Abstract
Deleted in AZoospermia Associated Protein 1 (DAZAP1) is a ubiquitous hnRNP protein that has been implicated in RNA transcription, splicing, and translation. It is highly expressed in testes, predominantly in late stage spermatocytes and post-meiotic spermatids. Dazap1 deficiency in mice results in growth retardation and spermatogenic arrest. The gene produces two major transcripts of 2.4 and 1.8 kb, designated Dazap1-L and Dazap1-S, respectively. Results of our previous RNA in situ hybridization and immunostaining suggested translational regulation of the Dazap1 transcripts during spermatogenesis. The main objectives of the study were to determine the origin of the two Dazap1 transcripts and to investigate whether they were similarly translated. Our Northern and 3′ RACE analyses showed that the two transcripts were generated through alternative polyadenylation. In mouse testes, the levels of both transcripts were low at postnatal day 12 (P12), increased significantly at P18, and reached maximum at P27. Sucrose gradient analyses showed that at P12 both transcripts were actively translated. Afterward, an increasing portion of Dazap1-S became associated with the translationally inactive mRNPs, and the translational repression was accompanied by an increase in the length of its poly(A) tail. A much smaller portion of Dazap1-L was also sequestered to mRNPs as testes matured, but there was no changes in its poly(A) tail length. Using RNA pull-down followed by mass spectrometry, we identified DAZL, a germ-cell specific translation regulator, as one of the proteins that bound to the 3′UTR region specific for Dazap1-L. We further showed that DAZL preferentially bound to Dazap1-L in testis lysates and stimulated the translation of a reporter gene carrying Dazap1-L 3′UTR. In summary, our study shows that the translation of the two Dazap1 transcripts is differentially regulated. It also provides a new example of translational repression associated with poly(A) tail elongation during spermatogenesis.
Introduction
Most eukaryotic mRNAs are translated soon after they are processed and exported to the cytoplasm. However, some mRNAs are stored in the cytoplasm as inert messenger ribonucleoprotein particles (mRNPs) for a substantial period of time before they are translated. Such translation repression or mRNA masking is most frequently observed during gametogenesis and early embryogenesis [1]. A classic example of translational regulation during spermatogenesis is the Prm1 gene (encoding protamine 1) which is transcribed in round spermatids and translated several days later in elongated spermatids [2]. Storage of Prm1 mRNA for later translation is a necessary step for male germ cell development because protamine is needed at a stage when the nuclear transcription has been shut down. Premature translation of Prm1 mRNA in round spermatids results in precocious condensation of nuclear DNA and dominant male sterility [3]. Translational repression of Prm1 mRNA is mediated through its 3′UTR, and several proteins that bind to Prm1 3′UTR have been identified although how they regulate the translation remains unclear [4], [5], [6], [7]. Additional patterns of gene repression are also observed in spermatogenic cells [8]. Some mRNAs appear to be partially repressed at all time, while others are almost totally inactivated in free mRNPs and generate little proteins. Recent profiling of testicular mRNAs in polyribosomes (polysomes) and mRNP fractions of prepuberal and adult mice identified over seven hundreds of transcripts that are differentially translated during testis development [9].
Deleted in AZoospermia (DAZ) proteins are a family of RNA-binding proteins that regulate mRNA translation in germ cells [10]. They include DAZ, a spermatogenic factor encoded by the AZFc region of the Y chromosome that is frequently deleted in azoospermic men [11], and the autosomal encoded DAZL and BOULE [12], [13], [14]. DAZ proteins are expressed predominantly, if not exclusively, in germ cells and their deficiencies impair reproduction [15], [16]. A role of DAZ proteins in mRNA translation is supported by their presence on polysomes [17], [18] and their effects on translation in cultured cells [18], intact oocytes [19], [20] and zebrafish embryos [21]. A recent study using tethered function assay in Xenopus oocytes suggests that DAZL facilitates translation initiation by binding to the 3′UTR of its target mRNAs to recruit poly(A) binding proteins (PABPs) and stimulate the formation of 80 S ribosomes [19]. DAZL recognizes GUU triplet in different sequence context [18], [22], [23], and several putative natural substrates of DAZL have been identified though some of them need further verification in vivo [20], [24], [25], [26].
DAZ Associated Protein 1 (DAZAP1) is a DAZ interacting protein that is expressed ubiquitously but most abundantly in the testis [27], [28]. It is a component of the hnRNP particles [29] and has been implicated in several cellular processes including RNA transcription [30], [31], splicing [32], [33], [34], and translation [35]. Very recently, DAZAP1 was suggested as a potential disease gene for amyotrophic lateral sclerosis [36]. Most mice deficient in DAZAP1 die perinatally, and the few survivors manifest growth retardation and spermatogenic arrest [37]. In mouse testes, DAZAP1 first appears abundantly in the nuclei of mid-pachytene spermatocytes, whereas our previous RNA in situ hybridization detected high levels of Dazap1 transcripts in spermatogonia and early spermatocytes [38]. We thus set out to investigate possible delayed translation of Dazap1 transcripts during spermatogenesis. Our results showed that two Dazap1 transcripts were generated through alternative polyadenylation and their differential translation during spermatogenesis involved poly(A) tail length alteration and DAZL binding.
Results
Two Dazap1 transcripts are generated through alternative polyadenylation
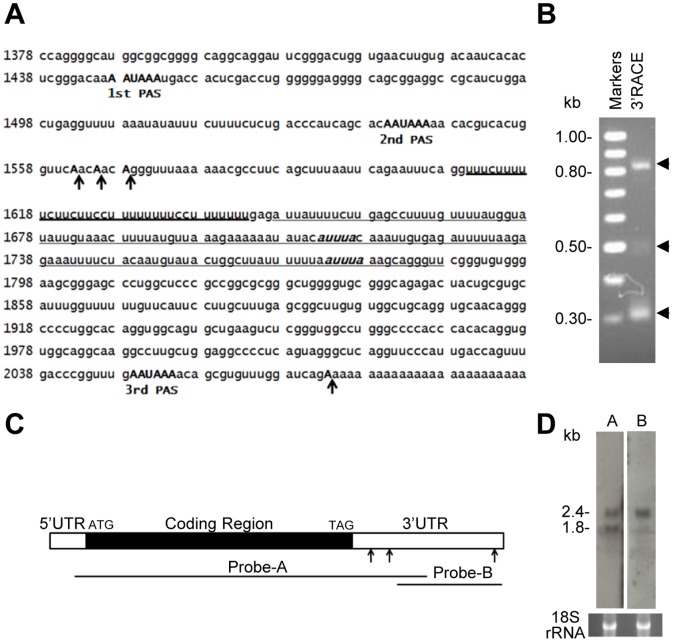
Mouse testes express two major Dazap1 transcripts of 2.4 and 1.8 kb, designated Dazap1-L and Dazap1-S, respectively [28]. We failed to detect alternative splicing and promoter usage by RT-PCR and 5′ Rapid Amplification of cDNA Ends (RACE), respectively, suggesting that the two Dazap1 transcripts were generated through alternative polyadenylation (data not shown). The 3′UTR of Dazap1 contains three canonical AATAAA polyadenylation signals (PASs) (Fig. 1A), but only the second and the third ones are evolutionally conserved throughout vertebrates (data not shown). To identify the polyadenylation sites of Dazap1 transcripts, we performed 3′ RACE of adult mouse testis RNA and detected two major fragments around 0.8 and 0.3 kb (Fig. 1B). Sequencing of the fragments revealed that polyadenylation of the 0.3 kb fragment initiated from three CA sites downstream of the second PAS, whereas that of the 0.8 kb product started downstream of the third PAS (Fig. 1A). The minor 0.5 kb fragment represented a PCR artifact amplified from the 0.8 kb fragment since it was also produced by PCR amplification of a clone containing the 0.8 kb fragment. Thus the two Dazap1 transcripts were produced through alternative polyadenylation utilizing the second and the third PASs. This was further confirmed by Northern blot (Fig. 1C, D). Probe-A which covers a large portion of the Dazap1 cDNA detected both transcripts, whereas probe-B which contains a segment downstream of the second PAS hybridized only to Dazap1-L.
Figure 1. Alternative polyadenylation of the mouse Dazap1 transcripts.
