches.(Dominguez, Boelens et al. 2003; van Dijk, Boelens et al. 2005) Recently, a cross-saturation method was developed,(Takahashi, Nakanishi et al. 2000; Shimada 2005) which enables one to unambiguously identify amide (Takahashi, Nakanishi et al. 2000) or methyl (Takahashi, Miyazawa et al. 2006) protons located near the interface of the target protein. However, this rather sparse information does not allow one to identify the interface with high spatial resolution. A third approach may combine several methods including the measurement of residual dipolar couplings in weakly oriented samples, (Clore 2000) the anisotropy of diffusion tensors, (Fushman, Xu et al. 1999; Fushman and Cowburn 2003; Fushman, Varadan et al. 2004; Ryabov and Fushman 2007) and pseudo-contact shifts in paramagnetic molecules (Guiles, Sarma et al. 1996; Gaponenko, Altieri et al. 2002; Pintacuda, Park et al. 2006) Such data can provide constraints pertaining to the relative orientation of the binding partners in a complex, and can be combined with intermolecular NOE’s (Clore 2000) or chemical shift perturbation data (van Dijk, Boelens et al. 2005) to implement protein-protein docking.
Here, we introduce an NMR protocol for identifying biomolecular interfaces based on the study of a REDuced/Standard PRoton density INTerface (REDSPRINT). In this approach, the source protein (or other ligand) is prepared without any isotopic enrichment, thus having a high proton density (HIPRO), whereas the target protein is isotopically labeled (13C and/or 15N and ~85% 2H) so as to have a reduced proton density (REDPRO).(Shekhtman, Ghose et al. 2002) Cross-relaxation from the high to the low proton-density molecule is monitored by using a modified version of isotope-filtered nuclear Overhauser spectroscopy (NOESY) (Breeze 2000) which allows one to identify the interface.
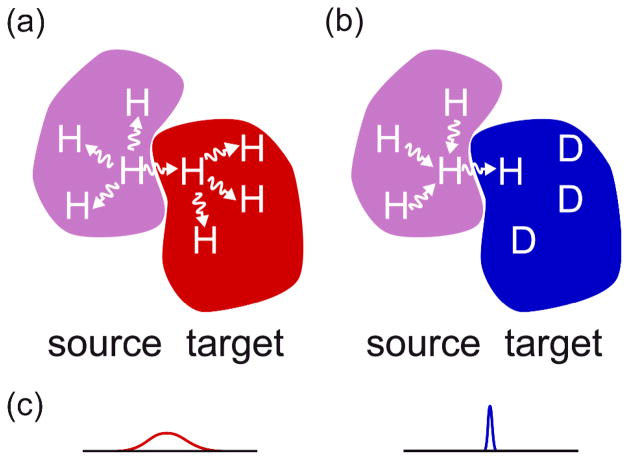
The methods that rely on intermolecular dipolar interactions,(Breeze 2000; Hamel and Dahlquist 2005; Kiihne, Creemers et al. 2005; Shimada 2005; Sui, Xu et al. 2005) and specially those employing intermolecular NOE’s are prone to spin diffusion. Spin diffusion is a potential drawback since it limits the accuracy of identifying both the source and the target protons. However, in the REDSPRINT protocol, and in other similar approaches (Eichmuller, Tollinger et al. 2001; Zangger, Oberer et al. 2003), spin diffusion within the source protein turns out to be beneficial whereas spin diffusion in the target protein is decreased because of its reduced proton density.(Gross, Gelev et al. 2003; Shimada 2005) One advantage of this methodology is that no chemical shift assignment of the source protein/ligand is required and one can probe various NMR probes (amide, aliphatic and aromatic protons) using a single sample. As shown in Fig. 1b, source protons that are not located at the interface constitute a polarization reservoir that is connected to the interface via spin-diffusion.
Figure 1.
Schematic figure illustrating the REDSPRINT methodology. The target protein is shown in red at the right (a) when the proton density is high (HIPRO) and in blue (b) when the proton density is low (REDPRO). The source protein (or ligand) with high proton density is shown in magenta. Two experimental approaches are compared: (a) in a traditional filtered NOESY experiment where both the target and the source have a high proton density: cross-relaxation is efficient but spin-diffusion within the target and the source affects the accuracy and the sensitivity of the experiment; (b) when the target has low proton density and the source has a high proton density, proton-dilution reduces the sensitivity of the experiment but spin-diffusion ensures that the source protein (or ligand) acts as a large polarization reservoir (provided chemical shift labeling is avoided), while spin diffusion in the target is reduced. (c) In addition, the traditional labeling scheme (a) makes observation difficult because of fast transverse relaxation resulting in broad signals, while in the REDPRO target (b) the signals are narrow.
This methodology is demonstrated on a protein-ligand complex involving human ubiquitin as target protein and the ubiquitin-interacting motif of ataxin 3 (AUIM) as the source protein. This technique was also applied to map the interface between the Src homology 3 domain (SH3) (target) of C-terminal Src Kinase (CSK) and the 25 residue peptide from the proline-enriched tyrosine phosphatase (PEP) (source), the putative interaction motif perturbed in the autoimmune disease-related single nucleotide polymorphism in PTPN22.(Bottini, Musumeci et al. 2004) The Csk SH3-PEP system was studied both in 2H2O and in a viscous mixture of 2H2O and [2H8] glycerol to mimic the behavior of complexes with high molecular mass.
RESULTS AND DISCUSSION
Polarization transfer
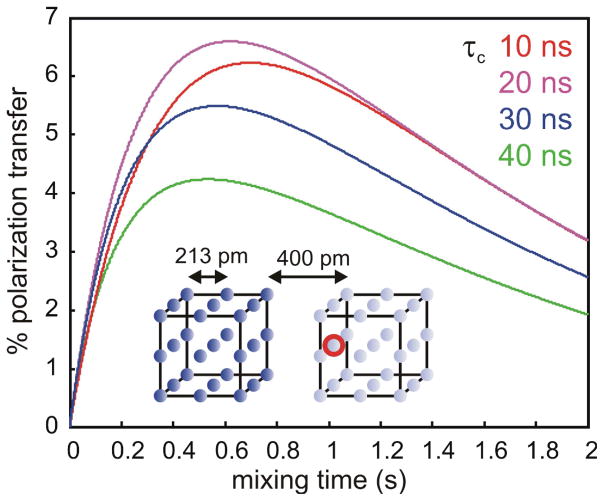
We have simulated the efficiency of polarization transfer during a REDSPRINT experiment on a simple system (Fig. 2). We considered the case where (i) the proton density of the source protein (or other ligand) is high (HIPRO, left-side cube) and (ii) the proton density of the target protein is low (REDPRO, right-side cube). In the initial state, right after the isotope filter (see Figure S1 and S2),(Zwahlen, Legault et al. 1997) the polarization in the right-hand REDPRO cube vanishes, while the equilibrium polarization of the left-hand HIPRO cube is attenuated by transverse relaxation during the isotope filter. The cross-peak amplitudes arising from the polarization transfer to the observed proton at the interface in the REDPRO protein is shown in Figure 2. The efficiency of the polarization transfer shows little dependence on the global correlation time (τc) (i.e. on the size of the system) in the 10–40 ns range, corresponding to about 20 to 80 kDa. This is due to the combination of two effects: with increasing size, (i) transverse proton relaxation in the HIPRO protein during the isotope filter decreases the amount of polarization available before the mixing time and (ii) the transfer itself is more efficient (simulations have shown that the maximum transfer efficiency is twice as large for τc = 40 ns than for τc = 10 ns). For large complexes (MW ≥ 80 kDa), polarization transfer within the target protein limits the ability to detect the protons at the interface of the target protein except for systems, which are in fast exchange between free and bound states.
