Abstract
Both underweight and obesity have been associated with increased mortality1,2. Underweight, defined as body mass index (BMI) ≤ 18,5 kg/m2 in adults 3 and ≤ −2 standard deviations (SD) in children4,5, is the main sign of a series of heterogeneous clinical conditions such as failure to thrive (FTT) 6–8, feeding and eating disorder and/or anorexia nervosa9,10. In contrast to obesity, few genetic variants underlying these clinical conditions have been reported 11, 12. We previously demonstrated that hemizygosity of a ~600 kb region on the short arm of chromosome 16 (chr16:29.5–30.1Mb), causes a highly-penetrant form of obesity often associated with hyperphagia and intellectual disabilities13. Here we show that the corresponding reciprocal duplication is associated with underweight. We identified 138 (132 novel cases) duplication carriers (108 unrelated carriers) from over 95,000 individuals clinically-referred for developmental or intellectual disabilities (DD/ID), psychiatric disorders or recruited from population-based cohorts. These carriers show significantly reduced postnatal weight (mean Z-score −0.6; p=4.4×10−4) and BMI (mean Z-score −0.5; p=2.0×10−3). In particular, half of the boys younger than 5 years are underweight with a probable diagnosis of FTT, while adult duplication carriers have an 8.7-fold (p=5.9×10−11; CI_95=[4.5–16.6]) increased risk of being clinically underweight. We observe a significant trend towards increased severity in males, as well as a depletion of male carriers among non-medically ascertained cases. These features are associated with an unusually high frequency of selective and restrictive feeding behaviours and a significant reduction in head circumference (mean Z-score −0.9; p=7.8×10−6). Each of the observed phenotypes is the converse of one reported in carriers of deletions at this locus, correlating with changes in transcript levels for genes mapping within the duplication but not within flanking regions. The reciprocal impact of these 16p11.2 copy number variants suggests that severe obesity and being underweight can have mirror etiologies, possibly through contrasting effects on eating behaviour.
Keywords: Adolescent; Adult; Aged; Aging; Body Height; genetics; Body Mass Index; Case-Control Studies; Child; Child, Preschool; Chromosomes, Human, Pair 16; Cohort Studies; Comparative Genomic Hybridization; Developmental Disabilities; genetics; Energy Metabolism; Europe; Female; Gene Dosage; Gene Duplication; Gene Expression Profiling; Genetic Predisposition to Disease; Genome-Wide Association Study; Head; anatomy & histology; Heterozygote; Humans; Infant; Infant, Newborn; Male; Mental Disorders; Middle Aged; Mutation; North America; Obesity; Phenotype; RNA, Messenger; analysis; Sequence Deletion; Thinness; Transcription, Genetic; Young Adult
Copy number variants at the 16p11.2 locus have been associated with cognitive disorders including autism (deletions) and schizophrenia (duplications)14–19, conditions that have been suggested to lie at opposite ends of a single spectrum of psychiatric phenotypes20. We and others have reported that deletion of this region spanning 28 genes (Supplementary Table S1) increases the risk of morbid obesity 43-fold (Supplementary Figure S1)13,21. We hypothesized that the reciprocal duplication, with its resulting increase in gene dosage, may influence BMI in a converse manner. The duplication was identified in 73 out of 31,424 patients with DD/ID, a frequency consistent with previous reports17 (Table 1). Four additional cases were identified among 1,080 patients affected by bipolar disease or schizophrenia. Compared to its prevalence in seven European population-based GWAS cohorts22–24 (31 out of 58,635 individuals), the duplication was significantly more frequent in both the DD/ID cohorts (p=4.23×10−13; OR=4.4, CI_95=[2.9–6.9]) and the psychiatric cohorts (p=3.6×10−3; OR=7.0, CI_95=[1.8–19.9]) (Table 1) strengthening previous reports of similar associations16,17. Our data do not support a two-hit model25 for the effects of 16p11.2 duplications or deletions (Supplementary Text and Table S2).
Table 1.
16p11.2 rearrangements in cases and controls.
| Ascertainment | Cohorts | Dup | Del | Total | ||
|---|---|---|---|---|---|---|
| n | p 1 | n | p 1 | |||
| Neuro-developmental disorders | Unspecified DD/ID* from 28 cytogenetic centers | 72 | 113 | 30323 | ||
| ADHD 4, deCODE 4 | 0 | 1 | 591 | |||
| Childhood Autism 4, deCODE | 0 | 2 | 159 | |||
| Childhood Autism spectrum disorder 4, deCODE | 1 | 3 | 351 | |||
| TOTAL | 73 | 4.23e-13 | 119 | 5.43e-32 | 31424 | |
| Rearrangement frequency (95% CI) | 0.23% (0.18–0.29) | 0.38% (0.31–0.45) | ||||
|
| ||||||
| Family history | 1st degree relatives of probands | 30 | 35 | 43/622 | ||
|
| ||||||
| Adult psychiatric symptoms | Schizophrenia, deCODE | 0 | 1 | 657 | ||
| Bipolar disease, Rouen | 1 | 0 | 156 | |||
| Schizophrenia, Schizo-affective, Rouen. | 3 | 0 | 267 | |||
| TOTAL | 4 | 3.57e-03 | 1 | 3.78e-01 | 1080 | |
| Rearrangement frequency (95% CI) | 0.37% (0.01–0.73) | 0.09% (0–0.27) | ||||
|
| ||||||
| Underweight | Eating disorder, Spain | 1 3 | 0 | 441 | ||
|
| ||||||
| Obesity | Obesity, Spain | 0 | 2 | 653 | ||
| Adult Obesity, France | 0 | 4 | 705 | |||
| Childhood Obesity, France & UK | 0 | 7 | 1574 | |||
| Obesity Bariatric Surgery, France | 0 | 2 | 141 | |||
| Obesity Discordant Siblings, Sweden | 0 | 2 | 159 | |||
| Obesity and Cognitive Delay, France & UK | 0 | 9 | 312 | |||
| TOTAL | 0 | 4.21e-01 | 26 | 2.52e-19 | 3544 | |
| Rearrangement frequency (95% CI) | 0 | 0.73% (0.45–1.01) | ||||
|
| ||||||
| Population-based cohorts | NFBC1966 Finnish | 4 | 3 | 5319 | ||
| CoLaus Swiss | 5 | 0 | 5612 | |||
| EGCUT Estonian | 2 | 1 | 2994 | |||
| deCODE Iceland | 17 | 18 | 36601 | |||
| SHIP Germany | 1 | 2 | 4070 | |||
| KORA F3+F4 Germany | 2 | 1 | 3458 | |||
| Pediatric family study | 0 | 0 | 581 | |||
| TOTAL | 31 | 25 | 58635 | |||
| Rearrangement frequency (95% CI) | 0.05% (0.03–0.07) | - | 0.04% (0.03–0.06) | - | ||
: Not a disease-specific cohort. Detailed distribution is provided in the online methods.
DD/ID: developmental delay/intellectual disabilities
Fisher’s exact test, compared to the combined frequency in general population groups.
total number of parental pairs tested for dup/del. 13 out of 43 duplications and 27 out of 62 deletion cases were de novo.
atypical duplication (see Supplementary Figure S5)
There was no overlap between these 3 cohorts
We compared available data on weight, height and BMI for 106 independent duplication carriers (including published cases) to those in gender-, age- and geographically-matched reference populations (Table 2, Supplementary Tables S3 and S4, Methods). The duplication was strongly associated with lower weight (mean Z-score −0.56; p=4.4×10−4) and BMI (mean Z-score −0.47; p=2.0×10−3) (Table 2, Supplementary Table S5). Birth parameters (n=48) were normal indicating a postnatal effect. Adults carrying the duplication had a relative risk (RR) of being clinically underweight (BMI < 18.5)26 of 8.7, CI_95=[4.5–16.6] (p=5.9×10−11) (Methods). Concordantly, none of the 3,544 patients in our obesity cohorts13,21 carried the duplication (Table 1).
Table 2. Comparisons of the height, weight and BMI distributions in duplication carriers and controls.