(A) Sequence of the Dazap1 3′ UTR (NM_133188, nt1378–2093). The polyadenylation signals (PASs) are in bold capital letters, and the polyadenylation sites determined by 3′RACE are marked with arrows. The CU-rich region is indicated with heavy underline, and the AU-rich region with two AUUUA motifs (in bold italic) is indicated with light underline. (B) 3′RACE products of Dazap1 transcripts in adult mouse testes. (C) Diagram of the Dazap1 cDNA. The locations of Northern hybridization probes are indicated. The arrows point to the three PASs. (D) Northern analyses of Dazap1 transcripts in adult mouse testes. Ethidium bromide (EtBr) staining of the 18 S rRNA is shown at bottom as a loading control.
The Dazap1 transcripts are differentially expressed during spermatogenesis
We next studied the levels of the two Dazap1 transcripts in prepuberal and adult mouse testes. Spermatogenesis is initiated soon after birth, and day 8 mouse testes contain only type A and type B spermatogonia [39]. Spermatocytes start to appear afterward. In postnatal day 12 (P12) testes, germ cells make up 60% of the total cell population and the most advanced germ cell is zygotene spermatocytes. At P18, pachytene spermatocytes dominate the germ cell population, and haploid spermatids start to appear after day 20. P27 testes are enriched in late round spermatids [40], and P34 and adult testes contain spermatids as the major cell types. Northern analyses of mouse testes at different ages showed that the levels of Dazap1 transcripts were low at P12, increased significantly at P18, and reached the highest levels at P27 (Fig. 2A, B), consistent with Dazap1 expression mainly in late pachytene spermatocytes and spermatids. The ratio of the two transcripts also changed as spermatogenesis progresses (Fig. 2B). The two transcripts were present in similar levels at P12 and P18, but Dazap1-S became the dominant form at P27 and after.
Figure 2. Dazap1 expression during mouse testicular development.
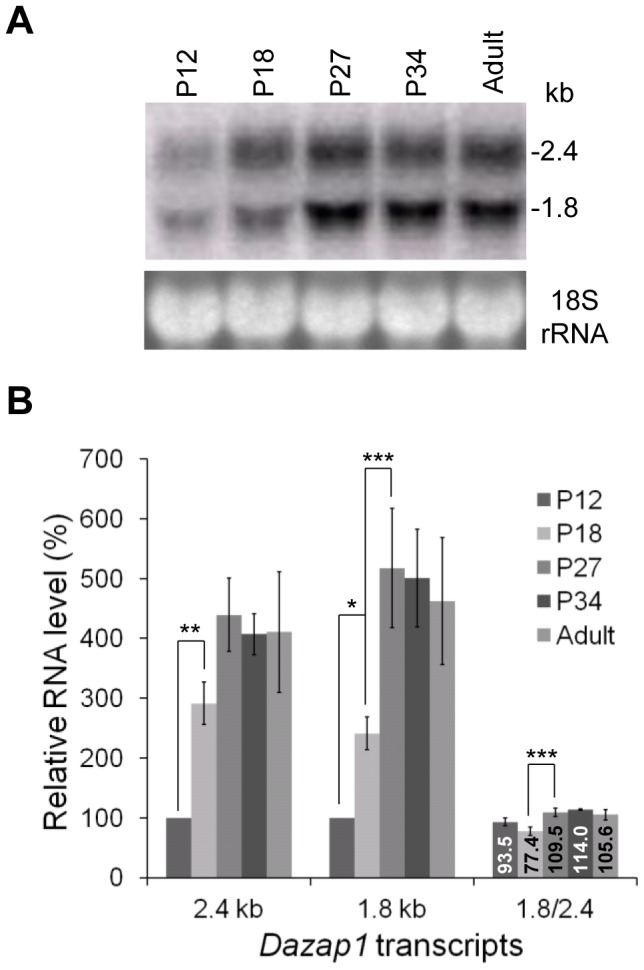
(A) Northern blot of total RNAs isolated from mouse testes at different postnatal ages. EtBr staining of the 18 S rRNA is shown as a loading control. (B) Quantification of the Northern blot signals. Radioactive bands on the hybridization members were excised and counted. Signals at P12 were used as the reference (100%). The results represent the averages of three independent experiments. Statistical significance of the differences was determined using one-way ANOVA. * denotes p<0.05, ** denotes p<0.01, *** denotes p<0.001. The relative levels (%) between 1.8 and 2.4 kb transcripts are given inside the bars.
The Dazap1 transcripts are differentially translated in the testis
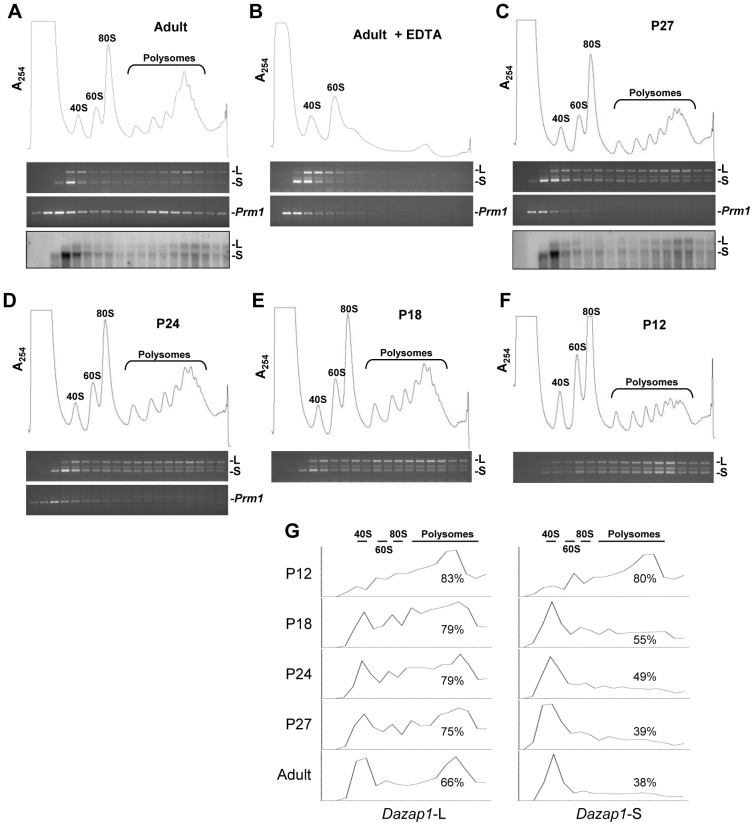
To investigate whether both Dazap1 transcripts are equally translated during spermatogenesis, we fractionated mouse testis lysates on 10–50% sucrose gradients and monitored the distribution of Dazap1 transcripts by 3′RACE as well as Northern blot when Dazap1 transcripts were sufficiently abundant for detection (Fig. 3). Both methods showed that in adult testes Dazap1-L had a bimodal distribution, with a large fraction associated with the polysomes, whereas a vast majority of Dazap1-S was present near the top of the gradient in the mRNP fraction (Figs. 3A, G). As a control, we also determined the distribution of the Prm1 transcript and found it in both mRNP and polysomal fractions as expected [9]. Upon EDTA treatment, the polysomes dissociated and all Dazap1 and Prm1 transcripts were found near the top of the gradient (Fig. 3B). The polysomal profiles of Dazap1 mRNAs in P27, P24, and P18 testes were similar to that of the adult testes, with a large fraction of Dazap1-S near the top of the gradients and most Dazap1-L associated with polysomes (Figs. 3C, D, E; G). On the other hand, the Prm1 transcripts in P27 and P24 testes were present only in the mRNP fractions because the most advanced spermatogenic cells in these testes are round spermatids. At P18 and P12, Prm1 transcripts were undetectable. There was a major change in the distribution of Dazap1 transcripts in P12 testes, with both transcripts mainly associated with polysomes being actively translated (Figs. 3F, G). Since the major difference in germ cell constitution between P12 and P18 testes is the appearance of pachytene spermatocytes in the latter, our results suggest that after the meiotic germ cells reach the pachytene stage, a large fraction of Dazap1-S and a much smaller fraction of Dazap1-L start to be sequestered to mRNPs and become translationally inactive.