Figure 2.
Predicted efficiency of the polarization transfer. The dark blue spheres indicate the high probability (P = 1.0) of finding protons on the HIPRO source protein or ligand (left cube) whereas the the light blue spheres represent the low probability (P = 0.1) of finding protons in the REDPRO target protein (right cube). The red circle represents the observed proton near the interface in the target protein. The cross-peak amplitudes were calculated using Equations 1 (see text and Suppl, Material) and plotted for a global rotational correlation time τc = 10 ns (red); 20 ns (magenta); 30 ns (blue); and 40 ns (green).
Fast mapping of the interface
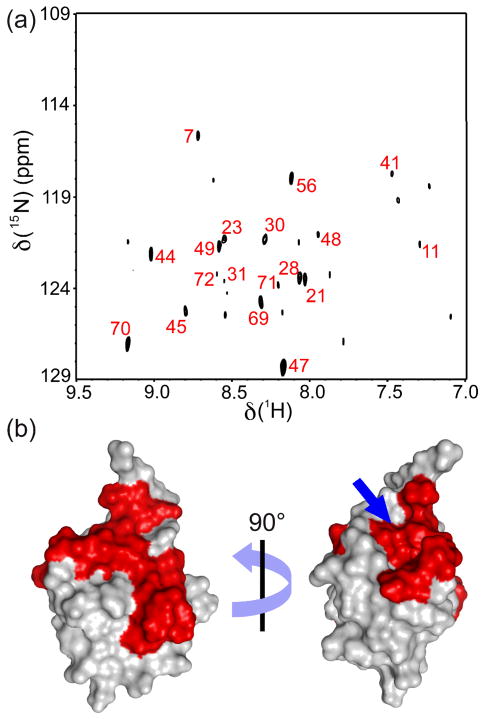
This REDSPRINT protocol utilizes a fast and straightforward mapping of the interface based on isotope-filtered NOESY is to compute the difference between the spectra acquired with and without a mixing time as shown by Zangger et al. (Zangger, Oberer et al. 2003) Using this approach we suppress the signal from residual polarization at the end of the filter with sufficient accuracy, as shown in Figures 3a and 4a. For each system of interest, a threshold is defined to distinguish the actual transfer of polarization from noise and artifacts. The assignment of protons receiving polarization from the HIPRO source protein (or ligand) permits identification of the interface on the target protein. The residues above the threshold are mapped on to the surface plot of Ubiquitin (Figure. 3b). The interface on ubiquitin is a continuous surface (shown in red) except a few residues on the opposite side (Asp21, Ile23, Ala28, Ile30 and Gln31). Indicated by a blue arrow in Figure 3b, is an unusual extension of the interface around the methyl groups of Ile36 and Leu71.
Figure 3.
(a) REDSPRINT spectrum of ubiquitin with reduced proton density (REDPRO) bound to the HIPRO source peptide AUIM. The peaks labeled with their residue numbers have an intensity of at least one-third of the peak height of Gly47, which is the most intense. (b) These residues are mapped on the surface of ubiquitin (PDB code 1D3Z). The blue arrow points toward an atypical extension of the interface. The filtered-NOESY spectra at mixing time of 1 ms, 300ms and the difference spectra are shown in Figure S3 a–f.
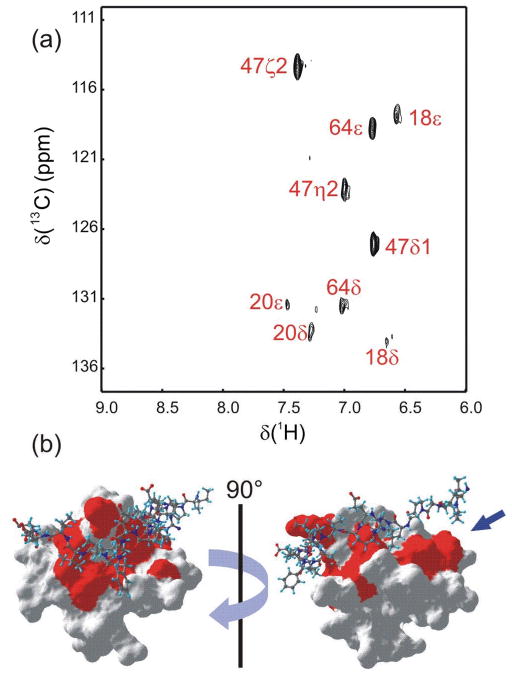
Figure 4.
(a) REDSPRINT spectrum recorded with 13C decoupling, showing the aromatic region of REDPRO Csk SH3 (target protein) bound to HIPRO PEP (source peptide). All peaks appearing in the spectrum are labeled. The residues that show cross-relaxation from PEP to Csk SH3 are mapped on the surface of the Csk SH3-PEP complex (PDB code 1JEG) (b). The arrow indicates a part of the interface that was not identified in earlier studies (see text).(Ghose, Shekhtman et al. 2001) The filtered-NOESY spectra at mixing time of 1 ms, 300ms and the difference spectra are shown in Figure S3 g–l.
Similarly, figure 4b show the residues of the REDPRO Csk SH3 domain (in red) that are involved in the interaction with the HIPRO PEP source protein. A large “patch” (blue arrow) is found facing the Pro9-Pro10-Pro11 segment of PEP. The signals from this region of the source protein are difficult to assign using conventional 3D NMR experiments. Therefore, no distance restraints were used for these residues in the earlier structure calculation.(Ghose, Shekhtman et al. 2001) This result demonstrates that the REDSPRINT strategy is valuable to identify an interface of a ligand, domain or segment whose signals that cannot be assigned. These additional restraints can be used for structure refinement of Csk SH3-PEP complex to get a more accurate structure.
In our study, the fast mapping of the interface was successful for the ubiquitin-AUIM complex (overall correlation time ca. 10 ns) and the Csk SH3-PEP complex in 2H2O (overall correlation time 12.9 ns). Therefore, REDSPRINT provides with very accurate and complete information about the binding interface compared to the chemical shift mapping. However, the fast mapping approach for the slow-tumbling Csk SH3-PEP complex (overall correlation time 20 ns) was less successful. The signals from the aromatic side-chain were not unambiguously detected due to the combination of rather intense residual peaks remaining after the filter and fast transverse relaxation of the aromatic protons located near the interface. On the other hand all the residues with at least one methyl group that are located near the interface, e.g. Thr23, Ala24, Ala40, Val41, Thr42 and Ile59 (see Supporting Information, Fig. S11) were identified. The residues near the surface of the Csk SH3 domain that are facing the polyproline segment of the PEP ligand have no methyl groups. Therefore, calculating the normalized polarization transfer ratios (see below) will help in identifying this missing interface. Nevertheless, the results obtained from the methyl groups using the fast mapping approach provides accurate, if limited, information about the localization of the interface on the target protein of large complex.
Normalized transfer ratios
The suppression of residual magnetization after the filter in the fast mapping procedure is not 100 % efficient. The detection of protons near the interface may be complex because of fast transverse relaxation due to chemical exchange and/or intermolecular dipole-dipole interactions in large complexes. In such cases, the discrimination between the polarization transferred to a proton near the interface from residual polarization on other protons may be limited. A more elaborate analysis of the data may be carried out to extract quantitative information from the spectra. The normalized polarization transfer ratios for the amide protons in the ubiquitin-AUIM complex and the side-chain protons in the Csk SH3-PEP complex are computed using Eq. 8 (see Supporting Information Fig. S4 and Tables S1 and S2). According to Eq. 8, the normalized polarization transfer nearly vanishes for protons of the target protein that are far away from the interface, even in the presence of residual polarization after the isotope filter. For the protons near the interface, the normalized polarization transfer ratio is dominated by polarization transferred from the HIPRO source protein. The variations of the inherent sensitivity of each signal as well as auto-relaxation during the mixing time are also taken into account by the normalization procedure.
