Abstract
We evaluated the consequences of nutritional imbalances, particularly lipid/nitrogen imbalances, on wine yeast survival during alcoholic fermentation. We report that lipid limitation (ergosterol limitation in our model) led to a rapid loss of viability during the stationary phase of fermentation and that the cell death rate is strongly modulated by nitrogen availability and nature. Yeast survival was reduced in the presence of excess nitrogen in lipid-limited fermentations. The rapidly dying yeast cells in fermentations in high nitrogen and lipid-limited conditions displayed a lower storage of the carbohydrates trehalose and glycogen than observed in nitrogen-limited cells. We studied the cell stress response using HSP12 promoter-driven GFP expression as a marker, and found that lipid limitation triggered a weaker stress response than nitrogen limitation. We used a SCH9-deleted strain to assess the involvement of nitrogen signalling pathways in the triggering of cell death. Deletion of SCH9 increased yeast viability in the presence of excess nitrogen, indicating that a signalling pathway acting through Sch9p is involved in this nitrogen-triggered cell death. We also show that various nitrogen sources, but not histidine or proline, provoked cell death. Our various findings indicate that lipid limitation does not elicit a transcriptional programme that leads to a stress response protecting yeast cells and that nitrogen excess triggers cell death by modulating this stress response, but not through HSP12. These results reveal a possibly negative role of nitrogen in fermentation, with reported effects referring to ergosterol limitation conditions. These effects should be taken into account in the management of alcoholic fermentations.
Introduction
The nutrients available for yeasts in grape musts have a major impact on the kinetics of alcoholic fermentations. Yeast is more or less active (with variable fermentation rates) and is able or not able to withstand the stress of alcoholic fermentation (high ethanol concentrations, low pH), according to the availability of nutrients. Nitrogen is a particularly significant nutrient [1], [2], because its availability determines the fermentation rate and to a large extent, the fermentation duration [3]. Lipids are also key nutrients in alcoholic fermentation and oxygen is required for the synthesis of sterols and unsaturated fatty acids [4], [5]. Limitations of unsaturated fatty acids or sterols have negative effects on the maintenance of viability at the end of fermentations [6], [7]. Indeed, some wine making practices, such as strong clarification of musts, can lead to such compounds becoming limiting and are associated with a loss of yeast cell viability.
Little is known about the molecular mechanisms involved in yeast cell death under such conditions and yeast cell death has been only described in terms of the consequences of unsuitable membrane composition. Indeed, the plasma membrane is considered as the major target of ethanol toxicity, and both sterols and unsaturated fatty acids act as regulators of membrane function thereby protecting against the deleterious effects of ethanol [8], [9]. On another side, recent studies have reported that the ability of yeasts to survive starvation depends on how they enter into the state of starvation and on the nature of the limiting nutrient [10]. The adaptation to nutritional deficiencies is affected by the activity of various signalling pathways, including TOR [11], [12] or PKA [13], and pathways that initiate sensing of nutrients directly at the plasma membrane level [14]. In a nutrient-rich environment, the nutrient-sensing pathways TOR/Sch9p and RAS/PKA promote growth and repress the stress response and autophagy. At low ammonium or amino acid concentrations, cell survival, or chronological lifespan (CLS), is extended [15], [16]. It is unclear whether the activity of these signalling pathways affects cell behaviour in situations of nutrient disequilibrium in alcoholic fermentation.
Based on recent theoretical analyses of yeast starvation biology, we explored the molecular mechanisms and the signalling pathways involved in triggering cell-death in response to disequilibrium of lipid/nitrogen nutrients in alcoholic wine fermentation. We show that cell death in lipid-limited fermentations is strongly modulated by the availability of nitrogen. Molecular analyses indicated that TOR nitrogen cellular signalling is involved in triggering cell death in such conditions. Our results demonstrate that the nitrogen status of grape musts is a strong determinant of the outcome of alcoholic fermentations in conditions of lipid limitation.
Results
The yeast cell viability is modulated by nitrogen/lipid imbalances
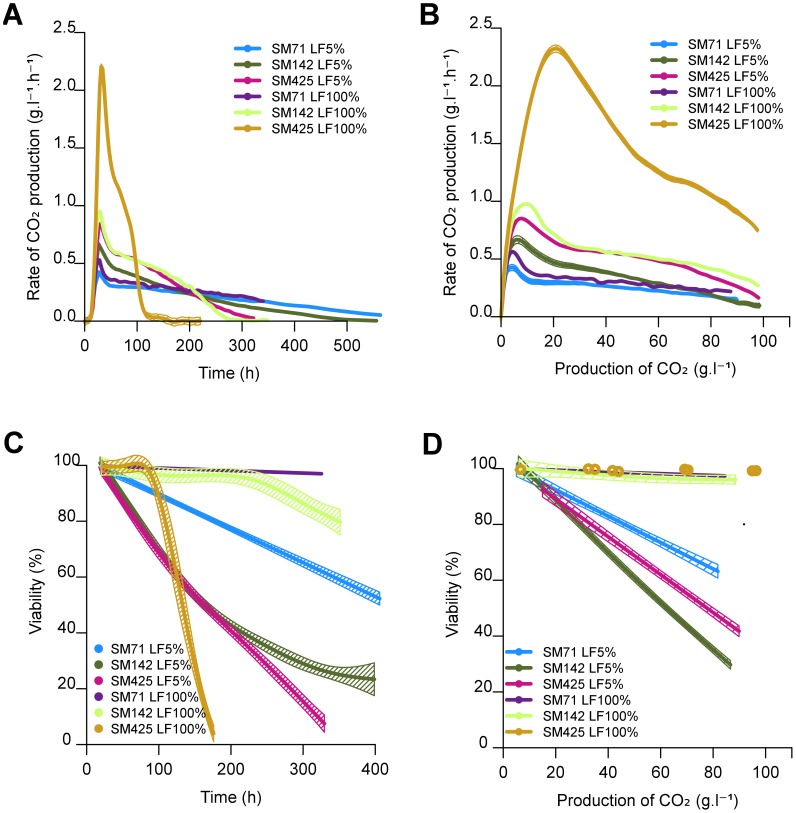
We examined the effects of the nitrogen and lipid status on the behaviour of the wine yeast EC1118 in alcoholic fermentation. The EC1118 strain was fermented in synthetic medium SM425, SM142 or SM71 (containing 425, 142 and 71 mg/L of assimilable nitrogen, respectively) with either high (LF100%) or low (LF 5%) levels of lipid factors (LF). As expected, when lipids were not limiting, the amount of nitrogen had a significant effect on the rate of fermentation, with a maximal peak of CO2 production at 2.25, 1 and 0.59 g/L/h, respectively, and a fermentation period lasting 110 to 340 h (Figure 1A and B). In lipid-limited medium (LF5%), the Vmax of the yeast fermentation was lower, from 0.8 to 0.4 g/L/h, depending on the nitrogen content.
Figure 1. Effect of nutrient imbalances during alcoholic fermentation.
Rate of CO2 production by S. cerevisiae EC1118 cultures at 24°C (A) at various times of fermentation or (B) CO2 production, and viability of S. cerevisiae EC1118 cells according to (C) time of alcoholic fermentation at 24°C and (D) CO2 production. The synthetic medium contained 71 mg/L (SM71) or 142 mg/L (SM142) assimilable nitrogen, and 5% or 100% lipid factors (LF5% or LF100%). Viability was measured by flow cytometry after propidium iodide staining. The graphs are the result of smoothing of measurements series (at least 3 repetitions) using the software R. Smoothing obtained is framed by a confidence interval calculated at 95%.
The cell population densities (Table 1) were between 39 and 49 106 cells/mL in lipid-limited conditions (LF5%), the lipid availability being the growth limiting factor (Figure S1). By contrast, the cell population density was far higher in SM425, SM142 and SM71 with LF100% (from 202 106 cells/mL to 68 106 cells/mL). A two-way ANOVA on the log population shows a high significant effect of lipid and of nitrogen, with a significant interaction effect (P-value < 0.001). Moreover, the Wilcoxon rank-sum test shows significant differences (P-value < 5%) for each condition taken two by two.