Figure 3. Sucrose gradient analyses of Dazap1 transcripts in mouse testis lysates.
Testis lysates were prepared from adult (A , B), P27 (C), P24 (D), P18 (E), and P12 (F) mice and fractionated on 10–50% sucrose gradients. The polysome profiles determined by optical absorbance at 254 nm (A254) are shown at top. Dazap1-L (L) and Dazap1-S (S) transcripts in the fractions were detected by 3′RACE (top gel panels) and Northern hybridization (bottom panels in A and C), and the Prm1 transcript was detected by RT-PCR. Prm1 expression in P18 and P12 testes was undetectable. Quantification of the 3′RACE signals is shown in (G), with the percentage of total signals in the 80 S and polysomal fractions indicated.
Translationally repressed Dazap1-S has longer poly(A) tails
It has been reported that mouse Prm1, Prm2, Tnp1, and Tnp2 transcripts being actively translated on polysomes have shorter Poly(A) tails than those that are translationally repressed [41]. To investigate whether translational regulation of Dazap1 transcripts also involved changes in poly(A) tail length, we used a recently developed extension Poly(A) Test (ePAT) method [42] to monitor poly(A) tail lengths of Dazap1 transcripts during mouse testis development and in sucrose-gradient fractions. Our initial test of ePAT on Prm1 transcripts confirmed previous Northern hybridization results, i.e., translationally active polysomal Prm1 transcripts had much shorter poly(A) tails than the major Prm1 transcript present in the free mRNP fraction (Fig. 4A and Fig. S1). It is noticed that a small fraction of Prm1 transcripts in the mRNPs contained short poly(A) tails. Based on the size of the TVN-PAT product that had an invariant poly(A) tail of 12 As as well as the sequences of five or more randomly selected ePAT clones (Table S1), the lengths of the poly(A) tails in the short and long Prm1 transcripts were determined to be approximately 25 and 120 bases, respectively, compared with previous estimations of 30 and 150 bases [41]. We also found that when Prm1 transcripts were first detected in P27 testes, the vast majority of them had long poly(A) tails (designated as LATs). As the testis matured to contain later stages of germ cells with active Prm1 translation, the fraction of Prm1 transcripts with short poly(A) tails (designated as SATs) increased progressively, consistent with translation activation of Prm1 in elongated spermatids.
Figure 4. Developmental changes in the lengths of poly(A) tails of Dazap1 and Prm1 transcripts.
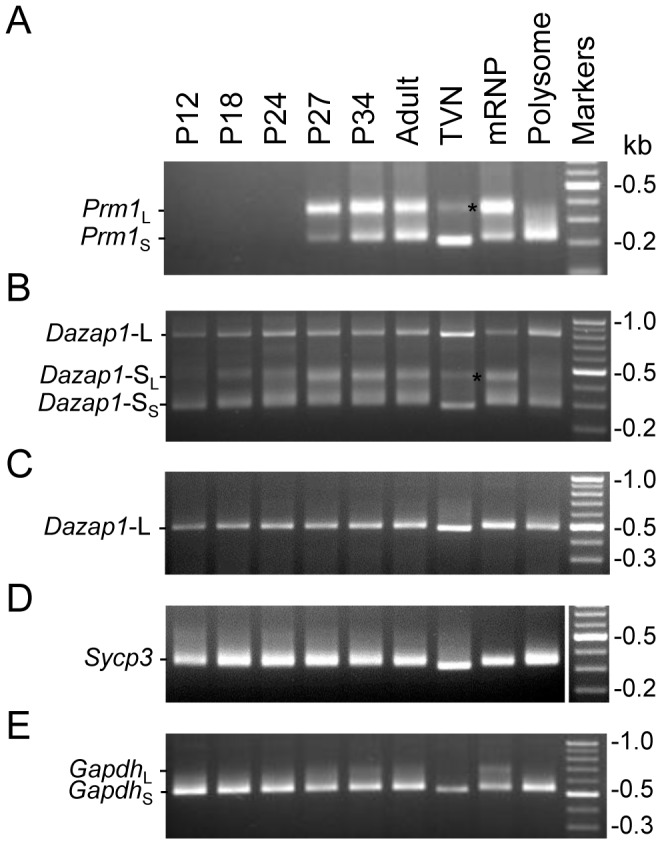
Total testicular RNAs isolated from mice at the various ages and RNAs extracted from the free mRNP and polysomal fractions of sucrose gradients of adult testis lysates were subjected to ePAT analysis using forward primers specific for the various genes as indicated. In the TVN lanes, the major products contain an invariant 12-(A) poly(A) tail whereas the minor products, marked with asterisks, likely came from nonspecific amplification. Transcripts with long and short poly(A) tails are indicated with L and S subscript, respectively.
We next used a forward primer common to both Dazap1 transcripts in the e-PAT reactions (Fig. 4B and Fig. S1). Dazap1-S also produced two major ePAT products with poly(A) tails approximately 30 and 170 bases long, respectively (Table S1). Before P27 most Dazap1-S had SATs. However, the level of Dazap1-S with LATs increased progressively as the mice grew older, and reached a level comparable to that of Dazap1-S with SATs at P27. Similar to the Prm1 transcripts, Dazap1-S with both SATs and LATs were present in the translationally inactive mRNP fraction whereas only Dazap1-S with SATs was present in the polysomal fraction. In contrast, the poly(A) tail length of Dazap1-L remained unchanged during testis development. The tail was determined to be approximately 30 bases long using a different Dazap1-L specific forward primer that produced shorter ePAT fragments (Fig. 4C and Table S1). Dazap1-L in both mRNP and polysomal fractions had the same SATs. Additional ePAT reactions showed that the germ-cell specific Sycp3 transcript also had constant SATs of approximately 30 bases (Fig. 4D and Table S1). On the other hand, the house-keeping Gapdh transcript produced a minor band representing LATs in the mRNP fraction, suggesting possible translation repression on a small fraction associated with poly(A) tail elongation (Fig. 4E). Our results therefore showed a correlation between poly(A) tail length and translational status for a subset of transcripts in the testis, with LATs of 120∼170 bases associated with translation repression.
Several RNA-binding proteins bind to Dazap1 3′UTR
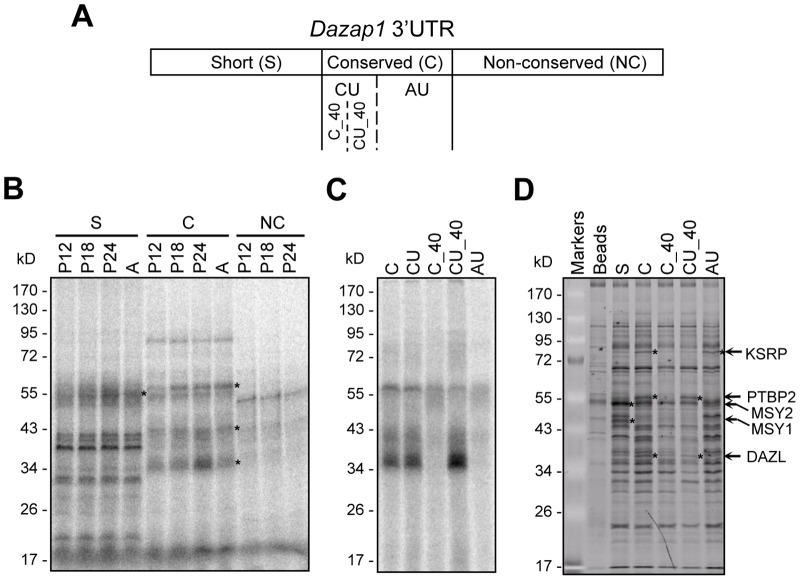
3′UTRs are known to contain cis-acting elements for the binding of translational regulators. We therefore searched for testicular proteins that bound to Dazap1 3′UTR. We separated Dazap1 3′UTR into three regions (Fig. 5A). The short (S) region (nt1378–1561) consists of the short 3′ UTR present in Dazap1-S; the conserved (C) (nt1568–1793) and the non-conserved (NC) (nt1788–2072) regions include the 5′ and 3′ segments, respectively, of the distal portion of Dazap1 3′UTR that is present only in Dazap1-L. RNA fragments corresponding to these regions were incubated with testis lysates prepared from mice at various ages, and the RNA-protein complexes were UV cross-linked and analyzed by SDS-PAGE (Fig. 5B). Probe S and probe C, but not probe NC, detected multiple proteins, and some of them showed increased binding during testis development.