The signals of 15N-bound protons in the ubiquitin-AUIM complex are sufficient to define the interface. In the Csk SH3-PEP complex, the aromatic side-chains permit us to identify the residues, which are located at the interface (see Supporting Information Fig. S4). Note that the side-chain of Trp47 shows very high polarization transfer, whereas the side-chain of the next residue, Tyr48, which points towards the core of the protein, receives no detectable polarization from PEP (Supporting Information, Fig. S5). Similarly, this approach when applied to the Csk SH3-PEP complex in [2H8] glycerol, mimicking a high molecular weight complex, one was able to identify several protons located near the interface (see Supporting Information Table S2), including the methyl protons identified by the fast mapping approach.
Nuclei Envelope Belonging to UnLabeled Additive (NEBULA) calculations
The large number of probes near the surface of the REDPRO target protein provides a sufficiently detailed picture of the proton density of the HIPRO source protein to determine the docking interface. The normalized polarization transfer ratio was used to evaluate the sum of the intermolecular dipolar cross-relaxation rates from the HIPRO source to an observed proton on the REDPRO target. Polarization transfer calculations using the same model system as presented in Fig. 2 were used to validate the use of Eq. 1.
Several factors were neglected in the semi-quantitative analysis of our data collected using the modified isotope-filtered NOESY experiment. The site-to-site variation of the transverse relaxation rates of the HIPRO source protein (e.g AUIM or PEP ligand) and the variations of the initial polarizations were not taken into account. In large molecules, these site-to-site variations tend to average out through intramolecular spin-diffusion during the mixing time. However, this approximation is less accurate in small-to-middle size systems such as the ubiquitin-AUIM complex. Nevertheless, one should note that an error of ~50% in the transferred polarization results in ~12 % error in the distance.
The uncertainty in the origin of the polarization from the source protein that is transferred to the target protein makes it difficult to carry out a site-by-site evaluation of the population probability. The polarization transferred to a proton of the source protein is a property of the configuration of the protons of the target within a 5 Å sphere around the source proton. Such a configuration can be generated in a docking protocol. We have chosen a Monte Carlo-based approach, that requires no prior knowledge about the target protein. This procedure provides an estimate of the population probabilities with no assumption about the structure of the target protein or the relative positions of the source and target proteins. The results were shown to be robust with respect to a limited number of inaccurate constraints. A steric exclusion criterion eliminates the constraints obtained from the protons that are deeply buried in the core of the target protein. Anti-constraints are also important because if a “parasitic” constraint (an outlier) is isolated then the sum of neighboring anti-constraints will lead to a low estimate of the proton density of surrounding sites.
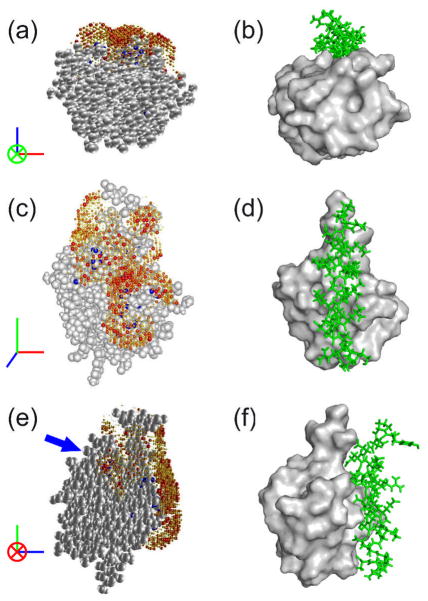
The results obtained after the REDSPRINT analysis and NEBULA calculations for the ubiquitin-AUIM complex are shown in Figure 5(a, c, e), thereof called NEBULA plots. The structure of another ubiquitin-UIM complex (Swanson, Kang et al. 2003) is shown in Fig. 5(b, d, f) for comparison. Examination of Figure 5 shows that the REDSPRINT analysis places the helix of AUIM in the correct groove (Figure 5a and b) and also in the correct orientation (Figure 5c and d), although another ‘transverse’ orientation cannot be ruled out. Apart from the higher probabilities computed for the longitudinal orientation, docking of an α helix in the groove on the surface of the ubiquitin would lead to a much larger contact area than a ‘bridge’ configuration in the ‘transverse’ orientation. The presence of proton density identified by an arrow in Figure 5e is strikingly different from a typical ubiquitin-UIM structure (Swanson, Kang et al. 2003). The fast mapping procedure and the normalized polarization transfer ratio show that the γ2 methyl group of Ile36 of ubiquitin is in contact with AUIM. This additional interaction surface may originate due to (i) the presence of an alternate transverse orientation (ii) an interaction with a part of the AUIM peptide that does not belong to the typical α-helix, (iii) spin-diffusion. Further investigation should discriminate between two possible binding modes, a real extension of the interface or an experimental artifact.
Figure 5.
(a, c, e) NEBULA plots showing the proton density of AUIM bound to ubiquitin represented by gray spheres, while the probability of AUIM proton densities are represented by spheres that are color-coded from white (zero) to red (maximum). The radii of the spheres are scaled with the corresponding proton density. Blue spheres represent the protons of ubiquitin having REDSPRINT constraints. (b, d, f) Representative ubiquitin-UIM complex (PDB code 1Q0W(Swanson, Kang et al. 2003)) with ubiquitin surface shown in gray and the UIM in green.
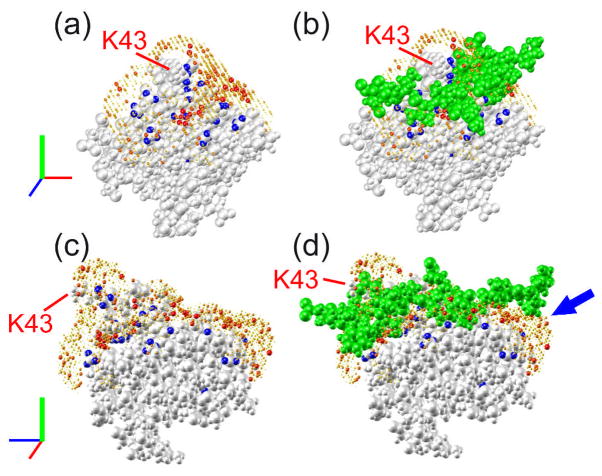
NEBULA calculations for the Csk SH3-PEP complex in a 2H2O/[2H8] glycerol mixture were carried out by using the structure of the Csk SH3 domain.(Ghose, Shekhtman et al. 2001) The NEBULA plots are displayed in Figure 6(a, c), while the NMR structure of the complex is shown in Figure 6(b, d), where the ligand (PEP) is shown in green spheres. It is clear from the figure that REDSPRINT analysis has predicted the interface quite accurately as the overall shape of the binding surface is very similar (Figure 6c and d). Similar to the results obtained from the fast mapping technique of the interface for Csk SH3-PEP complex in 2H2O (see Figure 4b), the binding surface further extends on one side of the peptide (Figure 6d and S10) to include the Pro9-Pro10-Pro11 motif of PEP. This feature is particularly noteworthy since this proline-rich segment was shown to be necessary for the interaction between the Csk SH3 domain and PEP. Even for this slow-tumbling system, the use of normalized polarization transfer ratios enables NEBULA plots to reveal the full extent of the interface. NEBULA calculations are more accurate than a fast mapping procedure, which is based on difference NOESY spectra (Figures 3 and 4). The inspection of the proton density around Lys43 (Figure 6) is noteworthy as the side-chain of Lys43 projects out from the surface into the solvent. The identification of non-vanishing normalized polarization transfer ratio for β and γ protons leads to a sampling of the proton density around this side-chain. The combination of anti- and positive constraints allows NEBULA calculations to unambiguously predict the highest proton density on one side of the side-chain, consistent with the NMR structure of the complex.