Table 1. Cell population densities determined by flow cytometry at stationary phase.
| Strain | Synthetic medium composition | Cell population (106 cell/mL) | |
| Assimilable nitrogen (mg/L) | Lipid factor (%) | ||
| EC1118 | 425 | 100 | 202.6 ± 9.9 |
| EC1118 | 425 | 5 | 49.2 ± 4.5 |
| EC1118 | 142 | 100 | 106.5 ± 12.3 |
| EC1118 | 142 | 5 | 44.6. ± 6.3 |
| EC1118 | 71 | 100 | 68.4 ± 4.6 |
| EC1118 | 71 | 5 | 39.5 ± 8.8 |
Cell viability (Figure 1C and D) was high (more than 98%) during the stationary phase in media rich in lipids (LF100%) irrespective of the initial nitrogen content (SM425, SM142 or SM71). However, yeast viability was lower in all the fermentations in lipid-limited medium (LF5%). As the fermentation rates were highly variable (Figure 1A), we compared cell viabilities according to the progress of the fermentation, i.e. the production of CO2 (Figure 1D), rather than to time.
In lipid-limited fermentations, cell viability decreased regularly with a constant slope for each condition (Figure 1D). This behaviour is consistent with an ethanol-related loss of viability and not chronological aging-related loss of viability. In fact in our conditions, time is not relevant, but ethanol increase (directly proportional to the amount of CO2 released during fermentation) is, likely because it is a well-known stressor. Therefore, these findings could indicate that cell death was a direct consequence of ethanol toxicity, although it is not excluded that other factors present in the medium could also be involved. The decrease in cell viability varied depending on the nitrogen content of the medium (Figure 1C and D). Viability remained high in SM71 (70% at 70 g/L of CO2 produced), but decreased to a lower level in SM142 (45% at 70 g/L of CO2 produced), demonstrating a negative effect of nitrogen availability on the cell viability in lipid-limited fermentations. High nitrogen effect on cell death is strongly dependent on the lipid content and it triggers cell death only when the lipid content is low. Surprisingly the highest nitrogen concentration (425 mg/L) led to an intermediate rate of cell death. Thus, the effect was not strictly dependent on the nitrogen concentration suggesting “complex effect” of nitrogen sources.
The nitrogen sources used in this study were a mixture of amino acids and ammonium, and mimicked a natural must. The nitrogen sources were monitored during the fermentations. All amino acids, except proline, were up taken from SM71 and SM142 at 20 g of CO2 (Table S1). As the populations were similar in all fermentations, these nitrogen sources were not used to make new cells. Note that, cell death differed between these cultures, with the lipid content supposed to be the same, excluding the involvement of lipids in the differences. However, in cultures in SM425, most of the amino acids were not totally metabolised (Table S1). This suggests that some residual nitrogen sources may trigger a protective effect in these conditions.
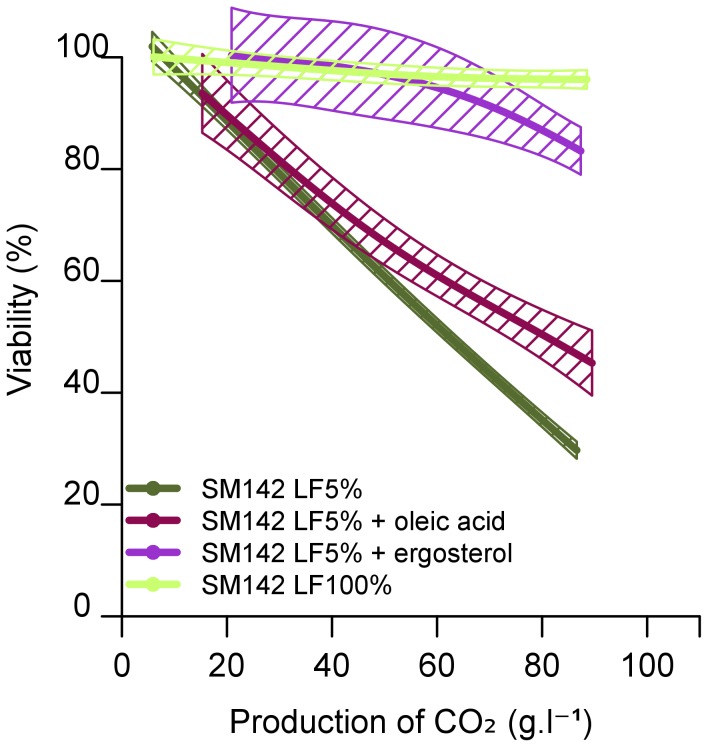
The source of LF used contained a mixture of ergosterol and oleic acid, and we investigated which of these lipids was the limiting factor for growth. Fermentation variables were measured in SM142 LF5% supplemented with oleic acid or ergosterol so that the final lipid concentration was similar to a non-limited medium (LF100%). The addition of oleic acid had only a small effect on the final population of cells, whereas the addition of ergosterol led to an increase in the population (77 106 cells/mL in SM142 LF5% plus ergosterol) (Table 2). The addition of ergosterol also increased cell viability (Figure 2) to close to 100%, and rate of CO2 production (Figure S2). Ergosterol was therefore the lipid limiting growth under our experimental conditions.
Table 2. Effect of treatment on cell population densities as determined by flow cytometry at stationary phase.
| Strain | Synthetic medium composition | Comparison low | Cell population (106 cell/mL) | Comparison high | |
| Assimilable nitrogen (mg/L) | Lipid factor (%) | ||||
| EC1118 | 142 | 100 | ** | 106.5±12.3 | high |
| EC1118 | 142 | 5 | low | 44.6±6.3 | ** |
| EC1118 hsp12::GFP | 142 | 5 | NS | 57.5±6.7 | NS |
| EC1118 | 142 | 5 + oleic acid | * | 36.6±0.6 | NS |
| EC1118 | 142 | 5 + ergosterol | * | 77.3±6.7 | NS |
| EC1118 | 142 + rapamycin | 5 | NS | 46.6±2.7 | NS |
| EC1118 | 142 | 5 | * | 44.6±6.3 | high |
| EC1118 | 71 | 5 | low | 39.5±8.8 | * |
| EC1118 hsp12::GFP | 71 | 5 | * | 48±2.2 | NS |
| EC1118 | 142 (71 mix + 71 | 5 | NS | 47.6 | NS |
| EC1118 | NH4 +) | 5 | NS | 42.1±1 | NS |
| EC1118 | 142 (71 mix + 71 Arg) | 5 | NS | 48.4±1.8 | NS |
| EC1118 | 142 (71 mix + 71 Glu) | 5 | NS | 44.3±4.1 | NS |
| EC1118 | 142 (71 mix + 71 Gln) | 5 | NS | 37.1±2.5 | * |
| EC1118 | 142 (71 mix + 71 His) | 100 | NS | 53.3 | NS |
| EC1118 | 142 (71 mix + 71 Pro) | * | 68.4±4.6 | * | |
| 71 | |||||
The log population of each condition is compared to the two standards conditions (indicated as “low” and “high”) with the Wilcoxon rank-sum test (*: P-value <5%; **: P-value < 1%).
Figure 2. Effect of lipid factors on the viability of S. cerevisiae EC1118 cells during alcoholic fermentation.
The synthetic medium contained 142 mg/L assimilable nitrogen (SM142) and 5% lipid factor (LF5%), with or without additional oleic acid or ergosterol (content as in LF100%). Viability was measured by flow cytometry after propidium iodide staining. The graphs are the result of smoothing of measurements series (at least 3 repetitions) using the software R. Smoothing obtained is framed by a confidence interval calculated at 95%.