Figure 5. Identification of mouse testicular proteins binding to Dazap1 3′UTR.
(A) Diagram of Dazap1 3′UTR showing the locations of the various RNA probes used in B-D. (B) UV cross-linking of 32P-labeled RNA probes from various regions of Dazap1 3′UTR and testicular proteins from mice at different ages. Asterisks point to proteins showing increasing expression during testis development. (C) UV cross-linking of probes from the C region and adult testis lysates. (D) RNA affinity pulldown assays of Dazap1 3′UTR fragments. Adult testis lysates were incubated with biotin-labeled RNAs and affinity purified by streptavidin-agarose beads. Bound samples were separated by 10% SDS-PAGE and proteins with specific binding were identified by LC/MS/MS spectrometry.
We further separated the C region into the CU region (nt1568–1646) that contains a noticeable CU-rich sequence and the AU region (nt1644–1793) that contains several AU-rich elements (AREs), and dissected the CU region again into C_40 (not CU-rich, nt1568–1610) and CU_40 (nt1607–1646) subregions (Figs. 1A, 5A). UV cross-linking assays showed that most proteins detected by probe C bound to the CU_40 region (Fig. 5C).
We next performed RNA affinity pull-down assays followed by LC/MS/MS spectrometry to identify proteins that bound specifically to Dazap1 3′UTR. Using this approach we found several RNA-bind proteins, including MSY1 (Mouse Y box protein 1) and MSY2 (Mouse Y box protein 2) which bound to the S region, and KSRP (KH-type Splicing Regulatory Protein), PTBP2 (Polypyrimidine Tract-Binding Protein 2), and DAZL (Deleted in AZoospermia-Like) which bound to the C region (Fig. 5D). Additional pull-down experiments with smaller probes in the C-region showed that KSRP bound to the AU region whereas PTBP2 and DAZL bound to the CU_40 region.
DAZL preferentially binds to Dazap1-L
DAZL has been reported to regulate the translation of several mRNAs in germ cells [10], and DAZL consensus binding sequences are present in different regions of Dazap1 3′UTR. We therefore performed RNA gel-shift assays using His-tagged recombinant DAZL to confirm the binding of DAZL to Dazap1 3′UTR (Fig. 6). We first mapped the DAZL binding site to the C region that is present only in Dazap1-L, and used smaller probes to narrow down the binding site to the CU_40 region which contains three partially overlapping copies of the DAZL consensus sequence (Fig. 6A). The binding between labeled CU_40 and DAZL could be competed out by increasing levels of cold CU_40 but not cold C_40, confirming the specificity of the binding (Fig. 6B).
Figure 6. Specific binding of DAZL to the CU rich region in Dazap1 3′UTR.
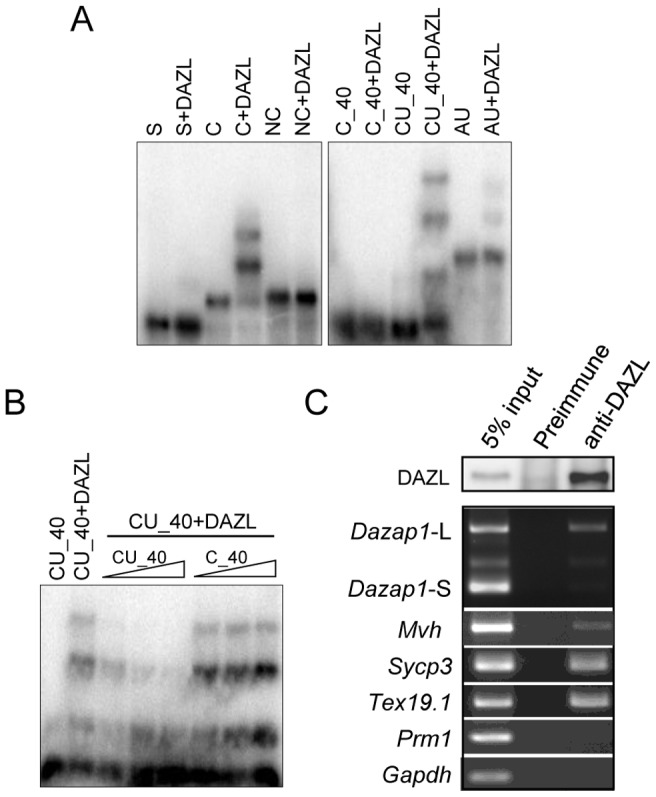
(A) RNA gel-shift assays. His-tagged DAZL was incubated with 32P-labeled RNA probes corresponding to various regions of Dazap1 3′UTR and analyzed on 1.5% agarose gels. DAZL binds to the CU_40 segment within the C region. (B) Competition of the binding between CU_40 and DAZL by increasing amounts (1, 5, 10 ng) of unlabeled RNA probes. (C) Preferential binding of DAZL to Dazap1-L in mouse testes. Adult mouse testes lysates were treated with a preimmune serum or an anti-DAZL antibody. The DAZL protein in the immunoprecipitates was detected by western blot with an anti-DAZL antibody (top panel). Dazap1-L and Dazap1-S in the precipitated RNAs were detected by 3′RACE, and the remaining transcripts were detected by RT-PCR with gene-specific primers (bottom panel).
DAZL enhances the translation of a reporter with Dazap1–L 3′UTR
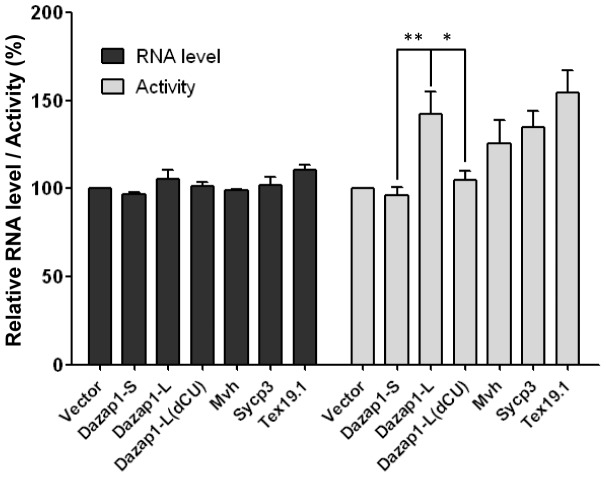
We used luciferase reporter assays to indirectly study the effect of DAZL on the translation of Dazap1 transcripts. We transfected 3T3 cells with a DAZL expression vector or the empty expression vector, together with the empty luciferase (Luc) reporter vector or a vector with Luc linked to the 3′UTRs of various genes. Western and RT-PCR analyses showed no significant differences in the levels of DAZL (Fig. S2) and Luc mRNA (Fig. 7) in the cells transfected with the DAZL expression vector and different Luc reporters. However, cells transfected with the reporter linked to the 3′UTR of Dazap1-L exhibited significantly higher luciferase activities than those transfected with the empty vector or the reporter linked to the 3′UTR of Dazap1-S (Fig. 7). Deletion of the entire CU-rich sequence from the Dazap1-L 3′UTR (Dazap1-L-dCU) abolished the translation enhancement. The degree of DAZL-induced translation enhancement of Dazap1-L was comparable to those of the reporters linked to the 3′UTRs of Mvh, Sycp3, and Tex19.1, which were much lower than what had previously been observed in other systems. DAZL was reported to increase approximately four-folds the translation of Mvh and Sycp3 luciferase reporters in Xenopus oocytes [25], [26] and about 10 folds that of a Tex19.1 reporter in mouse oocytes [20]. Differences in the expression levels of DAZL and other trans-acting factors in the various systems may account for the disparity in the observed translational enhancement.
Figure 7. Enhancement of the translation of luciferase reporters by DAZL.