The available BMI, weight and height data for duplication carriers were transformed to Z-scores using gender- and age-matched reference populations, and one-sided t-tests were carried out to determine whether the mean Z-scores deviate from zero. Significant differences were identified by reference to cutoffs controlling the false discovery rate (FDR) at 5% (see Methods): BMI – 0.022; weight – 0.032; height – 0.025 (significant results indicated in bold). Data were not available for all subjects.
| Combined2 | DD/ID or psychiatric2 | Non-medically ascertained3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Strata | Mean Z-score | p-value | N1 | Mean Z-score | p-value | N1 | Mean Z-score | p-value | N1 | |
| BMI | All | −0.47 | 2.0e-03 | 102 | −0.56 | 4.1e-03 | 76 | −0.45 | 6.0e-03 | 40 |
| Male | −0.54 | 2.1e-02 | 52 | −0.71 | 1.3e-02 | 43 | −0.31 | 2.0e-01 | 14 | |
| Female | −0.4 | 1.8e-02 | 50 | −0.37 | 8.3e-02 | 33 | −0.52 | 4.2e-03 | 26 | |
|
| ||||||||||
| Weight | All | −0.56 | 4.4e-04 | 104 | −0.65 | 1.3e-03 | 78 | −0.61 | 3.0e-03 | 40 |
| Male | −0.64 | 5.8e-03 | 53 | −0.79 | 4.4e-03 | 44 | −0.57 | 8.8e-02 | 14 | |
| Female | −0.47 | 1.7e-02 | 51 | −0.47 | 6.5e-02 | 34 | −0.63 | 8.6e-03 | 26 | |
|
| ||||||||||
| Height | All | −0.24 | 4.8e-02 | 103 | −0.33 | 3.6e-02 | 77 | −0.15 | 1.8e-01 | 40 |
| Male | −0.34 | 4.5e-02 | 52 | −0.4 | 4.6e-02 | 43 | −0.29 | 1.2e-01 | 14 | |
| Female | −0.14 | 2.6e-01 | 51 | −0.24 | 2.1e-01 | 34 | −0.07 | 3.7e-01 | 26 | |
Relatives of probands were excluded as required, to avoid including more than one member of the same family in a single analysis
Including 24 cases from the literature (Supplementary Table S3).
Population-based cases and first-degree relatives of probands.
To further investigate these associations, we carried out separate analyses of carrier patients (DD/ID and psychiatric) and non-medically ascertained carriers (population-based cohorts plus 11 transmitting parents and three other affected first-degree relatives for whom data were available) (Table 2). Each category exhibited significantly lower weight and BMI with similar effect sizes. However, the proportion of underweight cases (BMI ≤ −2 SD)26 was higher in the first than in the second group (17/76 compared to 2/40; p=0.017). Note that the impact of the duplication on underweight status might be understated due to prescription of antipsychotic treatments, often associated with weight gain27 (Supplementary Table S6)
Having demonstrated an association of the duplication with underweight, we investigated the contribution of gender to the resulting phenotypes (Figure 1, Supplementary Figure S2 and Table S7). In DD/ID patients, the impact of the duplication on being underweight is stronger in males – the effect in females is in the same direction, but is both smaller and statistically non-significant (Table 2). A similar and significant difference (p=0.0173) was observed in adult carriers (all groups combined): the RR of being underweight for males is 24.1 (CI_95=[9.5–61.2], p=2.2×10−11) and only 4.9 for females (CI_95=[2.0–12.3], p=6.7×10−4). A gender bias was also observed in the ascertainment of DD/ID duplication carriers, in which we have an excess of males (51M:33F, p=0.044). By contrast, carriers from the general population exhibited a strong overrepresentation of females (10M:21F, p=0.035) (Supplementary Text). A similar bias was observed among transmitting parents (7M:23F, p=5.53×10−4). Thus, there is an overrepresentation of males in the medically ascertained group and a depletion in the non-medically ascertained one. We suggest that males are more likely than females to present severe phenotypes, and that this may account for the observed gender bias, as severely affected males are less likely to be recruited to adult population cohorts or be reproductively successful.
Figure 1. Effect of the 16p11.2 duplication on BMI and head circumference.
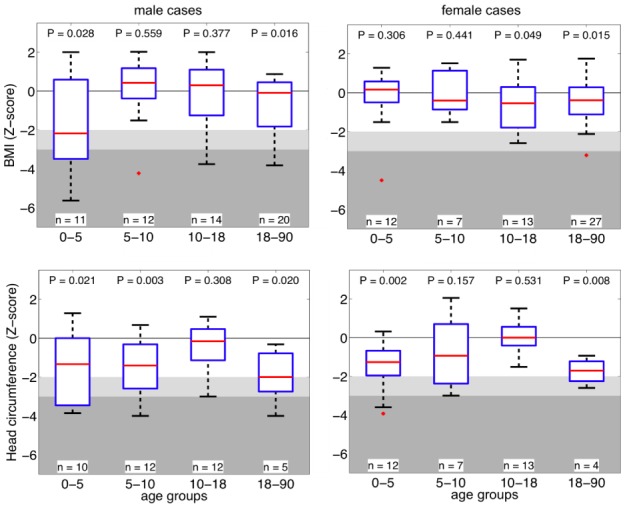
Z-score values of BMI (top row) and head circumference (bottom row) of 16p11.2 duplication carriers stratified by gender and age group. The most severe effect is observed in children 0–5 years of age. Abscise: age groups in years. Boxplots represent the 5th, 25th, median, 75th and 95th percentile for each age group. Light grey and dark grey background represent −2 and −3 SD respectively, which correspond to the WHO definition of moderate and severe underweight4. BMI is decreased in adolescent and adult females.
As previously reported28, the duplication was also associated with reduced head circumference (HC; mean Z-score −0.89; p=7.8×10−6) (Figure 1), 26.7% presenting with microcephaly, while carriers of the reciprocal deletion had an increased HC (mean Z-score +0.57; p=1.79×10−5) (Supplementary Table S8, Figure S3), demonstrating an additional instance of a mirror phenotype associated with reciprocal copy number changes at this locus. Notably, HC Z-scores correlate positively with those of BMI in carriers of both the duplication (rho=0.37; p=2.65×10−3) and the deletion (rho=0.42; p=1.9×10−5) (Supplementary Methods). This suggests that HC and BMI may be regulated by a common pathway or that a causal relationship exists between these two traits in these patients. A full list of malformations and secondary phenotypes reported in duplication carriers ascertained for DD/ID is available in Supplementary Table S9.
In view of their importance in obesity and underweight, the clinical reports of duplication carriers were screened for evidence of modified eating behaviours. Consequently, we carried out multiplex ligation-dependent probe amplification (MLPA, Supplementary Table S10) to screen for 16p11.2 rearrangements in 441 patients diagnosed with ED including anorexia nervosa (AN), bulimia and binge eating disorder (Table 1, Supplementary Text). No duplications of the entire region were identified, but one out of 109 AN patients carried an atypical 136 kb duplication that encompasses the SPN and QPRT genes (Supplementary Figure S4). This smaller duplication is currently the subject of further investigations, as it potentially delineates a critical region affecting eating behaviour.
Large genomic structural variants are known to affect the expression of genes not only within the affected region but also at a distance29–32. We therefore measured relative transcript levels in lymphoblastoid cell-lines of 27 genes mapping within the rearrangement or nearby (Supplementary Tables S1 and S11): six from deletion carriers, five from duplication carriers and ten from gender- and age-matched controls (Supplementary Table S12). Expression levels correlated positively with gene dosage for all genes within the copy number variable region (Figure 2) consistent with published partial results from adipose tissue13. Mean relative transcript levels in deletion and duplication carriers were, respectively, 67% and 214% of the levels measured in controls (Supplementary Table S13). While genes proximal (centromeric) to the rearrangement interval showed no significant variation in relative transcript levels between patients and controls (Figure 2), distal (telomeric) genes showed a significant alteration in relative expression. While lymphoblastoid cells may not recapitulate obesity-relevant tissues, previous experiments have shown a high degree of correlation between expression levels in different tissues/cell lines29, suggesting that the same pathways may be similarly disrupted in different cell lineages. Thus, the involvement of these distal genes in the control of BMI in our study subjects seems unlikely.