Figure 6.
NEBULA plots showing the proton density of the PEP peptide bound to the Csk SH3 domain in a 2H2O/[2H8]glycerol mixture. The Csk SH3 domain is represented by gray spheres while the color-coded spheres represent the probabilities of the presence of protons belonging to the source protein PEP. Blue spheres represent protons of the Csk SH3 domain with REDSPRINT constraints. (b, d) In addition to the NEBULA plot, the PEP ligand is shown as green spheres. The blue arrow indicates the extension of the interface that was not identified in the earlier studies while a red arrow designates the side-chain of Lys43.
NEBULA calculations for the Csk SH3-PEP complex in 2H2O are shown in the Supporting Information (Figure S10) and compared to the results obtained from 2H2O/[2H8] glycerol mixture (Figure 6a and c). The interacting surface obtained after the NEBULA calculations in both case are very similar. Some differences in the local proton densities may come from the inability to detect some polarization transfer functions in either one of the samples. In addition, more efficient intramolecular cross-relaxation within the source (PEP) protein may explain the higher homogeneity of the proton density observed for Csk SH3-PEP complex in 2H2O/[2H8] glycerol mixture. A low-proton density extension of the patch facing the poly-proline segment of PEP is due to an additional constraint obtained on the β protons of residue Asn19 in the slow-tumbling complex. The slight variations observed between the two NEBULA plots show that such an approach is a first step in the interpretation of the experimental polarization transfer. A thorough analysis employing the complete relaxation matrices and explicitly taking into account the fast exchange between free and bound forms using CORCEMA,(Jayalakshmi and Krishna 2002) combined with protein docking using HADDOCK(Dominguez, Boelens et al. 2003) should lead to models of the complex with improved precision and accuracy.
In the NEBULA plot of the rapidly-tumbling Csk SH3-PEP complex (Figure S10 (e and f)), one may notice a high proton density cluster (shown by circle) that lies in the vicinity of the PEP ligand, when NEBULA analysis is done using the first NMR model from the ensemble of 25 structures of the complex. After inspection of all models in the ensemble it is evident that the side-chain of Arg15 from PEP fills this space in 20 out of 25 models.47 Thus, our results support this statistically dominant orientation for the side-chain of Arg15 (see Supporting Information).
Comparison of REDSPRINT and other NMR approaches
The information obtained from REDSPRINT analysis is accurate but of medium-resolution and can be used as a valuable constraint for protein-protein or protein-ligand docking. To decide when REDSPRINT should be used, we compared the information content-to-cost ratio and the range of accessible complex sizes with other NMR techniques. The two filtered-NOESY spectra of the aromatic region recorded with the Csk SH3-PEP complex to obtain the difference spectrum took a total of 8 hours whereas to determine the normalized polarization transfer ratio, two non-filtered experiments were recorded for 4 hours, each with half as many transients. The same spectra of the aliphatic region required a total of 32 hours. The total duration of the experiments was 44 hours without optimization of the spectral windows and resolution. The number of transients recorded was doubled for the Csk SH3-PEP complex in 2H2O/[2H8] glycerol leading to 88 hours of experimental time. As a test for time-optimization, we have processed NMR data with half the resolution of the recorded spectra (i.e. with maximum evolution times t1max = 4.847 ms instead of 9.694 ms for the aliphatic spectra and 4.733 ms instead of 9.467 ms for the aromatic spectra). The signal-to-noise ratio slightly decreased for most peaks and the most notable decrease, of the order of 20% was noticed for methyl groups.
The normalized polarization transfer ratios were computed and compared with those used for the NEBULA calculations displayed in Figure 6(a–d). The error margins of the normalized polarization transfer ratios were comparable. In most cases the variation of the normalized polarization transfer ratio obtained from the low-resolution and the high-resolution data lie within the error margin with the exception of for two peaks (Hβ from Tyr18 and Hγ2 from Thr42) that leads to constraints in the NEBULA calculations. For these two peaks, the difference in the normalized polarization transfer ratios was around ~20 % which leads to a very small error in the evaluation of the distances between the protons of the source and the target proteins. In conclusion, it would have been possible to record aliphatic and aromatic region spectra within 44 hours, possibly less, for a sample with a low concentration (~400 μM) and an overall tumbling time of 20 ns. A similar optimization of the experimental conditions for the Csk SH3-PEP complex in 2H2O (tumbling time 13 ns) would lead to a total duration of 22 hours. The time required to collect data for REDSPRINT analysis is longer than for chemical shift perturbations and saturation transfer experiments. The requirement for complete deuteration (~100%) of the target protein makes saturation transfer studies more expensive and complicated. Under optimized conditions, the experimental time to collect REDSPRINT data is lower or comparable to the time necessary to record a single isotope-filtered NOESY experiment. However, for conventional structure determination of complexes based on isotope-filtered NOESY, the assignment of the source protein is necessary which may require days or even weeks and even require multiple samples with different isotope labeling.
Chemical shift perturbation studies, monitored by 1H{15N} HSQC spectra, is the most commonly used method for identifying the interface of the complexes. The information content obtained by saturation transfer experiments is low, since one can obtain only one constraint per residue, but attempts at protein docking based on saturation transfer experiments have been reported (Matsuda, Ikegami et al. 2004) On the other hand, REDPSRINT methodology is more accurate than chemical shift perturbations. In this method all sites with a residual protons can be probed (i.e. backbone amide, aliphatic and aromatic side-chains), therefore the number of constraints obtained is greater than those obtained from saturation transfer experiments. It seems that to obtain an accurate high-resolution structure of the complex one would require complete assignments of both the source and the target proteins and also generate distance constraints using isotope-filtered NOESY experiments. But sometimes it is difficult to obtain the distance constraints due to lack of assignment of the source protein which may lead to inaccurate interface mapping. In our study of Csk SH3-PEP we have illustrated that REDSPRINT can identify structural features, which were not accessible by conventional methods and thus aid in identifying the accurate binding interface and provide constraints for structural refinement.
As long as NMR signals can be observed, chemical shift perturbation studies will not be affected by the size of complexes (Fiaux, Bertelsen et al. 2002) although it should be noted that partial deuteration is sometime desirable. Since there are limited (Williams, Cai et al. 2005; Xu, Zheng et al. 2006) numbers of high molecular weight complex structures studied so far therefore it is difficult to assess the upper limit (>30 kDa) of the feasibility of structure determination using isotope-filtered NOESY experiments. Saturation transfer experiments can be performed on the larger systems but the transverse relaxation due to intermolecular dipole-dipole interactions may severely affect very large systems. Similarly, REDSPRINT data is also affected by intermolecular contributions to the transverse relaxation of interfacial protons. In this study we have demonstrate that REDSPRINT methodology can be used for identifying the interface of the complexes as large as 40 kDa (tumbling with ~20 ns). Simulations shown in Figure 2 suggest that larger complexes should be accessible, at least for favorable systems where transverse relaxation in the source protein is not too fast and the spectral overlap of the target protein is not severe. Further simulations (see Supporting Information Figure S9) have shown that polarizing transfer efficiency in REDSPRINT methodology is more sensitive than isotope filtered NOESY based experiments even with the high level of deuteration is due to the favorable relaxation properties and detects the sum of all intermolecular transfers after the long mixing times with the exception when a single intermolecular cross-relaxation pathway dominates.