The viability of wine yeasts under lipid limitation depends on the source of nitrogen
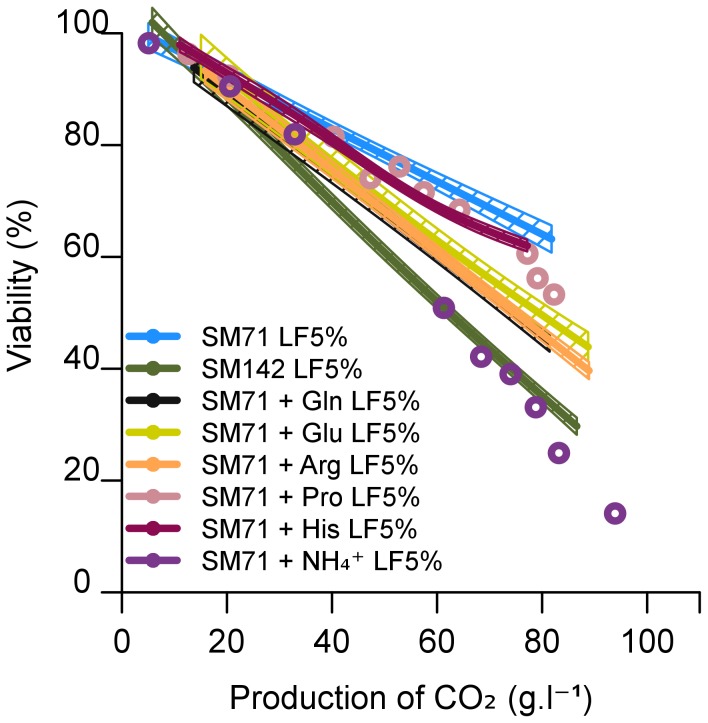
The combination of lipid limitation and nitrogen excess affected yeast cell viability. We therefore investigated the capacity of amino acids and ammonium to trigger cell death. We considered the amino acids most likely to be available on the basis of their natural abundance in musts and the ability of yeast to use them quickly (arginine, glutamine, glutamate, histidine or proline). SM71 was independently supplemented with each of various amino acids or ammonium, such that the final concentration of assimilable nitrogen was the same as that in SM142. Supplementation with histidine or proline had little or no effect on the rate of fermentation, consistent with the absence or only limited use of these amino acids. The other nitrogen sources triggered a significant increase in the rate of fermentation (Figure S3). Only small variations in final cell populations were observed (from 37 to 53 106 cells/mL) consistent with a tight control of growth by limiting ergosterol (Table 2). However, the addition of the nitrogen sources arginine, glutamine, glutamate or ammonium increased mortality (Figure 3). Thus, an excess of various nitrogen sources enhanced cell death under conditions of lipid limitation. The differences observed seem to be related to the ability of yeast cells to use the supplementary nitrogen source: ammonium was the most effective trigger of cell death with less than 20% viability at the end of fermentation. This effect is consistent with the observation that NH4 + is toxic for cells deprived for auxotrophy-complementing amino acids, although it is also toxic for cells grown in excess of amino acids. In addition, NH4+ reduced the yeast chronological life span by activating of the TOR/PKA pathways and by inhibiting Sch9 [17]. In fact, NH4 + like other nitrogen sources metabolized, triggers cell death in relation with its signaling capacity on TOR pathway. This leads to a reduction of the stress response and yeast protection against ethanol. Thus, there could be a combination effect of ethanol and NH4+, with ethanol acting as a permanent stressor but not being the initial trigger of cell death. However, several different sources of nitrogen increased cell death in our conditions, suggesting that the underlying mechanisms may be different.
Figure 3. Effect of nitrogen source on the viability of S. cerevisiae EC1118 cells during alcoholic fermentation.
The synthetic medium contained 71 mg/L assimilable nitrogen (SM71) and 5% lipid factors (LF5%), with or without additional arginine, glutamine, glutamate, histidine or proline (142 mg/L assimilable nitrogen final content as in SM142). Viability was measured by flow cytometry after propidium iodide staining. The graphs are the result of smoothing of measurements series (at least 3 repetitions) using the software R. Smoothing obtained is framed by a confidence interval calculated at 95%.
Stress response-dependent sugar storage is reduced in lipid-limited cells
Trehalose and glycogen accumulate in response to stress and can be used as indicators of the stress response in yeast. We therefore analysed the content of trehalose and glycogen in yeast stationary phase as markers of the stress response to nutrient limitations. Both compounds were more abundant in cells fermenting in SM71 LF5% than in SM142 LF5% (Figure 4). These results are consistent with a control of the stress response by nitrogen level and suggest that growth limitation by ergosterol does not trigger similar stress response.
Figure 4. Trehalose and glycogen contents at stationary phase of S. cerevisiae EC1118.
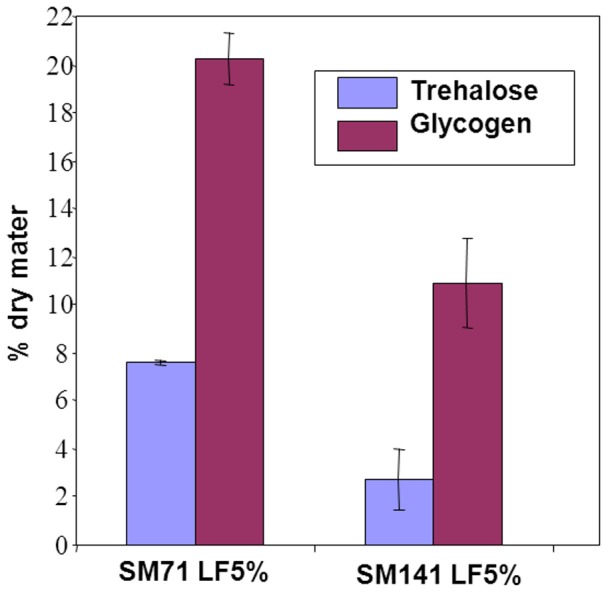
Cells were fermented in synthetic medium containing 71 mg/L (SM71) or 142 mg/L (SM142) assimilable nitrogen, and 5% or 100% lipid factors (LF5% or LF100%).
Nitrogen signalling is involved in triggering cell death in response to nitrogen/lipid imbalances
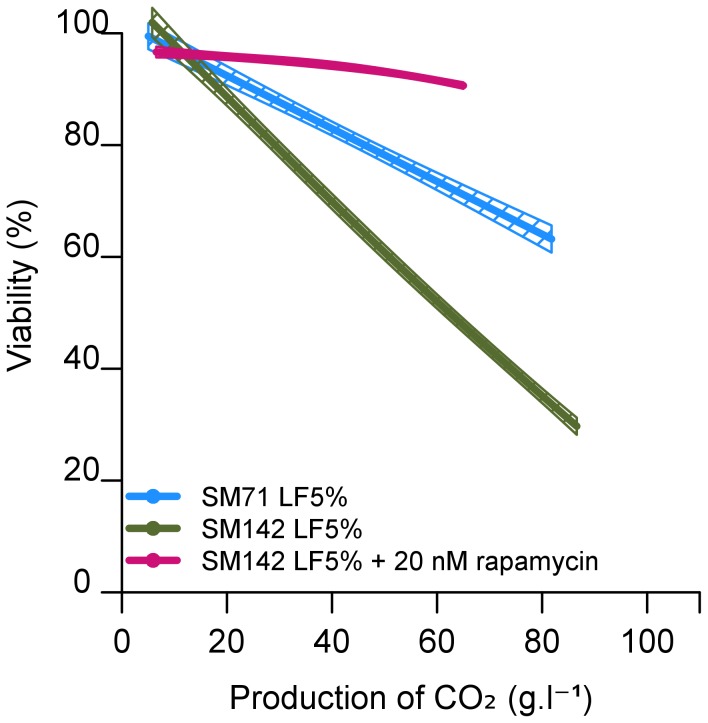
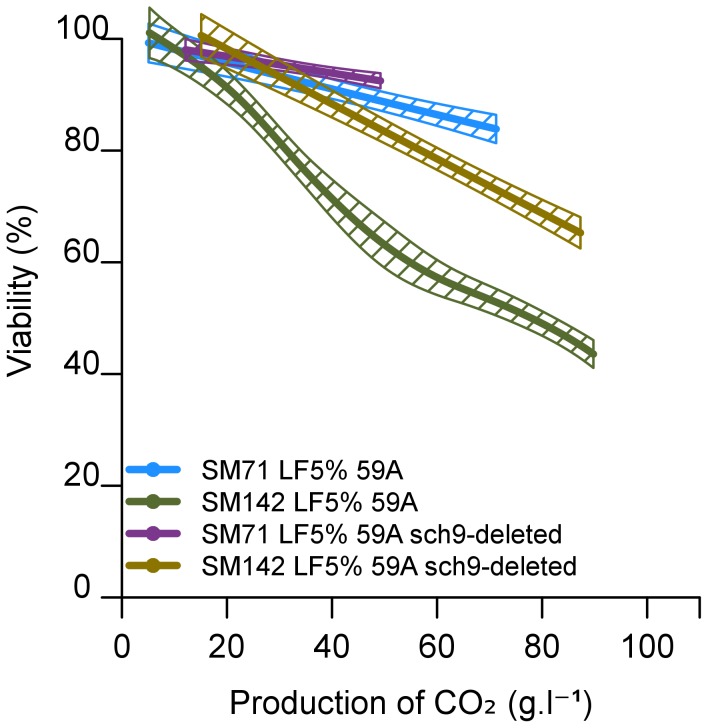
The TOR signalling pathway responds to nitrogen sources and controls various processes that affect the stress resistance and longevity of yeast cells. The TOR kinase, TORC1 (Target Of Rapamycin Complex 1), part of the TOR signalling pathway, is specifically inhibited by rapamycin [18]. The inclusion of 20 nmol/L rapamycin in cultures from inoculation of SM142 LF5% significantly increased the cell viability, to nearly 100 % (Figure 5). This implicates TOR signalling pathway in the cellular response leading to cell death. We then investigated whether SCH9 was involved. Sch9p is a protein kinase that plays a key role in controlling cell survival and integrates signals from TOR, PKA and other regulators [19], [20]. We assessed the consequences of deletion of the SCH9 gene from a haploid derivative of EC1118, 59A (strain sch9::KanMX 59A). We examined the response of the SCH9-deleted strain to nutritional imbalances and its effect on cell viability. In SM71 LF5% the production rate of CO2 by the SCH9-deleted strain was similar to that of the non-deleted controls (data not shown), and viability was similar. However, in SM142 LF5% the viability of the SCH9-deleted strain was significantly greater than that of the wild-type strain, reaching 80% versus 60% in the wild type strain at 60 g of CO2 produced (Figure 6). Thus, deletion of SCH9, like inhibition of TOR by rapamycin, favoured survival in conditions of lipid/nitrogen imbalance. This suggests that cell death in such nutritional conditions is controlled by Sch9p and TOR activity. Sch9p regulates the expression of a set of genes [21]–[23]. To identify target genes that may contribute to protect against cell death under our conditions, we evaluated the transcriptome changes associated with SCH9 deletion.