Figure 2. Transcript levels for genes within and near to the 16p11.2 rearrangements.
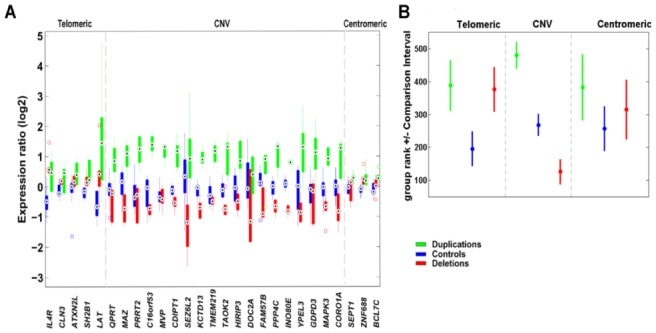
(A) Relative expression levels in boxplot format of 27 genes mapping to 16p11.2 in deletion and duplication carriers (red and green, respectively) and control cell lines (blue). Grey lines denote the extent of the 16p11.2 CNV (29.5–30.1 Mb). Complete lists of genes mapping within the rearranged interval and of the quantitative PCR assays can be consulted in Supplementary Table S1 and S11, respectively. Possible relevance of each of these genes to obesity/leanness and/or developmental delay/cognitive deficits can be consulted in 13.
(B) Rank comparison (Kruskal-Wallis test) between expression of 27 genes mapping to 16p11.2 in deletion and duplication carriers (red and green, respectively) and control cell lines (blue).
Distinction is made between genes mapping telomeric (or centromeric) to the rearranged interval and those within the rearranged interval. Dots correspond to the mean group rank and bars indicate the comparison interval, groups with non-overlapping intervals are significantly different (at α=5%).
Our study demonstrates the power of very large screens (>95,000 samples, the biggest of its kind so far) to characterize the clinical and molecular correlates of a rare functional genomic variant. We unambiguously demonstrate that carrying the 16p11.2 duplication confers a high risk of being clinically underweight and show that reciprocal changes in gene dosage at this locus result in multiple mirror phenotypes. As in the schizophrenia/autism20,34 and microcephaly/macrocephaly28 dualisms, abnormal eating behaviours, such as hyperphagia and anorexia, could represent opposite pathological manifestations of a common energy balance mechanism, although the precise relationships between these mirror phenotypes remain to be determined. We surmise that abnormal brain volume, thus neuronal circuitry, both cognitive function and eating behaviour, the latter possibly being the basis for the observed impact on BMI. Consistent with this are previous reports that a subgroup of children with microcephaly show concomitant reduction in weight percentile35. Our findings also support the observation that severe overweight and underweight phenotypes correlate with lower cognitive functioning7,36. Thus, abnormal food intake may be a direct result of particular neurodevelopmental disorders. Although it is possible that the 16p11.2 region encodes distinct genes specific for each trait, a more parsimonious hypothesis is that these different clinical manifestations of central nervous system dysfunction are all secondary to the disruption of a single gene-dosage-sensitive neurodevelopmental step. Further resolution of this issue may require identification of additional patients with rare atypical rearrangements in this region.
METHODS SUMMARY
Underweight is defined in adults and individuals of less than 18 years of age as BMI ≤ 18.5 and Z-score ≤ −2, respectively.
Statistics
Two-sided Fisher’s exact test was used to compare frequencies of the rearrangement in patients and controls. Z-scores were computed for all data using gender-, age- and geographically-matched reference populations. One-sided t-test was performed to test duplication carriers for lower than zero BMI, height, weight and HC Z-score values. We used Kruskal-Wallis to test differences in gene expression patterns. P-value thresholds were defined (by permutation) in order to control the false discovery rate at 5%. Relative risk of being underweight was calculated as the ratio of the fraction of underweight individuals among duplication carriers versus our control group.
CNV discovery
16p11.2 duplication and deletion carriers were identified through various procedures: (i) CGH with Agilent (Santa Clara, CA) 44K, 60K, 105K, 180K, 244K arrays; (ii) Illumina (San Diego, CA) Human317, Human370, HumanHap550, Human610 and 1M BeadChips; (iii) Affymetrix (Santa Clara, CA) 6.0, 250K genotyping arrays; (iv) QMPSF, (v) FISH and/or (vi) MLPA. CNV analyses of GWAS data were variously carried out using cnvHap, a moving window average intensity procedure, a Gaussian Mixture Model, Circular Binary Segmentation, QuantiSNP, PennCNV, BeadStudio GT module and Birdseed. At least two independent algorithms were used for each cohort.
Expression analyses
lymphoblastoid cell lines were established from carriers and controls. SYBR Green quantitative PCR was performed to assess relative expression of genes.
ONLINE METHODS
Patients referred for intellectual disabilities and developmental delay
Patients with cognitive deficits are routinely referred to clinical genetics for etiological work-ups including aCGH. We surveyed 28 cytogenetic centers in Europe and North America (Supplementary Table S3), identifying 31,424 patients ascertained for developmental delay, intellectual disabilities and/or malformations. Clinical ascertainment was as follows: developmental delay and mental retardation (MR): 51.0%, autism spectrum disorder (ASD) with or without MR: 14.4%, language delay with or without MR: 42.9%, malformations with MR: 27.6%, and malformations without MR: 4.8%.
These analyses were performed for clinical diagnostic purposes, all available phenotypic data being those provided anonymously and retrospectively by the clinician ordering the analyses. Consequently, research-based informed consent was not required by the Institutional Review Board of the University of Lausanne, which granted an exemption for this part of the study.
Patients referred for psychiatric disorders
Cohort from Rouen
Cases with schizophrenia and cases with bipolar disorder were ascertained at University Hospital, Rouen, France, from consecutive hospitalizations. All psychiatric diagnoses were established according to DSM-IV criteria following review of case notes and direct examination of cases. The Schedule for Affective Disorders and Schizophrenia was used for the clinical assessment of all cases with schizophrenia or schizoaffective disorder and the Diagnostic Interview for Genetic Studies (DIGS) was used in patients with bipolar disorder. The schizophrenia cohort was described before 37: 189 cases with schizophrenia and 47 cases with schizoaffective disorder. Post-morbid IQs were available for two-thirds of cases with schizophrenia; 18.0% of these cases had an IQ lower than 70. The bipolar cohort was made up of 150 patients with either bipolar disorder type I or type II. All subject have given written informed consent and this study was accepted by the local institutional review board.
General population cohorts
CoLaus
This prospective population cohort was described previously22: 6,188 white individuals aged 35–75 years were randomly selected from the general population in Lausanne, Switzerland. These individuals underwent a detailed phenotypic assessment, and were genotyped using the Affymetrix Mapping 500K array; 5,612 samples passed genotyping quality control. The institutional review board of the University of Lausanne approved this study, and written consent was obtained from all participants.
NFBC1966
The Northern Finland Birth Cohort 1966 is a prospective birth cohort of almost all individuals born in 1966 in the two northernmost provinces of Finland. Biochemical and DNA samples were collected with informed consent at age 31 years. Genotyping was done using the Illumina Infinium 370cnvDuo array and phenotypic characteristics of the cohort were as previously described23. Phenotypic and genotyping data was available for 5,246 subjects after quality control.