The results obtained from the chemical shift perturbation and REDSPRINT analyses were compared for the methyl groups of the ubiquitin-AUIM complex (Figure S6). In order to account for the residues involved at the interface (e.g. Leu8, Ile44, Val70, Thr7, Leu71, Ile36 and Ile61), a threshold of 0.01 ppm was chosen. On the other hand, the chemical shift perturbations of Ile30 and Ile50 are above the threshold even though they are not exposed at the surface of ubiquitin. Nevertheless, chemical shift perturbation studies provide a rather good characterization of the interface of ubiquitin (in agreement with the REDSPRINT results) but the small chemical shift changes make it possible to pick false positives.
Table S3 (see Supporting Information) lists the intermolecular NOEs obtained between HIPRO sample of ubiquitin (target protein) and AUIM (source protein) by using traditional isotope-filtered NOESY experiment. Both techniques identify the same protons located near the interface, with a few exceptions, assuming that the structure of the ubiquitin-AUIM complex is typical. Many intermolecular correlations are seen in the traditional isotope-filtered NOESY spectrum for the methyl groups of the hydrophobic patch (Leu8, Ile44 and Val70) that are also observed in the REDSRPINT spectrum. The presence of intense signals on the diagonal due to residual polarization after the filter prevents the identification of cross-peaks between protons with similar chemical shifts in the filtered NOESY experiment. The same residual polarization limits the accuracy of REDSPRINT since it necessitates a lower threshold limit in the analysis of the normalized polarization transfer. Hence it is possible that some small polarization transfer cannot be distinguished from an artifact of the filter. Therefore the limitation of REDSPRINT analysis is the efficiency of the “isotope filter” since it dictates the threshold limit to be chosen for the normalized polarization transfer.
Furthermore simulations were carried out to evaluate the necessity of deuteration in the REDSPRINT protocol. As expected, deleterious spin-diffusion within a HIPRO target protein is very efficient, so that deuteration is absolutely necessary to ensure the accuracy of REDSPRINT methodology for most systems. However, for small systems with global correlation time τc < 10 ns, spin-diffusion may be more tolerant. The use of very short mixing times, close to 50 ms, ensures a satisfactory accuracy and an acceptable signal-to-noise. For protein complexes that are amenable to isotope-filtered NOESY experiments, REDSPRINT analysis is useful to identify the interface and can also be combined with distance constraints to obtain more accurate information about the interface as illustrated in our study of Csk SH3-PEP complex. In cases where high-resolution structure of the complex is not required REDSPRINT analysis can identify the interface and help in docking. One can also use the isotope-filtered NOESY data to do NEBULA-based docking of the complex and simplify structure calculation.
CONCLUSIONS
We have presented an alternative method for identifying binding interfaces in protein complexes called REDSPRINT. This method requires a single NMR sample using the REDPRO isotopic labeling scheme. The easy experimental setup should make this approach applicable to large systems where high-resolution structures are not accessible by NMR. Whenever isotope-filtered NOESY methods are efficient, REDSPRINT should be a useful complement for structure refinement. It may also prove useful for in-cell STINT-NMR experiments.(Burz, Dutta et al. 2006) The analysis of REDSPRINT data not only allows one to define the interface, but also to map the proton density of one of the binding partners in the complexes. This information can be employed to assist the docking of two molecules. Applications to two different complexes show that the binding surfaces and NEBULA plots are well defined and could be used to complement high-resolution data, even when the assignment of the proton resonances of the binding partners is incomplete.
MATERIAL AND METHODS
Theory
Relaxation
NMR of biological macromolecules is often limited by fast transverse relaxation. Proton-detected experiments in large proteins are dramatically affected by rapid transverse relaxation due to strong proton-proton dipolar interactions, which results in the loss of signal-to-noise. This effect can be significantly reduced by deuteration, (Gardner and Kay 1998) although the sensitivity is also reduced in proportion to the concentration of the remaining protons. In protein-protein complexes, an asymmetric labeling scheme such as the one presented in Fig. 1b can be used in various strategies. (Fiaux, Bertelsen et al. 2002; Gross, Gelev et al. 2003; Shimada 2005) The benefits of partial deuteration can be lost in part because of intermolecular dipolar interactions that enhance transverse relaxation of protons located near the interface. Nevertheless, as demonstrated by studies on large complexes,(Shimada and Kawazoe 1984; Fiaux, Bertelsen et al. 2002; Gross, Gelev et al. 2003) this effect does not prevent one from investigating the interface.
In large biological macromolecules, dipolar cross-relaxation is very efficient thus making NOESY experiments (Neuhaus and Williamson 2000) particularly attractive for large systems with low proton density.(Horst, Fenton et al. 2007) In NOESY experiments, the initial chemical shift labeling of the proton polarization leads to shift-dependent polarization at the beginning of the mixing time. The effective longitudinal proton relaxation rates associated with the diagonal peaks correspond to selective relaxation rates, where only the protons of interest are inverted, so that the memory of the spin system is very short. The rapid transfer of the polarization towards neighboring protons determines the selective relaxation (Figure 1a). It is therefore necessary to keep the mixing time short to prevent extensive spin-diffusion. Under these conditions, a large number of accurate structural constraints can be obtained. However in protein complexes, fast intramolecular cross-relaxation within the source protein is an issue, which is not observed in filtered NOESY experiments. The ability to detect weak and long-range intermolecular dipolar cross-relaxation is limited by the short memory of the system. Therefore, to access longer mixing times and achieve sufficient accuracy, it is necessary to reduce the effects of intramolecular cross-relaxation.
In a modified filtered NOESY pulse sequence, (Zangger, Oberer et al. 2003) the initial polarization is not labeled by any chemical shift evolution before the mixing time. The information about the origin of the polarization is lost but the polarization of a given proton decays slowly because it is not affected by intramolecular cross-relaxation. It can be shown (see below) that for large protein complexes, the polarization transfer is more efficient.
As illustrated in Figure 1, the reduced proton density of the target protein greatly alleviates intramolecular spin-diffusion and leads to narrow signals. Simulations have shown that (Supporting Information Figures S7–S9) the accurate identification of the interface in a fully protonated system is difficult for mixing times τm ≥ 100 ms, and for systems whose global correlation times τc > 10 ns (Supporting Information Figures S1 and S2). Thus reducing the proton density results in a more efficient and accurate identification of the interface over a wider range of molecular masses.
An extensive network of cross-relaxing nuclei determines the longitudinal relaxation of protons in a macromolecule. Additional complexities may arise when one considers a REDPRO labeled protein. If a protein is deuterated to a level of 90%, then the probability of finding a proton in any particular site is only 10%. The multi-exponential relaxation behavior reflects an average over all possible proton distributions. To describe the longitudinal relaxation in a REDPRO sample, we define an average Liouvillian operator that accurately accounts for (i) the initial linear regime and (ii) the equilibrium polarization. A full treatment is available in Supplementary Material.