3T3 cells were transfected with a DAZL expression vector together with the empty Luc reporter vector (vector), or reporter vectors containing the 3′UTRs of the various transcripts. In Dazap1-L-dCU, the CU rich segment in the 3′UTR is deleted. The effects of DAZL on the luciferase activities in the transfected cells were determined. The Luc mRNA level and luciferase activity in cells transfected with the empty Luc vector were used as the reference (100%). The results shown represent the averages of three independent experiments. Statistical significance of the differences was determined using one-way ANOVA. * denotes p<0.05, ** denotes p<0.01.
Discussion
Alternative polyadenylation changes the content of the 3′UTR and offers a means to regulate gene expression at the post-transcriptional stage because most regulatory elements controlling the localization, stability and translation of a transcript are located in the 3′UTR [43]. Here we show that two Dazap1 transcripts were generated through alternative polyadenylation, and that they were differentially expressed and translated during spermatogenesis. In P12 testes that contain spermatogonia and spermatocytes up to the zygotene stage, the transcripts were present at low levels and were both actively translated. However, with increased Dazap1 expression at P18, there was a major shift in the polysomal profile of Dazap1-S, with an increasing portion of the transcript sequestered to translationally inactive mRNPs, whereas a large portion of Dazap1-L remained on polysomes. In P27 testes that contain mainly postmeiotic spermatids, the level of Dazap1-S increased significantly, consistent with previous EST analyses showing that round spermatids tend to have shorter truncated 3′UTRs [44]. Since most of the increased Dazap1-S was present in mRNPs, no increase in protein synthesis was expected and our previous immunostaining of mouse testicular sections also failed to detect significant increase in DAZAP1 signals in spermatids [38]. The situation is similar to that of the Rnf4 transcripts encoding SNURF/RNF4 transcription modulator [45]. Rnf4 has two transcripts of 3.0 and 1.6 kb that differ in their 3′UTRs. The 1.6 kb transcript is expressed abundantly in round spermatids but most of them are not associated with polysomes. It is unclear why germ cells spend their energy and resources to make mRNAs that are not used for protein production.
The Northern analyses on Dazap1 expression in prepuberal testes, presented here, suggest that the expression of Dazap1 expression in spermatogonia and early spermatocytes is much lower than that in pachytene spermatocytes. This contradicts the results of our previous RNA in situ hybridization on adult mouse testicular sections which showed highest levels of Dazap1 mRNA in spermatogonia and early preleptotene spermatocytes [38]. We have since repeated the in situ experiments and were able to reproduce the same hybridization pattern with the same 5′ probe, but not with a probe from a different region of the Dazap1 cDNA. We also purified different stage germ cells from adult mouse testes and found the highest levels of Dazap1 transcripts in spermatocytes (data not shown). In addition, previous microarray analyses on purified spermatogenic cells also showed almost three times as much Dazap1 transcripts in pachytene spermatocytes as in type A and type B spermatogonia [46]. Therefore our previous RNA in situ hybridization result likely represents an experimental artifact and there is no delayed translation of Dazap1 transcripts as we previously suggested.
We investigated why Dazap1-S was translated on polysomes in early germ cells and sequestered to mRNPS in later germ cells, and found that translational repression of Dazap1-S was associated with an increase in its poly(A) tail length. Translational regulation mediated through poly(A) tail is well documented for some maternal mRNAs during oocyte maturation and early embryogenesis [47]. In the oocytes, masked mRNAs usually have short poly(A) tails fewer than 20 bases, which elongate to about 80 to 150 bases before the mRNAs are recruited to the polysomes for translation. It is thought that longer poly(A) tails facilitate the binding of poly(A) binding protein (PABP) and subsequent recruitment of translational initiation factors. Some translating mRNAs with long poly(A) tails, on the other hand, undergo deadenylation during oocytes maturation, resulting in their translational repression. As first reported by Kleene [41], male germ cells apparently have a reverse pattern of poly(A) tail mediated translational regulation, with the translational activation of several post-meiotically expressed genes accompanied by poly(A) shortening instead of lengthening. The shortened poly(A) tails still contain about 30 As, sufficient for one PABP molecule which requires approximately 27 As for binding [48], [49]. Here we confirmed poly(A) shortening of the Prm1 transcript during translational activation using a recently developed ePAT method which offers higher sensitivity and resolution than the previously used Northern blot [41]. The Prm1 short poly(A) tails gave a band that was quite discrete, suggesting that they may not be as heterogeneous as previously suggested. In addition, we found that a small fraction of the mRNP-bound Prm1 transcripts contained SATs. Previous Northern analyses on sucrose gradient mRNAs enriched for transcripts with SATs did detect the presence of trace amount of Prm1, Prm2, Tnp1, and Tnp2 transcripts with SATs in the mRNP fractions [50]. Our ePAT results are likely biased for transcripts with SATs since it is well known that in PCR reactions short fragments are usually amplified with higher efficiency. It remains unclear whether poly(A) shortening is required for translational activation or it is merely a consequence of translation [49], [51]. Our observations that a small fraction of the mRNP-bound Prm1 transcript contained SATs whereas very little polysomal Prm1 transcript contained LATs would argue for the occurrence of poly(A) tail shortening at mRNPs instead of on polysomes as a consequence of translation. The poly(A) tail elongation of Dazap1-S accompanying its translational repression represents the other side of the coin. Similar to the Prm1 transcript, the mRNP-associated Dazap1-S had both LATs and SATs whereas the polysomal Dazap1-S contained only SATs. However for Dazap1-L which remained largely translationally active during spermatogenesis, there was no obvious changes in its poly(A) tail length. The transcript contained only SATs regardless whether it was sequestered to mRNPs or was actively translating on polysomes. Our results therefore showed that translational repression of Dazap1-S, but not Dazap1-L, was accompanied by poly(A) tail elongation. Whether the poly(A) tail plays an active role in modulating the translational activity of Dazap1-S remains to be investigated. Previous studies on testicular expression of Ldh-x and Rpl32 failed to find an association between poly(A) tail length and translational activity [52], [53]. However, the Northern blots in those studies were not of high resolution/quality and the results need to be confirmed with the new ePAT method.
We have identified several proteins, including KSRP, PTBP2, MSY2, MSY1, and DAZL, that bound to Dazap1 3′UTR. The only protein that we found to have any effects on Dazap1 expression when ectopically expressed in cultured cells was DAZL. DAZL is known to stimulate mRNA translation, and requires multiple DAZL binding sites for efficient translation activation [19]. DAZL consensus sequence U2–10[G/C]U2–10 is present in different regions of Dazap1 3′UTR. The S region contains a UUUCUUUU sequence, the CU_40 region contains three partially overlapping UUCUU sequences, and the AU region contains a UUUUCUU and a UUUUGUUUU sequence. Our RNA gel-shift assays showed that DAZL bound much stronger to the CU_40 region than the S or the AU region. Expression of luciferase reporters in 3T3 cells also indicated that the CU_40 region was responsible for the DAZL induced translation enhancement of the Luc gene linked to Dazap1-L 3′UTR. We also found that an anti-DAZL antibody preferentially precipitated Dazap1-L from mouse testis lysates. There is a good correlation between the expression of DAZL and DAZAP1 during spermatogenesis. Previous immunostaining of adult mouse testicular sections detected dramatic increases in the expression of both DAZAP1 and DAZL in pachytene spermatocytes [38], [54]. Our current analyses of Dazap1 expression during spermatogenesis showed that the level of Dazap1-L also increased significantly in pachytene spermatocytes. Therefore, increased expression of both Dazap1-L and DAZL in pachytene spermatocytes likely accounts for the robust translation of DAZAP1 in the cells.
Materials and Methods
Animals
C57BL/6 mice used in the study were housed in a specific pathogen-free animal facility. Adult mice were 2–4 months old. The experiments were approved by Academia Sinica's Institutional Animal Care & Utilization Committee.
3′Rapid Amplification of cDNA Ends (3′RACE)
3′RACE of mouse Dazap1 transcripts was carried out according Schaefer [55]. Testis RNA was reverse transcribed using the universal adapter primer PrRACE-dT: 5′-gctgtcaacgatacgctacgtaacgt24, and PCR amplified using primers PrDAP176: 5′-agtggcttcggacgcgggcagaaccacaac and Pr3′RACE: 5′-gctgtcaacgatacgctacgtaacg. The PCR products were cloned into pCRII-TOPO (Invitrogen, Carlsbad, CA) and sequenced.