EGCUT
The Estonian Genome Centre at the University of Tartu (EGCUT) maintains a general population based biobank, described in greater detail in 24. 2994 unrelated subjects were randomly selected among the 48,000 Estonian participants and genotyped using the IlluminaCNV370-Duo or –Quad BeadChips. EGCUT is conducted according to Estonian Gene Research Act. The project was approved by the Ethics Review Committee on Human Research of the University of Tartu. Written informed consent was obtained from all voluntary participants.
deCODE
Patients and controls were all Icelandic and were recruited from all over Iceland. All participants with ADHD met DSM-IV criteria for ADHD (477 combined type, 250 inattentive type, 58 hyperactive-impulsive type, 40 unspecified). ADHD subjects were recruited from outpatient pediatric, child, and adult psychiatry clinics in Iceland, and diagnoses had been made on the basis of standardized diagnostic assessments and had been reviewed by experienced clinicians 38,39. Autistic individuals (n=351) met Autism Diagnostic Interview-Revised (ADI-R) criteria and were ascertained through the State Diagnostic Counselling Center and the Department of Child and Adolescent Psychiatry in Iceland. schizophrenia diagnoses were assigned according to Research Diagnostic Criteria (RDC) 40 through the use of the Schedule for Affective Disorders and Schizophrenia Lifetime Version (SADS-L) 41. Schizophrenia patients (n=657) were recruited from outpatient pediatric, child, and adult psychiatry clinics, and diagnoses had been made on the basis of standardized diagnostic assessments and had been reviewed by experienced clinicians. Recruitment of the psychiatric patients at deCODE has been described in more details elsewhere, ADHD 42, ASD schizophrenia and the control population 17. All participants, cases and controls, returned signed informed consents prior to participation in the study. All personal identifiers associated with medical information, questionnaire results, and blood samples were encrypted according to the standards set by the Data Protection Committee of Iceland. All procedures related to this study have been approved by the Data Protection Authority and National Bioethics Committee of Iceland.
SHIP
The Study of Health in Pomerania (SHIP) is a cross-sectional survey in Western Pomerania, the north-eastern area of Germany 43,44. A sample from the population aged 20 to 79 years was drawn from population registries. 7,008 subjects were selected randomly from each community, proportional to community population size and stratified by age and gender. Exclusively, individuals with German citizenship and main residency in the study area were included. 4,308 individuals participated to the study and were genotyped on Affymetrix 6.0 SNP arrays. Both genotyped data (after QC filtering) and BMI were available for 4,070 individuals.
KORA F3/F4
The KORA study is a series of independent population-based epidemiological surveys of participants living in the region of Augsburg, Southern Germany 45. All survey participants are residents of German nationality identified through the registration office and were examined in 1994/95 (KORA S3) and 1999/2001 (KORA F4). In the KORA S3 study 4,856 subjects (response 75%), and in KORA F4 in total 4,261 subjects have been examined (response 67%). 3,006 subjects participated in a 10-year follow-up examination of S3 in 2004/05 (KORA F3). Individuals for genotyping in KORA F3 and KORA F4 were randomly selected. The age range of the participants was 25 to 74 years of recruitment. Age and sex were self-reported in a questionnaire survey. Height and Weight were measured following a standardized study protocol. Informed consent has been given by all participants. The study has been approved by the local ethics committee.
French paediatric cohort
The French paediatric cohort was previously published 46 and was genotyped using the Illumina Human CNV370-duo array. 581 non-obese children (BMI ≤ 90th percentile) passed quality control. All participants or their legal guardians gave written informed consent, and all local ethics committees approved the study protocol.
Stanislas French Cohort
This cohort was described elsewhere 47. This family cohort (1006 families) was recruited between 1993–1995 (first visit) at the Center for Preventive Medicine (CMP) of Vandoeuvre-lès-Nancy during a periodical health assessment. Inclusion criteria at the first visit were parents and grandparents of French origin; residence in the Lorraine region (north-east of France); nuclear families comprising two parents and at least two biological children over 6 years old; fidelity of the majority of the families coming for the second or third time. Exclusion criteria at that visit were chronic or acute disorders. The families were supposed healthy and free from any declared acute and/or chronic disease in order to be able to assess the effects of genetics on the variability of the intermediate phenotypes in physiological conditions without the influence of any medical treatment or disease. These data were used for the statistical analysis when comparing BMI of duplication cases ascertained in France.
Spanish control cohort, University Hospital Dr. Josep Trueta
The 335 Spanish control subjects were recruited between 2000 and 2006 from the general population at the University Hospital Dr. Josep Trueta of Girona as previously described in 48–51.
Obesity cohorts
Adult-obesity and child-obesity cohorts
These cohorts were described in a previous publication 13. The adult-obesity case-control groups and the child-obesity case control groups were as published previously46, and were genotyped with the Illumina Human CNV370-duo array. In all, 643 children with familial obesity (BMI≥97th centile corrected for gender and age, at least one obese first-degree relative, age less than 18 years), 581 non-obese children (BMI≤90th centile), 705 morbidly obese adults with familial obesity (BMI≥40kgm−2, at least one obese first-degree relative with BMI≥35kgm−2, age≥18 years) and 197 lean adults (BMI≤25kgm−2) passed quality control; All participants or their legal guardians gave written informed consent, and all local ethics committees approved the study protocol.
Bariatric surgery cohort
This cohort was described in a previous publication 13. Patients undergoing elective bariatric weight-loss surgery were recruited for the ABOS study at Lille Regional University Hospital. Genotyping was performed with the Illumina Human 1M-duo array, and data from 141 adults passed quality control. All participants gave written informed consent, and the study protocol was approved by the local ethics committee.
Swedish discordant sibling cohort
The SOS Sib Pair Study cohort was as published previously 52. It includes 154 nuclear families, each with BMI discordant sibling pairs (BMI difference>10kgm−2), giving a total of 732 subjects. Genotyping data with the Illumina 610K-Quad array was available for 353 siblings from 149 families. Expression data from subcutaneous adipose tissue (sampled after overnight fasting) were available for 360 siblings from 151 families. Subjects received written and oral information before giving written informed consent. The Regional Ethics Committee in Gothenburg approved the studies.
Obesity cohort, University Hospital of Dr. Josep Trueta, Girona, Spain
The clinical sample consisted of 285 male and 223 female Spanish subjects of Caucasian origin with morbid obesity or type 2 diabetes mellitus recruited between 2000 and 2006. The remaining subjects were from the general population. All subjects reported stable body weight for at least three months before the study. They had no systemic disease other than obesity and/or IGT. The mean BMI (kg/m2) was 32.0 (15.2–82.4). The average age at assessment was 46.1 years (range 18.0–79.1). The majority of the patients have been described in previous reports 48–51.
Obesity cohort, University Hospital Virgen de la Victoria, Malaga, Spain
The clinical sample consisted of 85 Spanish Caucasian patients with obesity (35% were men and 65 % women). The mean BMI (kg/m2) was 53,9 (40–73). The average age at assessment was 42.63 years (19–69). The majority of the patients have been described in previous reports.53
Obesity cohort, University Hospital Juan XXIII, Tarragona, Spain
The clinical sample consisted of 57 Spanish Caucasian patients with obesity (59% were men and 41 % women). The mean BMI (kg/m2) was 32,23 (27,01–48). The average age at assessment was 59,74 years (36–79). The cases have not previously been described.
Eating disorder cohort
Spanish Eating Disorder Cohort, University Hospital of Bellvitge-IDIBELL
The clinical sample consisted of 441 Spanish Caucasian patients with eating disorders (ED) consecutively admitted to the Eating Disorders Unit of the University Hospital of Bellvitge between 2000 and 2008. The majority of patients were female (94.6%), fulfilled DSM-IV criteria for ED, and were diagnosed using the structured clinical interview for mental disorders, research version 2.0 (SCID-I) 54. The sample consisted of n=109 anorexia nervosa (AN; 25%), n=193 bulimia nervosa (BN; 44%), n=111 ED not otherwise specified (EDNOS; 25%)) and n=28 binge eating disorder patients (BED; 6%). The mean lifetime minimum BMI (kg/m2) was 15.46 (SD 1.39) for AN patients, 19.89 (SD 2.95) for BN patients, 18.53 (SD2.75) for EDNOS patients and 24.31 (SD 4.57) for BED patients. The average age at assessment was 26.57 years (SD 7.67). The average age at onset of the disorder was 18.9 years (SD 4.53) for AN patients, 19.74 (SD 7.16) for BN patients, 18.94 years (SD 5.88) for EDNOS patients and 24.65 years (SD 9.59) for BED patients. The majority of the patients have been described in previous reports 55, 56, 57.