Figure 5. Effect of rapamycin on the viability of S. cerevisiae EC1118 cells during alcoholic fermentation.
The synthetic medium contains 142 mg/L assimilable nitrogen (SM142) and 5% lipid factors (LF5%), with or without 20 nmol/L rapamycin. Viability was measured by flow cytometry after propidium iodide staining. The graphs are the result of smoothing of measurements series (at least 3 repetitions) using the software R. Smoothing obtained is framed by a confidence interval calculated at 95%.
Figure 6. Effect of Sch9 deletion on the cell viability of haploid S. cerevisiae 59A during alcoholic fermentation.
The synthetic medium contains 71 mg/L (SM71) or 142 mg/L (SM142) of assimilable nitrogen, and 5% lipid factor (LF5%). Viability was measured by flow cytometry after propidium iodide staining. The graphs are the result of smoothing of measurements series (at least 3 repetitions) using the software R. Smoothing obtained is framed by a confidence interval calculated at 95%.
Global expression changes in the SCH9 mutant under lipid limitation
We performed a genome-wide analysis of the effect of the sch9-deletion on gene expression using DNA microarrays. We compared the expression profiles of wild-type cells (59A) and the sch9-deleted mutant during fermentation at 35 g CO2: 104 genes were found to be down-regulated (P-value < 0.05; log FC <−1) and 134 genes up-regulated (P-value < 0.05; log FC >1) in the sch9 mutant (Table S2 and Table S3). Most of the non-redundant GO terms that were significantly associated with the down-regulated gene list were related to ergosterol, sterol, steroid, lipid, isoprenoid and, isopentyl diphosphate biosynthetic processes (Table 3). Almost all the genes encoding proteins involved in ergosterol biosynthesis (ERG1, ERG2, ERG3, ERG5, ERG6, ERG8, ERG10, ERG11, ERG12, ERG13, ERG20, HMG1) and other lipid-related genes (12 genes), were down-regulated. Thus, sch9-deletion significantly affected lipid metabolism. Other down-regulated genes were ribosome biogenesis and rRNA processing genes (19 genes), nucleotide-related genes (6 genes), nitrogen-related genes (8 genes) and genes encoding permeases (9 genes). These findings confirm the interrelationship between the TORC1 signaling pathway and the lipid metabolism. They are consistent with the regulation of ribosome biogenesis and RNA processing by Sch9 [23], [24], but in contrast to the genes involved in the ergosterol biosynthesis pathway in non-lipid limited conditions [25]. Significant up-regulation of genes coding for proteins involved in the response to stress (SSA3, TIP1, HSP30, FMP45, SSA4, HSP12, CTT1, SKN7, XBP1, HSP104, DDR2, GRE1, HAL1), oxidoreductase activity (18 genes), cell adhesion molecule binding (11 genes), the MAPK signalling pathway and mating (8 genes) were observed in sch9-deleted mutant (Table 4). Therefore, deletion of SCH9 increased the stress response in lipid-limited conditions. Interestingly, most of the up-regulated genes encoded cell wall or plasma membrane proteins, HXT5, AGA1, SCW10 and HSP12 being among the most strongly over-expressed genes. It is also interesting to note that, related to the increase of HXT5 expression observed in the present study, this hexose transporter has also been described, among the most physiological-relevant transporters, as being the less inhibited by ethanol [26].
Table 3. GO term annotations for genes significantly down-regulated in the 59A sch9-deleted mutant.
| GO category | GO no. | P-value | GO description | k | f |
| Biological process | GO:0006696 | <1e-14 | ergosterol biosynthetic process | 12 | 23 |
| GO:0016126 GO:0006694 GO:0008610 GO:0008299 GO:0019287 | <1e-14 | sterol biosynthetic process | 14 | 29 | |
| GO:0008152 | <1e-14 | steroid biosynthetic process | 12 | 25 | |
| 6.403e-13 | lipid biosynthetic process | 13 | 52 | ||
| 1.457e-10 | isoprenoid biosynthetic process | 7 | 12 | ||
| 3.797e-06 | isopentenyl diphosphate biosynthetic process | 3 | 3 | ||
| 7.835e-06 | metabolic process | 20 | 425 | ||
| Cellular component | GO:0005730 | 3.722e-05 | nucleolus | 14 | 253 |
k: number of genes in the family identified as affected in the experiment; f: total number of genes in the family.
Table 4. GO term annotations of genes significantly up-regulated in the 59A sch9-deleted mutant.
| GO category | GO no. | P-value | GO description | k | f |
| Molecular function | GO:0016491 | 1.999e-06 | oxidoreductase activity | 19 | 272 |
| GO:0050839 | 8.175e-06 | cell adhesion molecule binding | 3 | 3 | |
| Biological process | GO:0007155 | 6.496e-08 | cell adhesion | 5 | 7 |
| Biological process | GO:0000755 | 1.704e-07 | cytogamy | 5 | 8 |
| GO:0000746 | 7.986e-07 | conjugation | 4 | 5 | |
| GO:0055114 | 1.999e-06 | oxidation-reduction process | 19 | 272 | |
| GO:0006950 | 1.094e-05 | response to stress | 13 | 152 | |
| Cellular component | GO:0005576 | 5.151e-10 | extracellular region | 15 | 95 |
| GO:0005886 | 5.299e-09 | plasma membrane | 26 | 350 | |
| GO:0001950 | 1.36e-08 | plasma membrane enriched fraction | 13 | 86 | |
| GO:0009277 | 9.462e-07 | fungal-type cell wall | 11 | 85 | |
| GO:0005618 | 7.976e-06 | cell wall | 9 | 68 | |
| GO:0031225 | 2.78e-05 | anchored to membrane | 8 | 61 | |
| GO:0000015 | 7.934e-05 | phosphopyruvate hydratase complex | 3 | 5 |
k: number of genes in the family identified as affected in the experiment; f: total number of genes in the family.