Northern hybridization
Northern hybridization was performed according to the standard protocol [56] using Sure Blot nylon membranes (Intergen, Purchase, NY) and a Stratalinker UV cross-linker 1800 (Stratagene, La Jolla, CA). Blots were hybridized with radioactive probes at 68°C in the ExpressHyb solution (Clontech, Palo Alto, CA) and analyzed with the Typhoon 9410 Imager (GE Healthcare, Piscataway, NJ). For quantification, the hybridization membranes were exposed to X-ray films and radioactive bands were located, excised, and counted.
Sucrose gradient analysis
Post-mitochondrial fractions of mouse testis lysates were fractionated on 10–50% sucrose gradients as previously described [57]. Briefly, mouse testes were homogenized with a motor-driven glass–Teflon homogenizer (Wheaton, Millville, NJ) on ice in 0.5 ml lysis buffer (100 mM NaCl, 3 mM MgCl2, 20 mM Hepes, pH 7.4 and 0.5% Triton X-100). The nuclei and mitochondria were pelleted by centrifugation for 5 min at 12,000 g. The supernatants were loaded onto linear gradients of 10%–50% sucrose (w/w) in the lysis buffer without Triton X-100 and centrifuged for 2 h at 36,000 rpm in a SW-41 rotor (Beckman, Palo Alto, CA). The gradients were pumped through a flow cell (Teledyne Isco, Lincoln, NE) to record the absorbance at 254 nm and fractionated into 0.5-ml fractions. Each fraction was extracted twice with phenol:chloroform (1∶1) and the RNA was ethanol precipitated. Polysome profiles of the Dazap1 transcripts were analyzed by 3′RACE-PCR or Northern hybridization. Control gradients, containing 16 mM EDTA in the postmitochondrial supernatants, were centrifuged and analyzed in parallel with the polysome gradients.
UV cross-linking assays
Testis extracts were prepared from mice at indicated age according to Xu and Hecht [58]. 32P-labelled single stranded RNA probes were in vitro transcribed using Riboprobe systems (Promega) and purified on 4.5% denaturing polyacrylamide gels containing 8 M urea. UV cross-linking of RNA-protein complexes was performed according to Xu and Hecht [58] with minor modifications. RNA-protein binding was carried out at room temperature for 20 min in 20- µl reaction mixtures containing 20 mM Hepes [pH 7.6], 3 mM MgCl2, 40 mM KCl, 2 mM DTT, 5% [w/v] glycerol, 40 µg testis extracts, and 5×104 cpm of 32P-labelled RNA probes. Heparin was then added to a final concentration of 5 mg/ml to reduce nonspecific RNA-protein interactions. After10 min, the reaction mixtures were exposed to UV light (Stratalinker 1800) for 11 min on ice, followed by RNase A (0.5 mg/ml) digestion for 15 min at 37°C. The RNA-protein complexes were resolved by 10% SDS-PAGE. The gels were dried and visualized with the Typhoon 9410 Imager.
RNA affinity pulldown assays
RNA affinity pulldown assay was done according to a published protocol [58] with minor modifications. Biotinylated RNA probes were in vitro transcribed using the Riboprobe system containing biotin-11-UTP (PerkinElmer, Boston, MA). Testes extracts containing 1 mg total proteins were mixed with 20 µg biotinylated RNA in 400 µl binding buffer (BB) containing 20 mM Tris-HCl [pH 7.4], 2.5 mM MgCl2, 120 mM KCl, 1 mM DTT, 5% [w/v] glycerol, and1× protease inhibitor cocktail for 30 min at room temperature and treated with 5 mg/ml heparin for 15 min. Streptavidin-agarose beads were then added to the mixture and incubated for 60 min. After five washes with BB, bound proteins were released by boiling in 4× SDS sample buffer, resolved by 10% SDS-PAGE and visualized by SYPRO Ruby staining. The protein bands of interest were excised and analyzed by LC/MS/MS with a Thermo Scientific LTQ XL mass spectrometer (Thermo Scientific, Waltham, MA).
RNA immunoprecipitation assays
RNA immunoprecipitation assay was performed according to Xu and Hecht [58]. Two mg testes extracts were precleaned using 50 µl protein G beads (GE Healthcare) and 100 µg/ml yeast tRNA in 1 ml TKM buffer (20 mM Tris [pH 7.4], 120 mM KCl, 1.5 mM MgCl2, 0.1% NP-40) with 500 U/ml SUPERase-In (Ambion, Austin,TX) for 4 hours at 4°C. After spinning down the beads, the supernatants were incubated overnight at 4°C with 10 µg anti-DAZL-C antibody which was generated against an oligopeptide containing the last 21 amino acid residues of mouse DAZL and affinity purified using the services of Bethyl Laboratories, Inc (Montgomery, TX). The mixtures were then incubated with 20 µl protein G beads for 3 hours at 4°C. The beads were spundown and washed five times by the TKM buffer. Immunoprecipitated RNAs were extracted by Trizol (Invitrogen) and analyzed by RT-PCR. Sequences of the PCR primers used in RNA immunoprecipitation assays are given in Table S2.
RNA gel shift assays
Gel shift assays were carried out according to Venables et al [22] with some modifications. The 10- µl binding reactions contained 1 µg recombinant DAZL, 20 mM Tris [pH 7.4], 2.5 mM MgCl2, 60 mM KCl, 1 mM DTT, 5% [w/v] glycerol, and 104 cpm of 32P-labelled RNA probes. After incubation at room temperature for 5 min, the reactions were treated with 5 mg/ml heparin and 0.5 mg/ml yeast tRNA for another 15 min. The RNA-protein complexes were separated on 1.5% agarose gels in 0.5× TBE buffer and visualized with the Typhoon9410 Imager.
Plasmid construction
The entire 3′UTRs of Dazap1, Mvh, Sycp3 and Tex19.1 were PCR amplified from mouse genomic DNA and cloned into pCRII-TOPO. pcDNA-Luc was generated by inserting the Luc gene into pcDNA3.1 (Invitrogen). The 3′UTRs of the various genes in pCRII-TOPO were then cloned into pcDNA-Luc to generate the reporter vectors. Deletion of the CU-rich sequence in Dazap1 3′UTR was generated by a PCR-mediated deletion approach [59]. For in vitro transcription, different regions of Dazap1 3′UTR were generated by PCR amplification and cloned into pBluescript (Stratagene). Dazl coding region was RT-PCR amplified from mouse testis RNA and cloned into pCRII-TOPO for sequencing. It was subsequently cloned into p3xFLAG-CMV-14 (Sigma, Saint Louis, MI) to generate pDAZL-Flag, and into pET-28a (Novagen, Madison, WI) to generate pET-DAZL for the production of His-tagged DAZL protein in E. coli [22], [60].
Analysis of the translation of luciferase reporters
3T3 cells were maintained in DMEM (Hyclone, Fremont, CA) supplemented with 10% fetal bovine serum and 1× penicillin/streptomycin (GIBCO, Eggenstein, Germany). Transfection was performed using the TurboFect reagent (Fermentas, Burlington, Ontario, Canada). The pDAZL-Flag or the empty p3× FLAG-CMV-14 were co-transfected into 3T3 Cells with different firefly luciferase reporter constructs and pRL-TK (Promega, San Luis Obispo, CA) which encodes the Renilla luciferase. The cells were harvested after 24 hours, and luciferase activities in the cell lysates were measured using the dual-luciferase reporter assay system (Promega) and normalized to Renilla luciferase activities. Luciferase RNA levels were determined by semi-quantitative RT-PCR and normalized to the levels of Renilla luciferase transcripts. Luciferase activities and RNA levels in cells transfected with pDAZL-Flag were normalized to those in cells transfected with p3× FLAG-CMV-14 to reflect the effects of DAZL, and the results of cells transfected with different Luc reporter genes were compared using those of cells transfected with the empty Luc reporter as a reference (100%). DAZL expression levels were determined by Western blot and normalized to the levels of tubulin.