CNV detection
Cases ascertained for intellectual disabilities and developmental delay were identified through standard medical diagnostic procedures. CNV analyses of GWAS data were variously carried out using cnvHap 58; a moving window average intensity procedure; a Gaussian Mixture Model 59; Circular Binary Segmentation60,61; QuantiSNP 62; PennCNV 63; BeadStudio GT module (Illumina inc); and Birdseed 64 (see below). At least two independent algorithms were used for each cohort.
Patients referred for intellectual disabilities and developmental delay
All diagnostic procedures (aCGH, QPCR and/or Quantitative Multiplex PCR of Short Fluorescent fragments) were carried out according to the relevant guidelines of good clinical laboratory practice for the respective countries. All rearrangements in probands were confirmed by a second independent method and karyotyping was performed in all cases to exclude a complex rearrangement.
NFBC cohort
CNV calling was previously described in 13. In brief, data were normalized using Illumina BeadStudio, then GC effects on ratios were removed by regressing on GC and GC2, while wave effects were removed by fitting a loess function65. CNV analysis was done using cnvHap58. All called 16p11.2 duplications were validated by direct analysis of log2 ratios. Data for each probe were normalized by first subtracting the median value across all samples (so that the distribution of ratios for each probes was centered on zero), and then dividing by the variance across all samples (to correct for variation in the sensitivity of different probes to copy number variation). All CNV calls were confirmed by MLPA.
DeCODE
Illumina (San Diego, CA, USA) Human317, Human370, HumanHap550, Human610 and 1M BeadChips were used for CNV analysis. BeadStudio (version 2.0) was used to call genotypes, normalize the signal intensity data, and establish the log R ratio and B allele frequency at every SNP according to the standard Illumina protocols. All samples passed a standard SNP-based quality control procedure; all samples with a SNP call rate lower than 0·97. PennCNV63, a free, open-source tool, was used for copy number variation detection. The input data for PennCNV are log R ratio (LRR): a normalized measure of the total signal intensity for the two alleles of the SNP and B allele frequency (BAF): a normalized measure of the allelic intensity ratio of the two alleles. These values are derived with the help of control genotype clusters (HapMap samples), using the Illumina BeadStudio software. PennCNV employs a hidden Markov model (HMM) to analyze the LRR and BAF values across the genome. CNV calls are made, based on the probability of a given copy state at the current marker, as well as on the probability of observing a copy state change from the previous marker to the current one. PennCNV uses a built-in correction model for GC content66.
CoLaus
Data normalization and CNV calling was previously described in 13. Data normalization included allelic cross-talk calibration 67,68, intensity summarization using robust median average and correction for any PCR amplification bias. Wave effects were corrected by fitting a Loess function65. CNV calling was done using a Gaussian mixture model (GMM) 59, that fits four components (deletion, copy neutral, 1 and 2 additional copy) to CN ratios. The final copy number at each probe location is determined as the expected (dosage) copy number. The method has been validated by comparing test datasets with results from the CNAT 69 and CBS 60,61 algorithms and by replicating a subset of CoLaus subject on Illumina arrays. Only duplications found by both GMM and CBS were considered.
EGCUT
Genotypes were called by BeadStudio software GT module v3.1 or GenomeStudio GT v1.6 (Illumina Inc). Log R Ratio and B Allele Frequency (BAF) values produced by the BeadStudio were formatted for further CNV analysis and break-point mapping with Hidden Markov Model based softwares QuantiSNP (ver.1.1) 62 and PennCNV 70 or CNVPartition 2.4.4 (Illumina Inc). All analyses were carried out using the recommended settings, except changing EMiters to 25 and L to 1,000,000 in QuantiSNP. For PennCNV, the Estonian population specific SNP allele frequency data was used. All detected duplications were confirmed by quantitative PCR.
SHIP
Raw intensities were normalized using Affymetrix Power Tools (Affymetrix inc), CNV analysis was done using Birdseye from the Birdsuite software package64 and PennCNV63. PennCNV predictions with confidence score less than 10 were removed. Birdsuite predictions were filtered as in 21: CNVs were kept if their LOD score was greater than 10, length greater than 1kb and number of probes greater or equal to 5 and size per number of probes less than 10,000.
KORA F3/F4
Genotyping for KORA F3 was performed using Affymetrix 500K Array Set consisting of two chips (Sty I and Nsp I). The KORA F4 samples were genotyped with the Affymetrix Human SNP Array 6.0. For both studies genomic DNA from blood samples was used for analysis. Hybridisation of genomic DNA was done in accordance with the manufacturer’s standard recommendations. Genotyping was done in the Genome Analysis Centre (GAC) of the Helmholtz Centre Munich. Genotypes were determined using BRLMM clustering algorithm (Affymetrix 500K Array Set) and Birdseed2 clustering algorithm (Affymetrix Array 6.0). For quality control purposes, we applied a positive control and a negative control DNA every 48 (KORA F3) samples or 96 samples (KORA F4). On chip level only subjects with overall genotyping efficiencies of at least 93% were included. In addition the called gender had to agree with the gender in the KORA study database. After exclusions 1,644 individuals remained in KORA F3 and 1,814 in KORA F4 for further analysis.
MLPA analysis
We used Multiplex Ligation-dependent Probe Amplification (MLPA) to determine changes in the copy number of a region of around 2 Mb on chromosome 16p11.2. Briefly, we designed, using hg18, nine probes within the targeted region, one control probe outside the rearranged region and seven control probes targeting unique position in the genome (Supplementary Table S10). Assays were performed with MRC-Holland reagents according to the manufacturer’s protocol71. The analysis of the amplification products was performed by capillary electrophoresis in the DNA Analyzer 3730XL and using the GeneMapper software v3.7 (Applied Biosystems, Foster City, CA). The calculations were performed independently for each experiment: we first normalized the MLPA data to minimize the amount of experimental variations summing all the signal values of each control probe for each sample, and afterwards, dividing each signal value of each sample with the sum. Then, the normalized signal values were compared to signal values from all other samples in the same experiment dividing the normalized signal values by the average calculated from all the samples in the same experiment. The product of this calculation is termed dosage quotient (DQ). A calculated DQ value below 0.65 and above 1.25 was considered as copy-number loss and gain, respectively as described 72–74.
Short arm of chromosome 16 custom array-CGH
DNA samples were labelled with Cy3 and cohybridized with Cy5-labelled DNA from the CEPH cell line, GM12042, to custom-made Nimblegen arrays. These arrays contained 71,000 probes spread across the short arm of chromosome 16 from 22.0 to 32.7 Mb (at a median space of 45 bp between 27.5 and 31.0 Mb) and 1,000 control probes situated in invariable region of the X chromosome. DNA labelling, hybridization and washing were performed according to Nimblegen protocols. Scanning was performed using an Agilent G2565BA Microarray Scanner. Image processing, quality control and data extraction were performed using the Nimblescan software v.2.5.
Weight, height, BMI and head circumference (HC) Z-scores as a function of age
• Pediatric cases
Weight, height, BMI and HC Z-scores were determined for pediatric cases (0–18 years) using clinical growth charts specific to the country of origin. Children were ascertained from 9 different countries. If charts were only available in percentile, those measures were transformed in Z-scores (cf. Statistics).
For United-States and Canada, data from the Center for Disease Control and National Center for Health Statistics (CDC/NCHS) were used to calculate Z scores. 75.
For the French pediatric population we used French national growth charts 76, 77. For the Swiss pediatric population we used Swiss national growth charts 78. For Dutch participants Dutch national growth charts were available. 79 For Italian, German, Finnish, Austrian cases (n= 6) height, weight and BMI Z-scores were estimated using WHO growth charts. 80
To check for discrepancies generated by the use of different growth charts, height, weight and BMI Z-scores were recalculated using WHO growth charts, for all cases less than 5 years, regardless of origin (http://www.who.int/childgrowth/standards/en/80). Z-scores obtained using the WHO data were not significantly different. These growth standards, developed by the World Health Organization Multicentre Growth Reference Study, describe normal child growth from birth to 5 years under optimal environmental conditions. These standards can be applied to all children everywhere, regardless of ethnicity, socioeconomic status, and type of feeding 81, 82.