Is the stress response an adaptation to nitrogen/lipid imbalance?
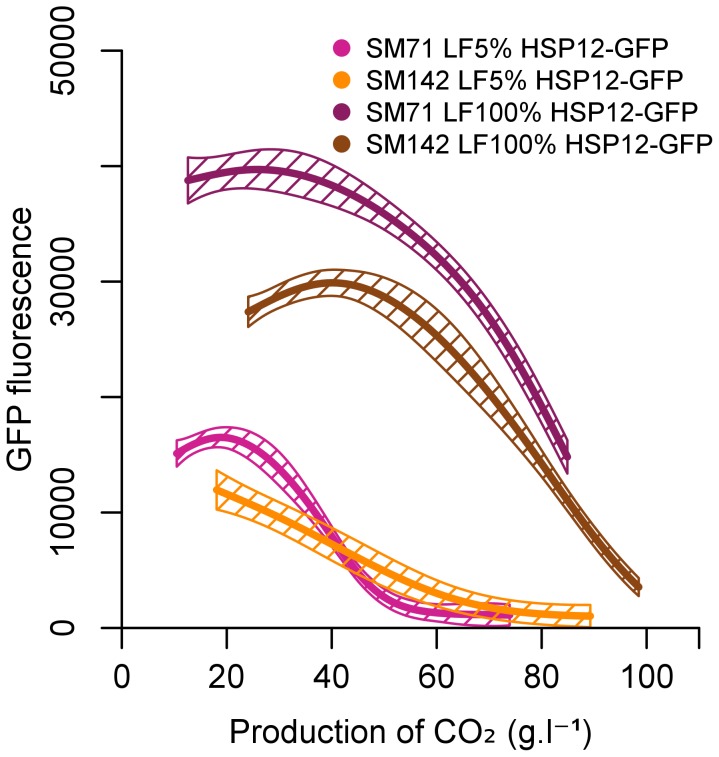
Since HSP12 was strongly overexpressed in the sch9-deleted strain and was shown to play a role in cell protection, we aimed to monitor its response in our conditions. We constructed a derivative of the strain EC1118 producing GFP under the control of the HSP12 promoter. We checked that this hsp12::GFP strain had a rate of CO2 production (Figure S4), a cell population (data not shown) and a viability (Figure S5) similar to the native strain during fermentation in each SM142 LF5% and SM71 LF5%. Then we used flow cytometry to monitor the activity of the HSP12 gene promoter, during fermentation under various conditions of nutritional imbalance (Figure 7). The HSP12-gene promoter was mainly active at the beginning of the stationary phase, decreasing thereafter in all the nutrient conditions tested: its activity was independent of nitrogen source, but depended on the lipids in the medium, as previously observed by Rossignol [27]. In our conditions, cells with greater viability had higher HSP12 gene promoter activity, consistent with the protective function of HSP12 under stress conditions [28]. However, deletion of the HSP12 gene has no effect on the mortality of the wine yeasts when both lipid and nitrogen were limited (data not shown). In our study, the absence of phenotype for HSP12 deletion is in line with several other papers that show no phenotype for deletion or overexpression of this gene under other stress conditions [29], [30].
Figure 7. Effect of the nutrient imbalances on HSP12 stress gene expression in S. cerevisiae EC1118 during alcoholic fermentation.
The synthetic medium contained 71 mg/L (SM71) or 142 mg/L (SM142) assimilable nitrogen, and 5% or 100% lipid factors (LF5% or LF100%). Fluorescence of the GFP reporter protein, produced under the control of the HSP12 promoter, was measured by flow cytometry. The graphs are the result of smoothing of measurements series (at least 3 repetitions) using the software R. Smoothing obtained is framed by a confidence interval calculated at 95%.
Discussion
Alcoholic fermentations of white and some rosé wines take place after clarification steps which decrease the lipid content of the must. Alcoholic fermentations are performed in conditions of strong anaerobiosis which prevents biosynthesis of sterols and unsaturated fatty acids. Several studies have reported the negative effect on wine fermentations of excessive clarifications of musts [6], [31], [32]. They lead to sluggish fermentations associated with high rates of yeast cell death and often to stuck fermentations. Yeast cell death in such conditions has been considered to be the direct outcome of membrane lipid changes and the inability of yeast to cope with ethanol toxicity. Indeed, we observed in our experiments that ergosterol availability had a major effect on yeast viability during fermentation, consistent with the findings of Luparia et al. [7]. The protective effect of ergosterol may be related to a substantial restoration of the rigidity of the plasma membrane [33], ergosterol being the main sterol in yeast: it plays important roles in the construction and maintenance of membrane structures and properties, such as integrity, fluidity, permeability, ethanol resistance and H+-ATPase activity (for a review, see [9]). The relevance of other nutrients on yeast cell death in such conditions has not previously been considered. Here, we show that yeast cell death in lipid-limited fermentations is strongly modulated by the nitrogen content of the medium, with high nitrogen availability leading to high rates of cell death. The triggering of yeast cell death was not restricted to a particular nitrogen source but was caused by various sources, including amino acids and ammonium, albeit to different extents. When included in SM71, only those amino acids that are metabolised were toxic: arginine, glutamate and glutamine caused cell death whereas proline and histidine did not. This suggests that the uptake of amino acids is required for this toxicity. We show that this toxic effect is dependent on the TOR nitrogen signalling pathway, and inhibition of the TOR kinase by rapamycin increased viability. The TOR kinase TORC1 is involved in sensing the nitrogen status of the cell and controls various cellular processes in response to nitrogen [34]. We also found that deletion of the gene encoding the TOR-associated protein Tco89, member of the TORC1 multiprotein complex required for TOR signalling, restored high viability in lipid-limited fermentations, providing further evidence of TOR involvement (data not shown). Our data therefore implicate TOR nitrogen signalling in the triggering of yeast cell death in lipid-limited fermentations. We report the first evidence of a relationship between the TORC1 nitrogen signalling pathway and lipid limitation in alcoholic fermentations. The involvement of nitrogen in wine yeast cell death has been recently reported in other situations [15]–[17], [35], for example in aging yeast cells. Ammonium is toxic for yeast and TOR signalling appears to be involved, but SCH9 deletion decreased yeast survival in these conditions [17]: the mechanism of the phenomenon we report is likely to be different because SCH9 deletion improved yeast cell viability in our model. Most studies have analysed cell death as a chronological ageing process, dependent on time [21], [22], [35], [36]. Here, we show that cell death during fermentation was controlled by the progress of fermentation, i.e. correlated with the ethanol content of the medium, consistent with a central role for ethanol.
Our experiments reveal a previously unsuspected role for nitrogen sources in wine fermentation: they have toxic effects when associated with low availability of lipids and specifically sterols. The role of nitrogen in cell death is obviously complex because the highest nitrogen levels were not found to be as toxic as intermediate concentrations. Many residual amino acids were detected in the medium in these conditions suggesting that they may act through other sensing pathways that interact with the TOR signalling outputs. The apparent complexity of the effects of nitrogen sources has probably contributed to obscuring their role in triggering yeast cell death, because high nitrogen levels are not always necessarily associated with high cell death rates in lipid-limited musts. Our data provide a basis for further analysis of the mechanisms and conditions which favour nitrogen toxicity in wine alcoholic fermentations.