Poly(A) tail length determination
A newly developed extension poly(A) test (ePAT) method was used to measure the poly(A) tail of mRNA [42]. This method makes use of the ability of the Klenow polymerase to extend the 3′ ends of RNA molecules with dNTPs from an annealed DNA template in standard reverse transcription buffers. The PAT-anchor primer 5′-cgctacgtaacggcatgacagtgt22 contains a unique universal DNA segment plus 22 Ts at the 3′ end and anneals to the end of poly(A) tails to serve as the DNA template for the Klenow polymerase. After the initial extension, the PAT-anchor primer also serves as the primer for the standard reverse transcription. The incubation was carried out at 55°C to eliminate unwanted priming from internal poly(A) tracts. The protocol started with heating 1 µg RNA together with 1 µl of 100 µM PAT-anchor primer in a volume of 8 µl at 80°C for 5 min followed by cooling down to 37°C. A master mixture containing 4 µl DEPC-treated water, 4 µl 5× Superscript III buffer (Invitrogen), 1 µl 100 mM DTT, 1 µl 10 mM dNTPs, 1 µl RNaseOUT (Invitrogen), and 1 µl (5 U) Klenow polymerase (New England Biolabs, Ipswich, MA) was added and the solution was incubated at 37°C for 1 hour. The reaction mixture was then heated at 80°C for 10 min to inactivate the polymerase and cooled down to 55°C. One µl (200 U) Superscript III (Invitrogen) was added and incubation was kept at 55°C for 1 hour. The reverse transcriptase was inactivated at 80°C for 10 min at the end of the reaction. PCR amplification of the 3′ ends of specific genes was performed using a universal reverse primer 5′-cgctacgtaacggcatgacagtg complementary to the 5′ end of the PAT-anchor primer and gene-specific forward primers, the sequences of which are provided in Table S3. A separate TVN-PAT reaction was also performed to generate a fragment with a defined poly(A) tail of 12 As. It served as a size standard for other PAT products that were amplified from the same gene specific forward primer. The TVN-PAT primer has an identical sequence as the PAT anchor primer except the addition of two 3′ variable bases V and N (V is A, G or C and N is any base) which fix the primer to the polyadenylation site during reverse transcription. The e-PAT products were analyzed on 2% agarose gels. Fragments of interest were purified from the gels, cloned into pCRII-TOPO and sequenced to determine the lengths of the poly(A) tails.
Supporting Information
Poly(A) tail length determination of Prm1 and Dazap1 transcripts in sucrose gradient fractions. Adult mouse testis lysate was fractionated on a 10–50% sucrose gradient and the same proportions of RNAs isolated from alternating fractions were subjected to ePAT and analyzed on a 2% agarose gel. The type of ribosome present in each fraction is indicated at the top. The major bands in the TVN lanes contain an invariant 12-(A) poly(A) tail. Transcripts with long and short poly(A) tails are indicated with L and S subscript, respectively.
(DOCX)
DAZL expression level in luciferase reporter assays. The upper panels show western blots of lysates of 3T3 cells cotransfected with pDAZL-Flag and different 3′UTR luciferase reporters. DAZL-Flag was detected with an anti-Flag antibody. The α-tubulin level is used as a loading control. The bottom panel shows quantification of the western blot signals. DAZL-Flag signals were normalized to the tubulin signals. The results represent the averages of three independent experiments.
(DOCX)
Poly(A) tail lengths of ePAT clones.
(DOCX)
PCR primers for RNAs precipitated from mouse testis lysate with an anti-DAZL antibody.
(DOCX)
Gene-specific ePAT forward primers.
(DOCX)
Acknowledgments
We thank Hsiang-Ying Chen for mouse breeding and handling and our institutional proteomic core for protein identification.
Funding Statement
This work was supported by the National Science Council (grant number 96-2311-B-001-026-MY3) and the Academia Sinica in Taiwan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kuersten S, Goodwin EB (2003) The power of the 3′ UTR: translational control and development. Nat Rev Genet 4: 626–637. [DOI] [PubMed] [Google Scholar]
- 2. Braun RE (1998) Post-transcriptional control of gene expression during spermatogenesis. Semin Cell Dev Biol 9: 483–489. [DOI] [PubMed] [Google Scholar]
- 3. Lee K, Haugen HS, Clegg CH, Braun RE (1995) Premature translation of protamine 1 mRNA causes precocious nuclear condensation and arrests spermatid differentiation in mice. Proc Natl Acad Sci U S A 92: 12451–12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braun RE, Peschon JJ, Behringer RR, Brinster RL, Palmiter RD (1989) Protamine 3′-untranslated sequences regulate temporal translational control and subcellular localization of growth hormone in spermatids of transgenic mice. Genes Dev 3: 793–802. [DOI] [PubMed] [Google Scholar]
- 5. Braun RE (2000) Temporal control of protein synthesis during spermatogenesis. Int J Androl 23 Suppl 292–94. [DOI] [PubMed] [Google Scholar]
- 6. Zhong J, Peters AH, Lee K, Braun RE (1999) A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet 22: 171–174. [DOI] [PubMed] [Google Scholar]
- 7. Davies HG, Giorgini F, Fajardo MA, Braun RE (2000) A sequence-specific RNA binding complex expressed in murine germ cells contains MSY2 and MSY4. Dev Biol 221: 87–100. [DOI] [PubMed] [Google Scholar]
- 8. Kleene KC (2003) Patterns, mechanisms, and functions of translation regulation in mammalian spermatogenic cells. Cytogenet Genome Res 103: 217–224. [DOI] [PubMed] [Google Scholar]
- 9. Iguchi N, Tobias JW, Hecht NB (2006) Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc Natl Acad Sci U S A 103: 7712–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brook M, Smith JW, Gray NK (2009) The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction 137: 595–617. [DOI] [PubMed] [Google Scholar]
- 11. Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, et al. (1995) Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet 10: 383–393. [DOI] [PubMed] [Google Scholar]
- 12. Cooke HJ, Lee M, Kerr S, Ruggiu M (1996) A murine homologue of the human DAZ gene is autosomal and expressed only in male and female gonads. Hum Mol Genet 5: 513–516. [DOI] [PubMed] [Google Scholar]
- 13. Yen PH, Chai NN, Salido EC (1996) The human autosomal gene DAZLA: testis specificity and a candidate for male infertility. Hum Mol Genet 5: 2013–2017. [DOI] [PubMed] [Google Scholar]
- 14. Xu EY, Moore FL, Pera RA (2001) A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci U S A 98: 7414–7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yen PH (2004) Putative biological functions of the DAZ family. Int J Androl 27: 125–129. [DOI] [PubMed] [Google Scholar]
- 16. Vangompel MJ, Xu EY (2011) The roles of the DAZ family in spermatogenesis: More than just translation? Spermatogenesis 1: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsui S, Dai T, Warren ST, Salido EC, Yen PH (2000) Association of the mouse infertility factor DAZL1 with actively translating polyribosomes. Biol Reprod 62: 1655–1660. [DOI] [PubMed] [Google Scholar]
- 18. Maegawa S, Yamashita M, Yasuda K, Inoue K (2002) Zebrafish DAZ-like protein controls translation via the sequence 'GUUC'. Genes Cells 7: 971–984. [DOI] [PubMed] [Google Scholar]
- 19. Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK (2005) The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J 24: 2656–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen J, Melton C, Suh N, Oh JS, Horner K, et al. (2011) Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev 25: 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zeng M, Lu Y, Liao X, Li D, Sun H, et al. (2009) DAZL binds to 3′UTR of Tex19.1 mRNAs and regulates Tex19.1 expression. Mol Biol Rep 36: 2399–2403. [DOI] [PubMed] [Google Scholar]
- 22. Venables JP, Ruggiu M, Cooke HJ (2001) The RNA-binding specificity of the mouse Dazl protein. Nucleic Acids Res 29: 2479–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jenkins HT, Malkova B, Edwards TA (2011) Kinked beta-strands mediate high-affinity recognition of mRNA targets by the germ-cell regulator DAZL. Proc Natl Acad Sci U S A 108: 18266–18271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiao X, Trifillis P, Kiledjian M (2002) Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol Reprod 66: 475–485. [DOI] [PubMed] [Google Scholar]