Intermolecular cross-relaxation
In large biomolecules, the spectral density functions have the following trend Jij(0)≫Jij(ω0)> Jij(2ω0), so that longitudinal relaxation of protons is largely dominated by cross-relaxation. In a conventional NOESY experiment, the polarizations are ‘labeled’ by the chemical shifts of the protons after the evolution interval. The effective relaxation rate associated with a diagonal peak during the mixing time is described by the selective relaxation rate (Macura and Ernst 1980) . This has dramatic effect on systems with high proton density. According to our calculations for a 30 kDa protein, almost 95 % of the intensity of a diagonal peak can be lost after a sort mixing time (τm) of 100 ms. When intramolecular nOe’s are to be detected, cross-relaxation leads to the build-up of desired off-diagonal peaks in the NOESY spectrum whereas intramolecular cross-relaxation does not lead to any cross peak in the filtered NOESY experiments. As illustrated by Figure 1, if the polarization is initially labeled by chemical shifts, intramolecular cross-relaxation results in a loss of polarization. Weak intermolecular nOe’s are strongly attenuated because of this fast relaxation process.
On the other hand, for t1 = 0 ms in the filtered NOESY experiments, all polarizations are in-phase so that the outcome is not affected by intramolecular cross-relaxation i.e. all intramolecular cross-peaks merge with the diagonal peak. The global decay of the longitudinal polarization can be approximated by a non-selective relaxation rate: , which is small and decreases with increasing protein size. Long-range dipolar cross-relaxation rates are proportional Jij (0), and have an efficiency that increases with molecular weight. As shown in Figure 1, intramolecular spin-diffusion within the source protein (or ligand) is beneficial since it constantly fuels the transfer of polarization to the target by providing the protons near the interface of the source with additional polarization. In such a situation the source protein behaves as a polarization reservoir.
The amount of polarization that is received by target proton i from the source protein reservoir is proportional to the population of site i, so that intermolecular polarization transfers are affected to the same extent as the intramolecular transfers within the low-density target protein. In a filtered NOESY experiment, the suppression of the initial proton polarization of the target protein boosts the relative polarization of the high-density source protein thus favoring the observation of intermolecular polarization transfer. The product of the probabilities for finding a proton at each site determines the probability of having an uninterrupted source of protonated sites. Therefore, in a REDPRO labeled sample, where the protonation probability is ~10%, spin diffusion is significantly attenuated (Figure 1).
A simple three spin-system model was used to simulate polarization transfer. In this model, the first spin represents the high-density proton reservoir, the second spin is the one that is observed and the third spin is used to account for intramolecular spin-diffusion within the REDPRO protein. In the slow tumbling limit, (i.e. when the spectral density J(0) dominates) the dipolar cross-relaxation rate between two spins is close to 40 % of the contribution of the dipolar interaction to transverse relaxation. When mixing time is shorter than 600 ms the estimate of the overall rotational correlation time is sufficient to evaluate the sum of all intermolecular dipolar cross-relaxation rates of the normalized polarization transfer. The expression employed to correlate the sum of intermolecular nOe’s to the normalized polarization intensity (Inorm) is given as:
| (1) |
where S = λτc is the sum of all intramolecular cross-relaxation rates, λ = 0.2×109 s−2 for a proton density of 10 %, while the polarization is of a proton from the REDPRO target and the cross-relaxation rates are between the observed proton in the target protein and all the protons in the HIPRO source protein. Details about the relaxation matrix used to derive Equation 7 can be found in the Supporting Information.
Preparation of the samples
The DNA sequence coding for the amino acid 221 to 251 of the human Ataxin 3 Ubiquitin Interacting Motif (AUIM) was cloned into the pTM expression vector.(Staley and Kim 1994) The AUIM peptide was over-expressed in the LB medium and purified as described elsewhere.(Staley and Kim 1994) The triple labeled [13C, 15N, 2H]-Ubiquitin using the REDPRO(Shekhtman, Ghose et al. 2002) labeling scheme was prepared as described previously (You, Cohen et al. 1999). The final [13C, 15N, 2H]-ubiquitin- [1H]-AUIM [0.5 : 1 mM] complex was made in 50 mM ammonium acetate, pH 4.5, 0.1% NaN3 in 90% 2H2O. All NMR data for the Ubiquitin-AUIM complex were recorded at 300 K. The detailed expression and purification protocols for Csk SH3 and the 25 residues long peptide from the tyrosine phosphatase (PEP) are given elsewhere.(Ghose, Shekhtman et al. 2001) For the present study the [13C, 15N, 2H]-Csk SH3 was prepared using the REDPRO labeling scheme and unlabeled PEP was grown in LB media. The final NMR sample of the [13C, 15N, 2H]-Csk SH3- [1H]-PEP [0.4 : 1.2 mM] complex was prepared in 98% 2H2O buffer (20 mM Tris-d11, 150 mM NaCl, 0.1% NaN3 and pH 7.2). The Csk SH3-PEP complex designed to mimic a high molecular weight protein was prepared by adding 21.7% (w/w) [2H8 12C] glycerol to the above NMR sample. All NMR data for the Csk SH3-PEP complex were collected at 298 K.
NMR spectroscopy
All experiments were carried out on Bruker Avance spectrometer with a proton Larmor frequency of 700 MHz, equipped with a TXI cold probe with z-axis gradients. The pulse sequences for the polarization transfer experiments were adapted for use on a cold probe from the pulse sequences previously developed by Zwahlen et al.(Zwahlen, Legault et al. 1997) (Figs. S1 and S2). The selective saturation of the solvent polarization at the beginning of the mixing time, or alternatively, the selective inversion of the solvent magnetization during the mixing time, suppresses the cross-peaks originating from exchange or cross-relaxation with the solvent. The cost of such a procedure is an increase of the longitudinal relaxation rates of protons that are exposed to the solvent. The complete backbone and side-chain carbon and proton assignments of Ubiquitin-AUIM and Csk SH3-PEP complexes were done using CBCACONH, HNCACB, CCCONH, HCCONH and HBHACONH experiments.(Ghose, Shekhtman et al. 2001; Muralidharan, Dutta et al. 2006) The aromatic side-chain assignments were based on constant-time (CT) 1H{13C} HSQC, 3D-aromatic NOESY-HSQC and the Hδ/Hγ −Cβ correlation experiment developed by Yamazaki et. al. (Yamazaki, Formankay et al. 1993). All spectra were processed and analyzed by using NMRPipe (Delaglio, Grzesiek et al. 1995) and viewed by NMRView (Johnson and Blevins 1994). The increase in the viscosity from pure 2H2O to the glycerol/2H2O mixture was monitored with a 13C version of the X-STE experiment. (Ferrage, Zoonens et al. 2003; Ferrage, Eykyn et al. 2004)
Polarization transfer calculations
The model used in the simulations consists of two cubes of length 4.36 Å, each comprising of 27 protons. The distance between the two cubes is 4 Å. We have employed a model-free spectral density function with a local order parameter S2 = 0.7 and a local correlation time τe = 0.1 ns for all nuclear pairs. The size of the cubes was adjusted so that the average transverse proton relaxation rate in the HIPRO cube followed the empirical rule R2/τc = 5×109 s−2. Numerical calculations were carried out using MATLAB 7 (Mathworks, Inc).
Normalized polarization transfer ratio
The polarization transfer was normalized to take into account the effects of (i) residual polarization after the isotope filter and (ii) site-to-site sensitivity differences due to variations in proton density and relaxation rates. Four HSQC-edited experiments were carried out: with and without the filter and with NOESY mixing times (τm) of 0 and 300 ms. The normalized polarization transfer Inorm is defined as:
| (8) |
where IF and INF are the intensities in the filtered and non-filtered experiments.