Sch9p is a serine/threonine protein kinase localised at the vacuole surface; it is a target of TORC1 and is central to nutrient-mediated signalling [19]. TORC1-independent functions of Sch9p, with a direct link between Sch9p and nitrogen metabolism, have also been suggested [22]. In addition, Sch9p integrates also stress signal from sphingolipids, whose synthesis was related to lifespan in yeast [37]. But in our conditions, only limited changes in expression of genes involved in sphingolipid biosynthesis were observed. In fact, we found 6 genes related to sphingolipid biosynthesis, metabolism or signaling whose expression was down-regulated in the SCH9 mutant vs wt. However, they have a log FC between −0.86 and −0.39 and only genes with log2 fold change greater than 1 (positive or negative) were considered. We show that SCH9 is involved in the control of cell death in alcoholic fermentation conditions. Our transcriptional analysis of a SCH9-deleted strain suggests that Sch9p may act through a modulation of the expression of many genes in our conditions. Indeed, several genes whose expression is controlled by Sch9p are expected to play a role in yeast cell protection and may contribute to cell survival. The deletion of SCH9 did not, however, exactly recapitulate the effect of nitrogen source modulation as exemplified by the response of HSP12 which is strongly regulated by SCH9 deletion but only weakly by nitrogen availability. In addition, the response of HSP12 highlights the existence of specific control of the expression of a stress gene by lipids and in particular down-regulation of its expression by low lipid availability.
The overall stress response during fermentation, as evaluated from the storage glycogen and trehalose, was higher when yeast cells were starved of nitrogen than when starved of ergosterol in lipid-limited-fermentations. Yeast cells are obviously unable to elicit an appropriate stress response when growth is stopped by limiting sterol availability. Such limitations are probably uncommon in natural environments since only trace amounts (10 mg/L) of oxygen permit the production of sufficient sterols [38]. Therefore, the conditions encountered by yeasts in wine alcoholic fermentations are extreme conditions for which yeasts have not developed an adequate adaptation system. Indeed, our experimental model may be considered to impose nutrient restrictions unlikely to be encountered in nature and can be related to auxotrophic mutants that die rapidly upon starvation [10]: yeast is auxotrophic for sterols and unsaturated fatty acids when no oxygen is available.
Our data show, for the first time, that nitrogen excess in situation of low lipid, and low oxygen availability, as is common in wine making processes, may lead to yeast cell death and potentially to stuck fermentations. This work provides new notions to consider when defining nutrient management strategies for wine alcoholic fermentations. Nitrogen has been considered to be a positive factor in the outcome of alcoholic fermentations and nitrogen (in ammonium form) addition to musts or fermenting wines is a common practice. We demonstrate that nitrogen sources can potentially have a negative impact due to their signalling effects and this need to be taken into account. An in-depth understanding of the specific effects of each nitrogen source under such conditions is required to improve the prediction of the risks associated with nitrogen excess in lipid-limited fermentations.
Materials and Methods
Yeast strains
Lalvin EC1118 is a S. cerevisiae wine yeast isolated in Champagne (France) and manufactured by Lallemand (Montreal, Canada): 59A is a strain generated from a meiotic haploid spore isolated from Lalvin EC1118, and selected for its similar fermentation performance and metabolite production [39]. The EC1118 strain with HSP12 promoter-driven GFP expression was constructed by inserting the GFP coding sequence at the 3′-end of the HSP12 promoter by standard homologous recombination with a yEGFP-KanMX cassette amplified from pKT127 (pFA6a-link-yEGFP-KAN, P30175 Euroscarf strain) using synthetic primers (1A and 1B in Table 5). A SCH9-deleted mutant of 59A was generated by PCR-mediated gene disruption using the loxP-KanMX-loxP cassette of the pUG6 vector amplified with synthetic primers (2A and 2B in Table 5), to replace the ORF in 59A strain with a gene that confers resistance to G418. Gene disruptions and constructs were confirmed by PCR (primers 1C and 1D for EC1118 hsp12::GFP, and 2C to 2F for 59A sch9::KanMX, respectively, Table 5).
Table 5. Synthetic oligonucleotides.
| Name | Sequence |
| 1A | AACTCAAAACAAAAAAAACTAAATACAACAATGTCTGACGCAGGTGACGGTGCTGGTTTA |
| 1B | AGAAAAAACCATGTAACTACAAAGAGTTCCGAAAGATTCGATGAATTCGAGCTCG |
| 1C | GTGGAGTGCGATTTGTTCGT |
| 1D | CCAACCAACGCATCAAGAGA |
| 2A | TTAGCTCTCAACACCAACATCCAAATGGACAGAACATTCGTACGCTGCAGGTCGAC |
| 2B | CTTCCACTGACAAATTCGTCATCCATGTGTTGGTCGCATAGGCCACTAGTGGATCTG |
| 2C | ATCGTCGAATCAGGATACTGGA |
| 2D | CAAGAGGAGCGATTGAGAAA |
| 2E | ATTACGGCTCCTCGCTGCAG |
| 2F | TGATTTTGATGACGAGCGTAAT |
Synthetic culture media
Unless otherwise specified, synthetic fermentation medium with 142 mg/L assimilable nitrogen (SM142) and 23% glucose + fructose (1/1), strongly buffered to pH 3.3 and simulating one third nitrogen and amino acid concentrations of a standard grape juice [40] was routinely used. This medium contained, per litre: 115 g glucose, 115 g fructose, 6 g citric acid, 6 g DL-malic acid, 750 mg KH2PO4, 500 mg K2SO4, 250 mg MgSO4.7H2O, 155 mg CaCl2.2H2O, 200 mg NaCl, 4 mg MnSO4.H2O, 4 mg ZnSO4. 7H2O, 1 mg CuSO4.5H2O, 1 mg KI, 0.4 mg CoCl2.6H2O, 1 mg H3BO3, 1 mg (NH4)6Mo7O24, 20 mg myo-inositol, 2 mg nicotinic acid, 1.5 mg calcium panthotenate, 0.25 mg thiamine- HCl, 0.25 mg pyridoxine and 0.003 mg biotin. It also contained ammoniacal nitrogen and amino acids as nitrogen sources (per litre): 153 mg NH4Cl, 204 mg L-proline, 169 mg L-glutamine, 60 mg L-tryptophane, 48 mg L-alanine, 40 mg L-glutamic acid, 26 mg L-serine, 25 mg L-threonine, 16 mg L-leucine, 15 mg L-aspartic acid, 15 mg L-valine, 13 mg L-phenylalanine, 125 mg L-arginine, 11 mg L-histidine, 11 mg L-isoleucine, 10 mg L-methionine, 6 mg L-glycine, 6 mg L-lysine, 6 mg L-tyrosine and 4 mg L-cysteine. The medium was heat-sterilized (100°C, 10 min). Lipid factors (LF) were added to the medium after sterilization. The LF final concentration in the fermentation medium (LF 100%) was 4.5 mg/L oleic acid and 15 mg/L ergosterol. To evaluate the adaptation to nutrient limitations, media containing different concentrations of nitrogen sources (SM142 and SM71) with two different LF contents (100% or 5%) were tested.
Fermentation conditions and kinetics
The yeast strains used in this study were precultured in a nutrient medium containing Yeast Nitrogen Base (YNB) without amino acids (6.7 g/L) and glucose (20 g/L), for 24 h at 28°C in Erlenmeyer flasks. The fermentation medium was inoculated with 1 106 cell/mL from preculture. Yeast cultures were carried out in fermenters (1.2 L, containing 1 L medium), with fermentation locks (CO2 bubbling outlets filled with water). Fermentation media were routinely de-aerated prior to inoculation by bubbling pure argon for 5 min. Filling conditions were controlled and fermentations were carried out under anaerobic and isothermal conditions (24°C), with permanent stirring (300 rpm).
The amount of CO2 released was calculated from automatic measurements (taken every 20 min) of fermenter weight [41]. The CO2 production rate was calculated by polynomial smoothing.
Establishment of the fermentation conditions
The aim of this study was to determine how S. cerevisiae wine yeast strains respond to lipid limitation in the presence of various nitrogen contents. This was done by monitoring CO2 production, cell counts and yeast viability in a synthetic grape juice mimicking an oenological environment. Two main fermentation conditions, with 230 g/L glucose + fructose (1/1), were established with different LF concentrations in the culture medium (LF100% and LF5%): SM142 and SM71 contain, respectively, one third and one sixth of the nitrogen concentration required for the yeast strain to complete fermentation. These N-limiting conditions were chosen to ensure sluggish fermentations. For rapamycin treatment, cells were grown in SM142 with LF5% and 20 nM rapamycin was added at T0. The conditions of growth of wt and sch9 strains used for microarray analysis were SM71 and SM142 with LF 5%.