If needed, Percentile values were transformed to Z-scores by the inverse-normal density function. When growth chart were unavailable we utilized reported LMS parameters (median (M), generalized coefficient of variation (S), and skewness (L)) to obtain Z-scores via the formula:
in which X is the genuinely observed value.
• Adults (>18 years)
When LMS parameters were unavailable we estimated them from the available sex-, age-, and origin-matched Swiss- (CoLaus), Estonian-, or French control populations. For cases identified from population based cohorts, Z-scores were directly inferred from the cohort.
Gene expression
We established lymphoblastoid cell lines from deletion and duplication carriers, as well as controls (Supplementary Table S12), by transforming peripheral blood mononuclear cells with EBV. Patients and controls were enrolled after obtaining appropriate informed consent by the physicians in charge and approval by the ethics committee of the University of Lausanne. More control cell lines were obtained from Coriell Institute for Medical Research (http://www.coriell.org/) (Supplementary Table S12). SYBR Green real-time quantitative polymerase chain reaction was performed as published 29,83. Briefly, 1μg of total RNA from lymphoblastoid cell lines was converted to cDNA using Superscript VILO (Invitrogen) primed with a mix of oligo(dT) and random hexamers. Oligos were designed using the PrimerExpress program (Applied Biosystem) with default parameters (Supplementary Table S11). Non intron-spanning assays were tested in standard +/− RT reactions of RNA samples for genomic contamination. The amplification efficiency of each primer pair was tested in a cDNA dilution series as described 84. A full list of genes mapping within the rearranged interval and exclusion criteria are presented in Supplementary Table S1. All RT-PCR reactions were performed in a 10μl final volume and triplicates per sample. The set up in a 384 wells plate format was performed using a Freedom EVO robot (TECAN) and assays run in an ABI 7900 Sequence Detection System (Applied Biosystems) with the following amplification conditions: 50°C for 2 min, 95°C for 10 min, and 45 cycles of 95°C 15 sec/60°C for 1 min. A final incubation of 95°C for 15 sec followed by 60°C for 15 sec was carried out in order to establish a dissociation curve. Each plate included the appropriate normalization genes to control for any variability between the different plate runs. Raw threshold cycles (Ct) values were obtained using SDS2.4 (Applied Biosystems). In order to calculate the normalized relative expression ratio of individuals carrying the copy number variant and of controls, we used the Biogazelle qBase Plus software 85 including geNorm 86. This program identified appropriate normalization genes (EEF1A1, RPL13, GUSB and TBP) having a gene-stability measure of M=0.25. We note that, one gene, namely LAT, showed a startlingly high expression profile in one of the duplication samples (DASYL, Supplementary Table S13), reaching a relative expression value of 27.3 (SE=1.37), compared to an average expression for other duplications of 1.89 (SE=0.51). Whilst at this point we cannot exclude that this finding is genuine (although confirmed it in a second experiment), it was excluded from further analyses as an outlier in order to give a more accurate overview of expression profiles for these genes.
In silico analysis was performed to check for brain, and specifically hypothalamus, expression of genes within the rearranged 16p11.2 interval (Supplementary Table S1). This was done using Allen Brain Atlas Resources, Seattle (WA): Allen Institute for Brain Science. ©2009. Available from: http://www.brain-map.org.
Cases with significant neurological signs
Significant neurological signs were defined by (i) the presence of neurological signs such as severe hypotonia, hypertonia, ataxia, severe spasticity, hypereflexia, hyporeflexia and/or extra-pyramidal signs, (ii) the severity of the developmental delay (e.g. no speech at age 5 and/or severe gross motor delay in walking acquisition >24 months); and (iii) the presence of epilepsy. Mental retardation, autism, psychiatric symptoms, unspecified hypotonia and mild spasticity were not considered.
Statistics
T-test
One-sided t-test was performed to test whether duplication carriers have lower than zero BMI, height, weight Z-score values. We found this analysis more suitable than linear regression analysis correcting for confounding factors such as sex and age, because these anthropometric traits have a highly non-linear dependence on these factors as can be observed in control population.
Kruskal-Wallis test
We used Kruskal-Wallis to test differences in the gene expression pattern between deletion and duplication carriers and control individuals. Since expression values are not necessarily normally distributed, this test is more adequate than a classical one-way ANOVA. To test pairwise differences, we computed the difference in mean group rank with its 95% confidence interval (as provided by the multcompare function in Matlab). Correction for multiple testing issues was done using a Bonferroni adjustment.
Multiple testing
We determined false discovery rate (FDR) based association p value thresholds for each phenotype to correct for multiple testing. For each phenotype we replaced the observed Z-scores with numbers randomly drawn from a standard normal distribution and performed the same t-tests for the same strata. The procedure was repeated 1,000 times. For various p value thresholds we asked how many test would be declared significant for the null set on average (over the 1,000 random draws). FDR was estimated as the fraction of this number and the actual number we obtained for the observed Z-scores. With this approach we controlled the dependence between the nested tests we carried out.
Relative risk
Among adults, we defined (the risk of) being underweight as having BMI below 18.5. The estimated Relative Risk (RR) is the ratio of the fraction of underweight individuals among duplication carriers versus our control group. Underweight adults were defined by the WHO criteria (BMI ≤ 18.5). The standard error of log(RR) and its significance were calculated as described in 87. In our control group (population based cohorts), the frequency of underweight is 1.9% (38 males and 148 females out of 9470).
Supplementary Material
Acknowledgments
We thank the Vital-IT high-performance computing centre of the Swiss Institute of Bioinformatics. SJ is recipient of a « bourse de relève académique de la Faculté de Biologie et Medecine de l’Universite de Lausanne ». This work was supported by the Leenaards Foundation Prize (SJ, DM and AR), the Jérôme Lejeune Foundation (AR), the Telethon Action Suisse Foundation (AR), the Swiss National Science Foundation (AR, JSB, SB and SEA), a SNSF Sinergia grant (SJ, DM, SB, JSB and AR), the European Commission anEUploidy Integrated Project grant 037627 (AR, SB, XE, HGB and SEA), the Ludwig Institute for Cancer Research (AV), the Swiss Institute of Bioinformatics (SB, ZK), an Imperial College Dept of Medicine PhD studentship (JSe-SM), the Comprehensive Biomedical Research Centre, Imperial College Healthcare NHS Trust, and the National Institute for Health Research (PE), the Wellcome Trust and the Medical Research Council (AIFB and PF), the Instituto de Salud Carlos III (ISCIII)-FIS, the German Mental Retardation Network funded through a grant of the German Federal Ministry of Education and Research (NGFNplus 01GS08160) to A Reis and European Union-FEDER (PI081714, PS09/01778), SAF2008-02278 (XE, MG, FFA), the Belgian National Fund for Scientific Research - Flanders (NVA, RFK), the Dutch Organisation for Health Research and Development (ZON-MW grant 917-86-319) and Hersenstichting Nederland (BBAdV), grant 81000346 from the Chinese National Natural Science Foundation (YGY), the Simons Foundation Autism Research Initiative, Autism Speaks and NIH grant GM061354 (JFG), and the OENB grant 13059 (AK-B). YS holds a Young Investigator Award from the Children’s Tumor Foundation and Catalyst Award from Harvard Medical School, and BLW, a Fudan Scholar Research Award from Fudan University, a grant from Chinese National “973” project on Population and Health (2010CB529601) and a grant from Science and Technology Council of Shanghai (09JC1402400). ERS and SL, recipients of the Michael Smith Foundation for Health Research Scholar award, acknowledge the CIHR MOP 74502 operational grant. EGCUT received support from the EU Centre of Excellence in Genomics and FP7 grants #201413 and #245536, from Estonian Government SF0180142s08, SF0180026s09 and SF0180027s10 (AM, KM, AK). The Helmholtz Zentrum Munich and the State of Bavaria financed KORA, also supported by the German National Genome Research Network (NGFN-2 and NGFNPlus: 01GS0823), the German Federal Ministry of Education and Research (BMBF), and the Munich Center of Health Sciences (MC Health, LMUinnovativ). CIBEROBN and CIBERESP are initiatives of ISCIII (Spain). SWS holds the GlaxoSmithKline-Canadian Institutes of Health (CIHR) Chair in Genetics, Genomics at the University of Toronto and the Hospital for Sick Children and is supported by Genome Canada and the McLaughlin Centre. deCODE was funded in part by NIH grant MH071425 (KS), EU grant HEALTH-2007-2.2.1-10-223423 (Project PsychCNV) and EU grant IMI-JU-NewMeds. We thank for their help M. Hass, Z. Jaros, M. Jussila, M. Koiranen, P. Rantakallio, MC. Rudolf, V. Soo, O. Tornwall, S. Vaara, T. Ylitalo and the French DHOS national CGH network, as well as all participating patients and clinicians. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author contributions
SJ, AR, PF and JSB wrote the manuscript with contributions from FZ, LH, RGW, NDB, ZK, AIFB and AV. LH, AV and AR produced and analyzed the expression data. ZK, AV, RGW and NDB conducted the statistical analyses guided by SJ, AR, PF and JSB. SJ, AR, FZ, LH, DM, YS, GT, MB, SB, DC, NdL, BBAdV, BAF, FFA, MG, AG, JH, AK, ClC, KM, KO, OSP, DS, MMVH, SVG, ATVvS, FW, BLW, YY, JA, XE, JFG, AM, SWS, KS, UT, AIFB, JSB, PF and all other authors phenotyped and/or genotyped patients and/or individuals of the general population. SJ, AR and JSB designed the study. All authors commented on and approved the manuscript.