NEBULA calculation
The normalized polarization transfer ratio was used to evaluate the sum of the dipolar cross-relaxation rates from the HIPRO source protein to a chosen proton on the low proton-density (REDPRO) target protein. The probability distribution was estimated using constraints derived from the observed cross relaxation effects, and anti-constraints reflecting the absence of cross relaxation. The space in which the calculations were performed is defined below. The coordinates of the target proteins (PDB codes 1D3Z and 1JEG for the NMR structures of ubiquitin (Cornilescu, Marquardt et al. 1998) and Csk SH3,(Ghose, Shekhtman et al. 2001) respectively) were the initial input data used for simulations of the proton density distribution. The origin of the frame was moved to the center of the protein structure. A three-dimensional grid with a resolution of 1 Å was defined, with edges located greater than 5 Å away from any hydrogen in the target protein. The next step of the simulation was to carve out a space around the target where the proton probabilities were computed. As a first approximation, only hypothetical proton coordinates lying within a radius of 5 Å from any given proton in the target protein with a detectable polarization transfer were retained. To verify that the selected coordinates were positioned at the exterior of the target protein, the following criterion was used: the distances to all atoms in the protein that lie within 7 Å of a proton carrying a positive constraint were calculated, and coordinates were kept only if all distances were larger than the sum of the van der Waals radii. Then a list of the dipolar cross-relaxation rates from each of the hypothetical proton coordinates to each hydrogen nucleus was computed. A Lipari-Szabo spectral density function was used so that the local dynamics and the high-frequency contributions to relaxation were not underestimated. In our analysis the methyl groups were treated as one entity, i.e. the average cross-relaxation rates were calculated between the three individual protons using the distances derived from the PDB file. The second approximation eliminates all hypothetical protons coordinates that have the predicted dipolar cross-relaxation rates higher than the experimental constraint. Finally, parasitic constraints associated with deeply buried protons (i.e. when the closest point on the grid is further than 5 Å) were eliminated.
A Monte Carlo simulation was performed by generating random distributions of protons at the coordinates of the 3D grid. The average population density of each configuration was set to 5.5% of the total number of points on the grid for the ubiquitin-AUIM complex, and 6.2% for the Csk SH3-PEP complex so that the source protein occupies most of the available space. The sum of all dipolar cross-relaxation rates from the source to the target protons was calculated and an experimental energy function E= Ec + Eac was determined for each configuration. Ec the energy from measured constraints is defined as: and Eac the energy from anti-constraints (corresponding to observable protons for which no cross-relaxation effects were observed) is defined as where and are the measured rate, the experimental threshold, the experimental error and the calculated sum of all cross-relaxation rates from the source to the target protons, respectively. The experimental threshold was defined in terms of normalized polarization transfer and was set to ~20% of the maximum value. Since the deuteration on Hα sites was close to 100%, these sites did not contribute to any anti-constraints. For the NEBULA calculations of Csk SH3-PEP complex, the exchangeable protons were systematically excluded so that they did not contribute to any anti-constraints. A set of configurations (between 300 and 1000 generated from 5×106 to 107 tests) was retained. Note that more than 107 configurations can be generated in an hour using an HP DL 145 SATA G2 Server with a DUAL AMD O280 processor at 2.4 GHz with 8 GB of RAM. Population probabilities were then derived for each site from a Boltzmann-weighted sum of populations of the selected configurations.
Supplementary Material
Acknowledgments
We thank Geoffrey Bodenhausen for his careful reading of the manuscript. Support by the grant GM 47021 from the National Institute of Health (DC) and 1-06-CD-23 from the American Diabetes Association (AS) is acknowledged. FF thanks the French Ministry of Foreign Affairs for a Lavoisier fellowship.
References
- Bottini N, Musumeci L, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36(4):337–8. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- Breeze AL. Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog NMR Spectrosc. 2000;36(4):323–372. [Google Scholar]
- Burz DS, Dutta K, et al. In-cell NMR for protein-protein interactions (STINT-NMR) Nat Protoc. 2006;1(1):146–52. doi: 10.1038/nprot.2006.23. [DOI] [PubMed] [Google Scholar]
- Burz DS, Dutta K, et al. Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR) Nat Methods. 2006;3(2):91–3. doi: 10.1038/nmeth851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM. Accurate and rapid docking of protein-protein complexes on the basis of intermolecular nuclear overhauser enhancement data and dipolar couplings by rigid body minimization. Proc Natl Acad Sci U S A. 2000;97(16):9021–5. doi: 10.1073/pnas.97.16.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Marquardt JL, et al. Validation of Protein Structure from Anisotropic Carbonyl Chemical Shifts in a Dilute Liquid Crystalline Phase. J Amer Chem Soc. 1998;120(27):6836–6837. [Google Scholar]
- Delaglio F, Grzesiek S, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, et al. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Eichmuller C, Tollinger M, et al. Mapping the ligand binding site at protein side-chains in protein-ligand complexes through NOE difference spectroscopy. J Biomol NMR. 2001;20(3):195–202. doi: 10.1023/a:1011299009214. [DOI] [PubMed] [Google Scholar]
- Ferrage F, Eykyn TR, et al. Frequency-switched single-transition cross-polarization: A tool for selective experiments in biomolecular NMR. Chemphyschem. 2004;5(1):76–84. doi: 10.1002/cphc.200300905. [DOI] [PubMed] [Google Scholar]
- Ferrage F, Zoonens M, et al. Slow diffusion of macromolecular assemblies by a new pulsed field gradient NMR method. Journal of the American Chemical Society. 2003;125(9):2541–2545. doi: 10.1021/ja0211407. [DOI] [PubMed] [Google Scholar]
- Fiaux J, Bertelsen EB, et al. NMR analysis of a 900K GroEL GroES complex. Nature. 2002;418(6894):207–11. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- Foster MP, Wuttke DS, et al. Chemical shift as a probe of molecular interfaces: NMR studies of DNA binding by the three amino-terminal zinc finger domains from transcription factor IIIA. J Biomol NMR. 1998;12(1):51–71. doi: 10.1023/a:1008290631575. [DOI] [PubMed] [Google Scholar]
- Fushman D, Cowburn D. Characterization of inter-domain orientations in solution using the NMR relaxation approach. In: Krishna NR, Berliner LJ, editors. Biological Magnetic Resonance. Vol. 20. New York: Kluwer Academic; 2003. pp. 53–77. [Google Scholar]
- Fushman D, Varadan R, et al. Determining domain orientation in macromolecules by using spin-relaxation and residual dipolar coupling measurements. Progress In Nuclear Magnetic Resonance Spectroscopy. 2004;44(3–4):189–214. [Google Scholar]
- Fushman D, Xu R, et al. Direct determination of changes of interdomain orientation on ligation: use of the orientational dependence of 15N NMR relaxation in Abl SH(32) Biochemistry. 1999;38(32):10225–30. doi: 10.1021/bi990897g. [DOI] [PubMed] [Google Scholar]
- Gaponenko V, Altieri AS, et al. Breaking symmetry in the structure determination of (large) symmetric protein dimers. J Biomol NMR. 2002;24(2):143–8. doi: 10.1023/a:1020948529076. [DOI] [PubMed] [Google Scholar]