Cell population densities, cell viability and GFP activity determinations
Cell population densities, cell viability and Hsp12 promoter activity were determined by flow cytometry using a C6 cytometer (Accuri, BD Biosciences). To determine cell population densities, the cell suspension was compared to a suspension of latex beads of known concentration. For cell viability analysis, propidium iodide (PI) (Calbiochem) was added to the cell suspension (5 µL of 0.1 mg/mL solution), and the samples mixed by gentle shaking. PI is a fluorescent nucleic acid stain (excitation 488 nm, emission 575 nm) which cannot penetrate intact cell membranes. PI flow cytometry analysis was performed 10 min after staining. Fluorescence data for cells stained by PI were collected in channel FL3. Viability was determined as the percentage of intact and fragile cells among all cells [42]. For Hsp12 promoter activity, GFP fluorescence (excitation 488 nm, emission 530 nm) was collected in channel FL2.
Determination of the trehalose and glycogen contents
Trehalose was extracted from cells with 0.5 M trichloracetic acid (TCA) and quantified with anthrone according to Rossignol [27]. Glycogen was extracted from the same sample with HCl-DMSO and treated with an amyloglucosidase enzyme. The glucose formed was assayed by colorimetry [43]. Results are expressed as % dry weight.
RNA extraction and DNA microarray analyses
Total RNAs were isolated from wild type 59A cells and sch9-deleted mutant cultures at 35 g of CO2 production, by the TRIzol® method according to Chomczynski and Sacchi [44]. Aliquots of 109 cells were harvested and quickly washed with 750 µL cooled (4°C) DEPC-treated water. Cells were pelleted and frozen in a −80°C methanol bath. Frozen cells were mechanically lysed by vortexing with glass beads (d = 0.3 mm) in 400 µL TRIzol® (GIBCO BRL) at 4°C for 15 min. The liquid phase was collected and TRIzol® added to give a final volume of 4 mL. The samples were mixed and incubated for 5 min at room temperature, and 800 µL chloroform was added. The mixture was vortexed and then incubated for 3 min and centrifuged (9,000 g for 15 min). The supernatant was centrifuged again (2,000 g for 2 min) in swing buckets. RNAs were pelleted from 2 mL aliquots of the supernatant by the addition of 2 mL cooled isopropanol (−20°C) and incubated for 10 min. The samples were centrifuged (9,000 g for 10 min) and the resulting nucleic acid pellet was washed twice with 750 µL 75% ethanol/DEPC-treated water and then dissolved in 150 µL of nuclease-free water (Qiagen).
Total RNA from 100 µg aliquots of these preparations was purified with a RNeasy® mini kit (Qiagen) following the RNA cleanup protocol, including membrane DNase digestion. RNAs were eluted with 2 × 30 µL of the provided RNAse-free water. RNA quality was verified by capillary electrophoresis with an RNA 6000 Nano LabChip Kit (Agilent Technologies). Samples of 100 ng of purified RNA were labelled with Low input Quick Amp Labelling one-colour kit (Agilent Technologies) according to manufacturer's recommendation (indirect labelling of mRNAs with Cyanin 3 dCTP dye). RNAs were hybridised on 8 × 15 k array Agilent standard Yeast V2 Gene Expression Microarrays (Agilent Technologies) for 17 h in a rotating oven at 65°C following the manufacturer's recommendation. A Genepix 4000B scanner was used for array digitalization: the laser voltage was set to avoid signal saturation and data was extracted with GenePix® Pro 7 software (Molecular Devices).
R.2.14.21. was used for statistical analysis [45]. The limma package [46]–[48] was used to normalise the microarray data (quantile method for normalisation between arrays). To analyse differential gene expression between experimental conditions, a modified t-test was used by filtering on confidence at p<0.05, using the Benjamini and Hochberg false discovery rate as multiple testing corrections of the t-test p-values [49]. Only genes with a log2 fold change greater than 1 (positive or negative) were considered. The complete data set is available through the Gene Expression Omnibus database (accession number GSE42027). For a statistical treatment of groups of genes, data were analysed using the web-based tool Funspec (http://funspec.med.utoronto.ca/; [50]) (P-value < 0.05 and Bonferroni correction) and genes were classified into functional categories, biological process and protein cellular localisations using the GO Database.
Supporting Information
Effect of nutrient imbalances on the cell population of S. cerevisiae EC1118 cultures during alcoholic fermentation at 24°C according to CO2 production. The synthetic medium contained 71 mg/L (SM71), 142 mg/L (SM142) or 425 mg/L (SM425) assimilable nitrogen, and 5% or 100% lipid factors (LF 5% or LF100%). The graphs are the result of smoothing of measurement series (at least 3 repetitions) using the software R.
(TIFF)
Effect of lipid factors on the rate of CO2 production of S. cerevisiae EC1118 cultures during alcoholic fermentation at 24°C. The synthetic medium contained 142 mg/L assimilable nitrogen (SM142) and 5% lipid factor (LF5%), with or without additional oleic acid or ergosterol (content as in LF 100%). The graphs are the result of smoothing of measurement series (at least 3 repetitions) using the software R.
(TIFF)
Effect of nitrogen sources on the rate of CO2 production of S. cerevisiae EC1118 cells during alcoholic fermentation at 24°C. The synthetic medium contained 71 mg/L assimilable nitrogen (SM71) and 5% lipid factors (LF5%), with or without additional arginine, glutamine, glutamate, histidine or proline (content as in SM142). The graphs are the result of smoothing of measurement series (at least 3 repetitions) using the software R.
(TIFF)
Effect of nutrient imbalances on the rate of CO2 production by cultures of S. cerevisiae EC1118 carrying the HSP12-GFP fusion during alcoholic fermentation at 24°C. The synthetic medium contained 71 mg/L (SM71) or 142 mg/L (SM142) assimilable nitrogen, and 5% or 100% lipid factors (LF5% or LF100%). The graphs are the result of smoothing of measurement series (at least 3 repetitions) using the software R.
(TIFF)
Effect of nutrient imbalances on the viability of S. cerevisiae EC1118 carrying the HSP12-GFP fusion during alcoholic fermentation at 24°C. The synthetic medium contained 71 mg/L (SM71) or 142 mg/L (SM142) assimilable nitrogen, and 5% lipid factors (LF5%). The graphs are the result of smoothing of measurement series (at least 3 repetitions) using the software R.
(TIFF)
Effect of nutrient imbalances on the amino acid content of S. cerevisiae EC1118 cells (µmol assimilable nitrogen/L).
(DOC)
Genes that were significantly down-regulated in the 59A SCH9 -deleted mutant.
(DOC)
Genes that were significantly up-regulated in the 59A SCH9 -deleted mutant.
(DOC)
Acknowledgments
We thank Isabelle Sanchez (INRA, UMR SPO) for the statistical analysis of the microarray data.
Funding Statement
Grants have been obtained from INRA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Agenbach WA (1977) A study of must nitrogen content in relation to incomplete fermentations, yeast production and fermentation activity. In: Proceedings South African Society of Enology and Viticulture, Cape Town, South Africa Stellenbosh SA: pp. 66–87.