References
- 1.Berrington de Gonzalez A, et al. Body-mass index and mortality among 1.46 million white adults. N Engl J Med. 2010;363:2211–9. doi: 10.1056/NEJMoa1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flegal KM, Graubard BI, Williamson DF, Gail MH. Cause-specific excess deaths associated with underweight, overweight, and obesity. JAMA. 2007;298:2028–37. doi: 10.1001/jama.298.17.2028. [DOI] [PubMed] [Google Scholar]
- 3.Physical status: the use and interpretation of anthropometry. Report of a WHO Expert Committee. World Health Organization technical report series. 1995;854:1–452. [PubMed] [Google Scholar]
- 4.de Onis M, Blossner M, Borghi E, Frongillo EA, Morris R. Estimates of global prevalence of childhood underweight in 1990 and 2015. JAMA. 2004;291:2600–6. doi: 10.1001/jama.291.21.2600. [DOI] [PubMed] [Google Scholar]
- 5.Mei Z, et al. Validity of body mass index compared with other body-composition screening indexes for the assessment of body fatness in children and adolescents. The American journal of clinical nutrition. 2002;75:978–85. doi: 10.1093/ajcn/75.6.978. [DOI] [PubMed] [Google Scholar]
- 6.Olsen EM, et al. Failure to thrive: the prevalence and concurrence of anthropometric criteria in a general infant population. Archives of disease in childhood. 2007;92:109–14. doi: 10.1136/adc.2005.080333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corbett SS, Drewett RF. To what extent is failure to thrive in infancy associated with poorer cognitive development? A review and meta-analysis. J Child Psychol Psychiatry. 2004;45:641–54. doi: 10.1111/j.1469-7610.2004.00253.x. [DOI] [PubMed] [Google Scholar]
- 8.Rudolf MC, Logan S. What is the long term outcome for children who fail to thrive? A systematic review. Arch Dis Child. 2005;90:925–31. doi: 10.1136/adc.2004.050179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant-Waugh R, Markham L, Kreipe RE, Walsh BT. Feeding and eating disorders in childhood. The International journal of eating disorders. 2010;43:98–111. doi: 10.1002/eat.20795. [DOI] [PubMed] [Google Scholar]
- 10.APA. Diagnostic and Statistical Manual of Mental Disorders, DSM-IV-TR. American Psychiatric Association; Washington, DC: 2000. [Google Scholar]
- 11.Scherag S, Hebebrand J, Hinney A. Eating disorders: the current status of molecular genetic research. Eur Child Adolesc Psychiatry. 19:211–26. doi: 10.1007/s00787-009-0085-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, et al. A genome-wide association study on common SNPs and rare CNVs in anorexia nervosa. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.107. [DOI] [PubMed] [Google Scholar]
- 13.Walters RG, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–5. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bijlsma EK, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet. 2009;52:77–87. doi: 10.1016/j.ejmg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez BA, et al. Phenotypic Spectrum Associated with De Novo and Inherited Deletions and Duplications at 16p11.2 in Individuals Ascertained for Diagnosis of Autism Spectrum Disorder. J Med Genet. 2009 doi: 10.1136/jmg.2009.069369. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy SE, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41:1223–7. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 18.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–73. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall CR, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–88. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crespi B, Stead P, Elliot M. Evolution in health and medicine Sackler colloquium: Comparative genomics of autism and schizophrenia. Proc Natl Acad Sci U S A. 2010;107 (Suppl 1):1736–41. doi: 10.1073/pnas.0906080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bochukova EG, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–70. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firmann M, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabatti C, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelis M, et al. Genetic structure of Europeans: a view from the North-East. PLoS One. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Girirajan S, Eichler EE. Phenotypic variability and genetic susceptibility to genomic disorders. Hum Mol Genet. 2010;19:R176–87. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO. Report of a WHO Expert Committee. 1995. Physical status: the use and interpretation of anthropometry. [PubMed] [Google Scholar]
- 27.Pramyothin P, Khaodhiar L. Metabolic syndrome with the atypical antipsychotics. Curr Opin Endocrinol Diabetes Obes. 2010;17:460–6. doi: 10.1097/MED.0b013e32833de61c. [DOI] [PubMed] [Google Scholar]
- 28.Shinawi M, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioral problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2009 doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merla G, et al. Submicroscopic deletion in patients with Williams-Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet. 2006;79:332–41. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrichsen CN, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41:424–9. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 32.Ricard G, et al. Phenotypic consequences of copy number variation: insights from Smith-Magenis and Potocki-Lupski syndrome mouse models. PLoS Biol. 2010;8:e1000543. doi: 10.1371/journal.pbio.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crespi B, Badcock C. Psychosis and autism as diametrical disorders of the social brain. Behav Brain Sci. 2008;31:241–61. doi: 10.1017/S0140525X08004214. discussion 261–320. [DOI] [PubMed] [Google Scholar]
- 35.Baxter PS, Rigby AS, Rotsaert MH, Wright I. Acquired microcephaly: causes, patterns, motor and IQ effects, and associated growth changes. Pediatrics. 2009;124:590–5. doi: 10.1542/peds.2008-2784. [DOI] [PubMed] [Google Scholar]
- 36.Li Y, Dai Q, Jackson JC, Zhang J. Overweight is associated with decreased cognitive functioning among school-age children and adolescents. Obesity (Silver Spring) 2008;16:1809–15. doi: 10.1038/oby.2008.296. [DOI] [PubMed] [Google Scholar]
- 37.Guilmatre A, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–56. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnusson P, Smari J, Gretarsdottir H, Prandardottir H. Attention- Deficit/Hyperactivity symptoms in Icelandic schoolchildren: assessment with the Attention Deficit/Hyperactivity Rating Scale-IV. Scand J Psychol. 1999;40:301–6. doi: 10.1111/1467-9450.404130. [DOI] [PubMed] [Google Scholar]
- 39.Magnusson P, et al. Validity of self-report and informant rating scales of adult ADHD symptoms in comparison with a semistructured diagnostic interview. J Atten Disord. 2006;9:494–503. doi: 10.1177/1087054705283650. [DOI] [PubMed] [Google Scholar]
- 40.Spitzer RL, Endicott J, Robins E. Research diagnostic criteria: rationale and reliability. Arch Gen Psychiatry. 1978;35:773–82. doi: 10.1001/archpsyc.1978.01770300115013. [DOI] [PubMed] [Google Scholar]
- 41.Endicott J, Spitzer RL. A diagnostic interview: the schedule for affective disorders and schizophrenia. Arch Gen Psychiatry. 1978;35:837–44. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- 42.Williams NM, et al. Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet. 2010;376:1401–8. doi: 10.1016/S0140-6736(10)61109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.John U, et al. Study of Health In Pomerania (SHIP): a health examination survey in an east German region: objectives and design. Soz Praventivmed. 2001;46:186–94. doi: 10.1007/BF01324255. [DOI] [PubMed] [Google Scholar]