- 25. Reynolds N, Collier B, Maratou K, Bingham V, Speed RM, et al. (2005) Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum Mol Genet 14: 3899–3909. [DOI] [PubMed] [Google Scholar]
- 26. Reynolds N, Collier B, Bingham V, Gray NK, Cooke HJ (2007) Translation of the synaptonemal complex component Sycp3 is enhanced in vivo by the germ cell specific regulator Dazl. RNA 13: 974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsui S, Dai T, Roettger S, Schempp W, Salido EC, et al. (2000) Identification of two novel proteins that interact with germ-cell-specific RNA-binding proteins DAZ and DAZL1. Genomics 65: 266–273. [DOI] [PubMed] [Google Scholar]
- 28. Dai T, Vera Y, Salido EC, Yen PH (2001) Characterization of the mouse Dazap1 gene encoding an RNA-binding protein that interacts with infertility factors DAZ and DAZL. BMC Genomics 2: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin YT, Yen PH (2006) A novel nucleocytoplasmic shuttling sequence of DAZAP1, a testis-abundant RNA-binding protein. RNA 12: 1486–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yuki Y, Imoto I, Imaizumi M, Hibi S, Kaneko Y, et al. (2004) Identification of a novel fusion gene in a pre-B acute lymphoblastic leukemia with t(1;19)(q23;p13). Cancer Sci 95: 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Prima V, Gore L, Caires A, Boomer T, Yoshinari M, et al. (2005) Cloning and functional characterization of MEF2D/DAZAP1 and DAZAP1/MEF2D fusion proteins created by a variant t(1;19)(q23;p13.3) in acute lymphoblastic leukemia. Leukemia 19: 806–813. [DOI] [PubMed] [Google Scholar]
- 32. Goina E, Skoko N, Pagani F (2008) Binding of DAZAP1 and hnRNPA1/A2 to an exonic splicing silencer in a natural BRCA1 exon 18 mutant. Mol Cell Biol 28: 3850–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Skoko N, Baralle M, Buratti E, Baralle FE (2008) The pathological splicing mutation c.6792C>G in NF1 exon 37 causes a change of tenancy between antagonistic splicing factors. FEBS Lett 582: 2231–2236. [DOI] [PubMed] [Google Scholar]
- 34. Pastor T, Pagani F (2011) Interaction of hnRNPA1/A2 and DAZAP1 with an Alu-derived intronic splicing enhancer regulates ATM aberrant splicing. PLoS One 6: e23349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith RW, Anderson RC, Smith JW, Brook M, Richardson WA, et al. (2011) DAZAP1, an RNA-binding protein required for development and spermatogenesis, can regulate mRNA translation. RNA 17: 1282–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, et al. (2011) A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A 108: 20881–20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hsu LC, Chen HY, Lin YW, Chu WC, Lin MJ, et al. (2008) DAZAP1, an hnRNP protein, is required for normal growth and spermatogenesis in mice. RNA 14: 1814–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vera Y, Dai T, Hikim AP, Lue Y, Salido EC, et al. (2002) Deleted in azoospermia associated protein 1 shuttles between nucleus and cytoplasm during normal germ cell maturation. J Androl 23: 622–628. [PubMed] [Google Scholar]
- 39. Bellve AR, Millette CF, Bhatnagar YM, O′Brien DA (1977) Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem 25: 480–494. [DOI] [PubMed] [Google Scholar]
- 40. Nguyen Chi M, Chalmel F, Agius E, Vanzo N, Khabar KS, et al. (2009) Temporally regulated traffic of HuR and its associated ARE-containing mRNAs from the chromatoid body to polysomes during mouse spermatogenesis. PLoS One 4: e4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kleene KC (1989) Poly(A) shortening accompanies the activation of translation of five mRNAs during spermiogenesis in the mouse. Development 106: 367–373. [DOI] [PubMed] [Google Scholar]
- 42. Janicke A, Vancuylenberg J, Boag PR, Traven A, Beilharz TH (2012) ePAT: a simple method to tag adenylated RNA to measure poly(A)-tail length and other 3′ RACE applications. RNA 18: 1289–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Moor CH, Meijer H, Lissenden S (2005) Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin Cell Dev Biol 16: 49–58. [DOI] [PubMed] [Google Scholar]
- 44. Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, et al. (2007) Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res 35: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pero R, Lembo F (2003) Chieffi P, Del Pozzo G, Fedele M, et al (2003) Translational regulation of a novel testis-specific RNF4 transcript. Mol Reprod Dev 66: 1–7. [DOI] [PubMed] [Google Scholar]
- 46. Shima JE, McLean DJ, McCarrey JR, Griswold MD (2004) The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 71: 319–330. [DOI] [PubMed] [Google Scholar]
- 47. Richter JD (1999) Cytoplasmic polyadenylation in development and beyond. Microbiol Mol Biol Rev 63: 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baer BW, Kornberg RD (1980) Repeating structure of cytoplasmic poly(A)-ribonucleoprotein. Proc Natl Acad Sci U S A 77: 1890–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yanagiya A, Delbes G, Svitkin YV, Robaire B, Sonenberg N (2010) The poly(A)-binding protein partner Paip2a controls translation during late spermiogenesis in mice. J Clin Invest 120: 3389–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kleene KC (1993) Multiple controls over the efficiency of translation of the mRNAs encoding transition proteins, protamines, and the mitochondrial capsule selenoprotein in late spermatids in mice. Dev Biol 159: 720–731. [DOI] [PubMed] [Google Scholar]
- 51. Kini HK, Vishnu MR, Liebhaber SA (2010) Too much PABP, too little translation. J Clin Invest 120: 3090–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fujimoto H, Erickson RP, Tone S (1988) Changes in polyadenylation of lactate dehydrogenase-X mRNA during spermatogenesis in mice. Mol Reprod Dev 1: 27–34. [DOI] [PubMed] [Google Scholar]
- 53. Kleene KC, Cataldo L, Mastrangelo MA, Tagne JB (2003) Alternative patterns of transcription and translation of the ribosomal protein L32 mRNA in somatic and spermatogenic cells in mice. Exp Cell Res 291: 101–110. [DOI] [PubMed] [Google Scholar]
- 54. Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, et al. (1997) The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 389: 73–77. [DOI] [PubMed] [Google Scholar]
- 55. Schaefer BC (1995) Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem 227: 255–273. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual.Cold Spring Harbor,NY :Cold Spring Harbor Laboratory Press. [Google Scholar]
- 57. Kleene KC, Distel RJ, Hecht NB (1984) Translational regulation and deadenylation of a protamine mRNA during spermiogenesis in the mouse. Dev Biol 105: 71–79. [DOI] [PubMed] [Google Scholar]
- 58. Xu M, Hecht NB (2007) Polypyrimidine tract binding protein 2 stabilizes phosphoglycerate kinase 2 mRNA in murine male germ cells by binding to its 3′UTR. Biol Reprod 76: 1025–1033. [DOI] [PubMed] [Google Scholar]
- 59. Hansson MD, Rzeznicka K, Rosenback M, Hansson M, Sirijovski N (2008) PCR-mediated deletion of plasmid DNA. Anal Biochem 375: 373–375. [DOI] [PubMed] [Google Scholar]
- 60. Fox M, Urano J, Reijo Pera RA (2005) Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics 85: 92–105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Poly(A) tail length determination of Prm1 and Dazap1 transcripts in sucrose gradient fractions. Adult mouse testis lysate was fractionated on a 10–50% sucrose gradient and the same proportions of RNAs isolated from alternating fractions were subjected to ePAT and analyzed on a 2% agarose gel. The type of ribosome present in each fraction is indicated at the top. The major bands in the TVN lanes contain an invariant 12-(A) poly(A) tail. Transcripts with long and short poly(A) tails are indicated with L and S subscript, respectively.
(DOCX)
DAZL expression level in luciferase reporter assays. The upper panels show western blots of lysates of 3T3 cells cotransfected with pDAZL-Flag and different 3′UTR luciferase reporters. DAZL-Flag was detected with an anti-Flag antibody. The α-tubulin level is used as a loading control. The bottom panel shows quantification of the western blot signals. DAZL-Flag signals were normalized to the tubulin signals. The results represent the averages of three independent experiments.
(DOCX)
Poly(A) tail lengths of ePAT clones.
(DOCX)
PCR primers for RNAs precipitated from mouse testis lysate with an anti-DAZL antibody.
(DOCX)
Gene-specific ePAT forward primers.
(DOCX)