- Gardner KH, Kay LE. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu Rev Biophys Biomol Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- Ghose R, Shekhtman A, et al. A novel, specific interaction involving the Csk SH3 domain and its natural ligand. Nat Struct Biol. 2001;8(11):998–1004. doi: 10.1038/nsb1101-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JD, V, Gelev M, et al. A sensitive and robust method for obtaining intermolecular NOEs between side chains in large protein complexes. J Biomol NMR. 2003;25(3):235–42. doi: 10.1023/a:1022890112109. [DOI] [PubMed] [Google Scholar]
- Guiles RD, Sarma S, et al. Pseudocontact shifts used in the restraint of the solution structures of electron transfer complexes. Nat Struct Biol. 1996;3(4):333–9. doi: 10.1038/nsb0496-333. [DOI] [PubMed] [Google Scholar]
- Hamel DJ, Dahlquist FW. The contact interface of a 120 kD CheA-CheW complex by methyl TROSY interaction spectroscopy. Journal of the American Chemical Society. 2005;127(27):9676–9677. doi: 10.1021/ja052517m. [DOI] [PubMed] [Google Scholar]
- Horst R, Fenton WA, et al. Folding trajectories of human dihydrofolate reductase inside the GroEL GroES chaperonin cavity and free in solution. Proc Natl Acad Sci U S A. 2007;104(52):20788–92. doi: 10.1073/pnas.0710042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M, Bax A. Isotope-Filtered 2D NMR of a Protein-Peptide Complex: Study of a Skeletal Muscule Myosin Light Chain Kinase Fragment Bound to Calmodulin. J Am Chem Soc. 1992;114:2433–2440. [Google Scholar]
- Jayalakshmi V, Krishna NR. Complete relaxation and conformational exchange matrix (CORCEMA) analysis of intermolecular saturation transfer effects in reversibly forming ligand-receptor complexes. J Magn Reson. 2002;155(1):106–18. doi: 10.1006/jmre.2001.2499. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kiihne SR, Creemers AF, et al. Selective interface detection: mapping binding site contacts in membrane proteins by NMR spectroscopy. J Am Chem Soc. 2005;127(16):5734–5. doi: 10.1021/ja045677r. [DOI] [PubMed] [Google Scholar]
- Kumar A, Ernst RR, et al. A Two-Dimensional Nuclear Overhauser Enhancement (2D NOE) Experiment for the Elucidation of Complete Proton-Proton Cross-Relaxation Networks in Biological Macromolecules. Biochem Biophys Res Commun. 1980;95:1–6. doi: 10.1016/0006-291x(80)90695-6. [DOI] [PubMed] [Google Scholar]
- Macura S, Ernst RR. Elucidation Of Cross Relaxation In Liquids By Two-Dimensional Nmr-Spectroscopy. Molecular Physics. 1980;41(1):95–117. [Google Scholar]
- Matsuda T, Ikegami T, et al. Model building of a protein-protein complexed structure using saturation transfer and residual dipolar coupling without paired intermolecular NOE. J Biomol NMR. 2004;29(3):325–338. doi: 10.1023/B:JNMR.0000032613.05864.87. [DOI] [PubMed] [Google Scholar]
- Muralidharan V, Dutta K, et al. Solution structure and folding characteristics of the C-terminal SH3 domain of c-Crk-II. Biochemistry. 2006;45(29):8874–84. doi: 10.1021/bi060590z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus D, Williamson MP. The Nuclear Overhauser Effect in Structural and Conformational Analysis. New York: John Wiley & Sons; 2000. [Google Scholar]
- Otting G, Wuthrich K. Heteronuclear filters in two-dimensional [1H,1H]-NMR spectroscopy: combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q Rev Biophys. 1990;23(1):39–96. doi: 10.1017/s0033583500005412. [DOI] [PubMed] [Google Scholar]
- Pintacuda G, Park AY, et al. Lanthanide labeling offers fast NMR approach to 3D structure determinations of protein-protein complexes. J Am Chem Soc. 2006;128(11):3696–702. doi: 10.1021/ja057008z. [DOI] [PubMed] [Google Scholar]
- Rain JC, Selig L, et al. The protein-protein interaction map of Helicobacter pylori. Nature. 2001;409(6817):211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Ryabov Y, Fushman D. Structural assembly of multidomain proteins and protein complexes guided by the overall rotational diffusion tensor. J Am Chem Soc. 2007;129(25):7894–902. doi: 10.1021/ja071185d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhtman A, Ghose R, et al. NMR structure determination and investigation using a reduced proton (REDPRO) labeling strategy for proteins. FEBS Lett. 2002;524(1–3):177–82. doi: 10.1016/s0014-5793(02)03051-x. [DOI] [PubMed] [Google Scholar]
- Shimada I. NMR techniques for identifying the interface of a larger protein-protein complex: Cross-saturation and transferred cross-saturation experiments. Nuclear Magnetic Resonance of Biological Macromolecules, Part C. 2005;394:483. doi: 10.1016/S0076-6879(05)94020-2. [DOI] [PubMed] [Google Scholar]
- Shimada I, Kawazoe Y. Quantitative-Analysis and Computer-Simulation of Feeding-Behavior of the Fruitfly, Drosophila-Melanogaster. Zoological Science. 1984;1(6):989–989. [Google Scholar]
- Staley JP, Kim PS. Formation of a native-like subdomain in a partially folded intermediate of bovine pancreatic trypsin inhibitor. Protein Sci. 1994;3(10):1822–32. doi: 10.1002/pro.5560031021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui XG, Xu YQ, et al. Mapping protein-protein interfaces on the basis of proton density difference. Angew Chem Int Ed. 2005;44(32):5141–5144. doi: 10.1002/anie.200501209. [DOI] [PubMed] [Google Scholar]
- Swanson KA, Kang RS, et al. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. Embo Journal. 2003;22(18):4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Miyazawa M, et al. Utilization of methyl proton resonances in cross-saturation measurement for determining the interfaces of large protein-protein complexes. J Biomol NMR. 2006;34(3):167–177. doi: 10.1007/s10858-006-0008-8. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Nakanishi T, et al. A novel NMR method for determining the interfaces of large protein-protein complexes. Nature Structural Biology. 2000;7(3):220–223. doi: 10.1038/73331. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403(6770):623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- van Dijk ADJ, Boelens R, et al. Data-driven docking for the study of biomolecular complexes. Febs Journal. 2005;272(2):293–312. doi: 10.1111/j.1742-4658.2004.04473.x. [DOI] [PubMed] [Google Scholar]
- van Dijk ADJ, Boelens R, et al. Data-driven docking for the study of biomolecular compexes. FEBS J. 2005;272:293–312. doi: 10.1111/j.1742-4658.2004.04473.x. [DOI] [PubMed] [Google Scholar]
- Williams DC, Jr, Cai M, et al. Solution NMR structure of the 48-kDa IIAMannose-HPr complex of the Escherichia coli mannose phosphotransferase system. J Biol Chem. 2005;280(21):20775–84. doi: 10.1074/jbc.M501986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zheng Y, et al. A new strategy for structure determination of large proteins in solution without deuteration. Nature Methods. 2006;33(11):931–937. doi: 10.1038/nmeth938. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Formankay JD, et al. 2-Dimensional Nmr Experiments for Correlating C-13-Beta and H-1-Delta/Epsilon Chemical-Shifts of Aromatic Residues in C-13-Labeled Proteins Via Scalar Couplings. Journal of the American Chemical Society. 1993;115(23):11054–11055. [Google Scholar]
- You J, Cohen RE, et al. Construct for high-level expression and low misincorporation of lysine for arginine during expression of pET-encoded eukaryotic proteins in Escherichia coli. Biotechniques. 1999;27(5):950–4. doi: 10.2144/99275st01. [DOI] [PubMed] [Google Scholar]
- Zangger K, Oberer M, et al. X-filtering for a range of coupling constants: application to the detection of intermolecular NOEs. J Magn Reson. 2003;160(2):97–106. doi: 10.1016/s1090-7807(02)00176-3. [DOI] [PubMed] [Google Scholar]
- Zwahlen C, Legault P, et al. Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy: Application to a bacteriophage lambda N-peptide/boxB RNA complex. Journal of the American Chemical Society. 1997;119(29):6711–6721. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.