- 44.Volzke H, et al. Cohort Profile: The Study of Health in Pomerania. Int J Epidemiol. doi: 10.1093/ije/dyp394. [DOI] [PubMed] [Google Scholar]
- 45.Wichmann HE, Gieger C, Illig T. KORA-gen--resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67 (Suppl 1):S26–30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 46.Meyre D, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–9. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 47.Visvikis-Siest S, Siest G. The STANISLAS Cohort: a 10-year follow-up of supposed healthy families. Gene-environment interactions, reference values and evaluation of biomarkers in prevention of cardiovascular diseases. Clin Chem Lab Med. 2008;46:733–47. doi: 10.1515/CCLM.2008.178. [DOI] [PubMed] [Google Scholar]
- 48.Gubern C, et al. Natural antibiotics and insulin sensitivity: the role of bactericidal/permeability-increasing protein. Diabetes. 2006;55:216–24. [PubMed] [Google Scholar]
- 49.Lopez-Bermejo A, et al. Alpha defensins 1, 2, and 3: potential roles in dyslipidemia and vascular dysfunction in humans. Arterioscler Thromb Vasc Biol. 2007;27:1166–71. doi: 10.1161/ATVBAHA.106.138594. [DOI] [PubMed] [Google Scholar]
- 50.Moreno-Navarrete JM, et al. Complement factor H is expressed in adipose tissue in association with insulin resistance. Diabetes. 2010;59:200–9. doi: 10.2337/db09-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreno-Navarrete JM, et al. Association of circulating lactoferrin concentration and 2 nonsynonymous LTF gene polymorphisms with dyslipidemia in men depends on glucose-tolerance status. Clin Chem. 2008;54:301–9. doi: 10.1373/clinchem.2007.095943. [DOI] [PubMed] [Google Scholar]
- 52.Jernas M, et al. Regulation of carboxylesterase 1 (CES1) in human adipose tissue. Biochem Biophys Res Commun. 2009;383:63–7. doi: 10.1016/j.bbrc.2009.03.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tinahones FJ, et al. Obesity and insulin resistance-related changes in the expression of lipogenic and lipolytic genes in morbidly obese subjects. Obes Surg. 2010;20:1559–67. doi: 10.1007/s11695-010-0194-z. [DOI] [PubMed] [Google Scholar]
- 54.First MB, Spitzer RL, Williams JBW. Structured Clinical Interview for DSM-IV (SCID-I): Users Guide and Interview, Research Version. Biometrics Research Department, New York Psychiatric Institute; New York: 1995. [Google Scholar]
- 55.Gabrovsek M, et al. Combined family trio and case-control analysis of the COMT Val158Met polymorphism in European patients with anorexia nervosa. Am J Med Genet B Neuropsychiatr Genet. 2004;124B:68–72. doi: 10.1002/ajmg.b.20085. [DOI] [PubMed] [Google Scholar]
- 56.Mercader JM, et al. Altered brain-derived neurotrophic factor blood levels and gene variability are associated with anorexia and bulimia. Genes Brain Behav. 2007;6:706–16. doi: 10.1111/j.1601-183X.2007.00301.x. [DOI] [PubMed] [Google Scholar]
- 57.Ribases M, et al. Association of BDNF with restricting anorexia nervosa and minimum body mass index: a family-based association study of eight European populations. Eur J Hum Genet. 2005;13:428–34. doi: 10.1038/sj.ejhg.5201351. [DOI] [PubMed] [Google Scholar]
- 58.Coin LJ, et al. cnvHap: an integrative population and haplotype-based multiplatform model of SNPs and CNVs. Nat Methods. 7:541–6. doi: 10.1038/nmeth.1466. [DOI] [PubMed] [Google Scholar]
- 59.Valsesia A, et al. Identification and validation of Copy Number Variants using SNP genotyping arrays from a large clinical cohort. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 61.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 62.Colella S, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–25. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korn JM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–60. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marioni JC, et al. Breaking the waves: improved detection of copy number variation from microarray-based comparative genomic hybridization. Genome Biol. 2007;8:R228. doi: 10.1186/gb-2007-8-10-r228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diskin SJ, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36:e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bengtsson H, Irizarry R, Carvalho B, Speed TP. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics. 2008;24:759–67. doi: 10.1093/bioinformatics/btn016. [DOI] [PubMed] [Google Scholar]
- 68.Bengtsson H, Ray A, Spellman P, Speed TP. A single-sample method for normalizing and combining full-resolution copy numbers from multiple platforms, labs and analysis methods. Bioinformatics. 2009;25:861–7. doi: 10.1093/bioinformatics/btp074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang J, et al. Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum Genomics. 2004;1:287–99. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schouten JP, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bunyan DJ, et al. Dosage analysis of cancer predisposition genes by multiplex ligation-dependent probe amplification. Br J Cancer. 2004;91:1155–9. doi: 10.1038/sj.bjc.6602121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fernandez L, et al. Comparative study of three diagnostic approaches (FISH, STRs and MLPA) in 30 patients with 22q11.2 deletion syndrome. Clin Genet. 2005;68:373–8. doi: 10.1111/j.1399-0004.2005.00493.x. [DOI] [PubMed] [Google Scholar]
- 74.Slater HR, et al. Rapid, high throughput prenatal detection of aneuploidy using a novel quantitative method (MLPA) J Med Genet. 2003;40:907–12. doi: 10.1136/jmg.40.12.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuczmarski RJ, et al. CDC growth charts: United States. Adv Data. 2000:1–27. [PubMed] [Google Scholar]
- 76.Sempé M, Pedron G, Roy-Pernot MP. Auxologie, méthode et séquences. Vol. 205. Paris: Théraplix; 1979. [Google Scholar]
- 77.Rolland-Cachera MF, et al. Body Mass Index variations: centiles from birth to 87 years. Eur J Clin Nutr. 1991;45:13–21. [PubMed] [Google Scholar]
- 78.Prader A, Largo RH, Molinari L, Issler C. Physical growth of Swiss children from birth to 20 years of age. First Zurich longitudinal study of growth and development. Helv Paediatr Acta Suppl. 1989;52:1–125. [PubMed] [Google Scholar]
- 79.Growth diagrams. Fourth dutch nation-wide survey. Bohn Stafleu Van Loghum; 1997. [Google Scholar]
- 80.de Onis M, et al. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ. 2007;85:660–7. doi: 10.2471/BLT.07.043497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Onis M, Garza C, Onyango AW, Borghi E. Comparison of the WHO child growth standards and the CDC 2000 growth charts. J Nutr. 2007;137:144–8. doi: 10.1093/jn/137.1.144. [DOI] [PubMed] [Google Scholar]
- 82.Mei Z, Ogden CL, Flegal KM, Grummer-Strawn LM. Comparison of the prevalence of shortness, underweight, and overweight among US children aged 0 to 59 months by using the CDC 2000 and the WHO 2006 growth charts. J Pediatr. 2008;153:622–8. doi: 10.1016/j.jpeds.2008.05.048. [DOI] [PubMed] [Google Scholar]
- 83.Molina J, et al. Abnormal social behaviors and altered gene expression rates in a mouse model for Potocki-Lupski syndrome. Hum Mol Genet. 2008;17:2486–95. doi: 10.1093/hmg/ddn148. [DOI] [PubMed] [Google Scholar]
- 84.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 85.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morris JA, Gardner MJ. Calculating confidence intervals for relative risks (odds ratios) and standardised ratios and rates. Br Med J (Clin Res Ed) 1988;296:1313–6. doi: 10.1136/bmj.296.6632.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