- 2. Cramer AC, Vlassides S, Block DE (2002) Kinetic model for nitrogen-limited wine fermentations. Biotechnol Bioeng 77: 49–60. [DOI] [PubMed] [Google Scholar]
- 3. Bely M, Sablayrolles JM, Barre PO (1990) Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in oenological conditions. J Ferm Bioen 70: 246–252. [Google Scholar]
- 4. Fornairon-Bonnefond C, Demaretz V, Rosenfeld E, Salmon JM (2002) Oxygen addition and sterol synthesis in Saccharomyces cerevisiae during enological fermentation. J Biosci Bioeng 93: 176–182. [DOI] [PubMed] [Google Scholar]
- 5. Zara G, Angelozz D, Belviso S, Bardi L, Goffrini P, et al. (2009) Oxygen is required to restore flor strain viability and lipid biosynthesis under fermentative conditions. FEMS Yeast Res 9: 217–225. [DOI] [PubMed] [Google Scholar]
- 6. Alexandre H, Nguyen VLT, Feuillat M, Charpentier C (1994) Contribution à l'étude des bourbes: influence sur la fermentescibilité des moûts. Rev Fr Oenol 146: 11–20. [Google Scholar]
- 7. Luparia V, Soubeyrand V, Berges T, Julien A, Salmon JM (2004) Assimilation of grape phytosterols by Saccharomyces cerevisiae and their impact on enological fermentations. Appl Microbiol Biotechnol 65: 25–32. [DOI] [PubMed] [Google Scholar]
- 8. van der Rest ME, Kamminga AH, Nakano A, Anraku Y, Poolman B, et al. (1995) The plasma membrane of Saccharomyces cerevisiae: structure, function and biogenesis. Microbiol Rev 59: 304–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Daum G, Lees ND, Bard M, Dickson R (1998) Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae . Yeast 14: 1471–1510. [DOI] [PubMed] [Google Scholar]
- 10. Boer VM, Amini S, Botstein D (2008) Influence of genotype and nutrition on survival and metabolism of starving yeast. Proc Natl Acad Sci 105: 6930–6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rohde JR, Bastidas R, Puria R, Cardenas ME (2008) Nutritional control via Tor signaling in Saccharomyces cerevisiae. . Curr Op Microbiol 11: 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smets B, Ghillebert R, de Snijder P, Binda M, Swinnen E, et al. (2010) Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae . Curr Genet 56: 1–32. [DOI] [PubMed] [Google Scholar]
- 13. Roosen J, Engelen K, Marchal K, Mathys J, Griffioen G, et al. (2005) PKA and Sch9 control a molecular switch important for the proper adaptation to nutrient availability. Mol Microbiol 55: 862–880. [DOI] [PubMed] [Google Scholar]
- 14. Shin CS, Kim SY, Huh WK (2009) TORC1 controls degradation of the transcription factor Stp1, a key effector of the SPS amino-acid-sensing pathway in Saccharomyces cerevisiae . J Cell Sci 122: 2089–2099. [DOI] [PubMed] [Google Scholar]
- 15. Fabrizio P, Battistella L, Vardavas R, Gattazzo C, Liou LL, et al. (2004) Superoxide is a mediator of an altruistic aging program in Saccharomyces cerevisiae . J Cell Biol 166: 1050–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, et al. (2010) A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet 6: e1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santos J, Sousa MJ, Leão C (2012) Ammonium is toxic for aging yeast cells, inducing death and shortening of the chronological lifespan. PLoS One 7: e37090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cardenas ME, Cutler NS, Lorenz MC, Di Como CJ, Heitman J (1999) The TOR signaling cascade regulates gene expression in response. Genes Dev 13: 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, et al. (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. . Mol Cell 26: 663–674. [DOI] [PubMed] [Google Scholar]
- 20. Zaman S, Lippman SI, Zhao X, Broach JR (2008) How Saccharomyces responds to nutrients. Ann Rev Genet 42: 27–81. [DOI] [PubMed] [Google Scholar]
- 21. Lavoie H, Whiteway M (2008) Increased respiration in the sch9D mutant is required for increasing chronological life span but not replicative life span. Euk Cell 7: 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smets B, Snijder PD, Engelen K, Joossens E, Ghillebert R, et al. (2008) Genome-wide expression analysis reveals TORC1-dependent and -independent functions of Sch9. FEMS Yeast Res 8: 1276–1288. [DOI] [PubMed] [Google Scholar]
- 23. Ge H, Wei M, Fabrizio P, Hu J, Cheng C, et al. (2010) Comparative analyses of time-course gene expression profiles of the long-lived sch9D mutant. Nucl Acid Res 38: 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huber A, Bodenmiller B, Uotilan A, Stahl M, Wanka S, et al. (2009) Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev 23: 1929–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wei M, Fabrizio P, Madia F, Hu J, Ge H, et al. (2009) Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet 5: e1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Santos J, Sousa MJ, Cardoso H, Inácio J, Silva S, et al. (2008) Ethanol tolerance of sugar transport, and the rectification of stuck wine fermentations. Microbiol 154: 422–430. [DOI] [PubMed] [Google Scholar]
- 27.Rossignol T (2004) Analyse de l'expression du génome des levures oenologiques en fermentation alcoolique par des approches post-génomiques. Montpellier: University of Montpellier II. 246 p.
- 28. Sales K, Brandt W, Rumbak E, Lindsey G (2000) The LEA-like protein HSP12 in Saccharomyces cerevisiae has a plasma membrane location and protects membranes against desiccation and ethanol-induced stress. Biochim Biophys Acta 1463: 267–278. [DOI] [PubMed] [Google Scholar]
- 29. Pacheco A, Pereira C, Almeida M J, Sousa MJ (2009) Small heat-shock protein Hsp12 contributes to yeast tolerance to freezing stress. Microbiol 155: 2021–2028. [DOI] [PubMed] [Google Scholar]
- 30. Shamrock VJ, Lindsey GG (2008) A compensatory increase in trehalose synthesis in response to desiccation stress in Saccharomyces cerevisiae cells lacking the heat shock protein Hsp12p. Can J Microbiol 54: 559–568. [DOI] [PubMed] [Google Scholar]
- 31. Groat M, Ough CS (1978) Effects of insoluble solids added to clarified musts on fermentation rate, wine composition, and wine quality. Am J Enol Vitic 29: 112–119. [Google Scholar]
- 32. Houtman AC, du Plessis CS (1986) Nutritional deficencies of clarified white grape juices and their correction in relation to fermentation. S Afr J Enol Vitic 1: 39–46. [Google Scholar]
- 33. Abe F, Hiraki T (2009) Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae . Biochim Biophys Acta 1788: 743–752. [DOI] [PubMed] [Google Scholar]
- 34.Loewith R (2010) TORC1 signaling in budding yeast. In: Hall M, Tamanoi F, editors. The enzymes: structure, function and regulation of TOR complexes from yeasts to mammals. Burlington: Academic Press. pp. 147–175. [Google Scholar]
- 35. Orozco H, Matallana E, Aranda A (2012) Wine yeast sirtuins and Gcn5p control aging and metabolism in a natural growth medium. Mech Ageing Develop 33: 348–358. [DOI] [PubMed] [Google Scholar]
- 36. Fabrizio P, Longo VD (2003) The chronological life span of Saccharomyces cerevisiae . Aging 2: 73–81. [DOI] [PubMed] [Google Scholar]
- 37. Huang X, Liu J, Dickson RC (2012) Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet 8: e1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sablayrolles JM (2009) Control of alcoholic fermentation in winemaking: current situation and prospects. Food Res Intern 42: 418–424. [Google Scholar]
- 39. Ambroset C, Petit M, Brion C, Sanchez I, Delobel P, et al. (2012) Deciphering the molecular basis of wine yeast fermentation traits using a combined genetic and genomic approach. G3 1: 263–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bely M, Sablayrolles JM, Barre PO (1990) Description of alcoholic fermentation: its variability and significance. Am J Enol Vitic 40: 319–324. [Google Scholar]
- 41. Sablayrolles JM, Barre P, Grenier P (1987) Design of a laboratory automatic system for studying alcoholic fermentations in anisothermal enological conditions. Biotech Technol 1: 181–184. [Google Scholar]
- 42. Delobel P, Pradal M, Blondin B, Tesnière C (2012) Fragile cell population revealed by preparation for cytometric assessment of Saccharomyces cerevisiae viability in lipid-limited alcoholic fermentation. Lett App Microbiol 55: 338–344. [DOI] [PubMed] [Google Scholar]
- 43. Roustan JL, Sablayrolles JM (2002) Trehalose and glycogen in wine-making yeasts: methodological aspects and variability. Biotech Lett 24: 1059–1064. [Google Scholar]
- 44. Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159. [DOI] [PubMed] [Google Scholar]
- 45.Team RDC (2008) A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing.
- 46. Smyth GK, Speed TP (2003) Normalization of cDNA microarray data. Methods 31: 265–273. [DOI] [PubMed] [Google Scholar]
- 47.Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3, Article 3. [DOI] [PubMed]
- 48. Smyth GK, Michaud J, Scott H (2005) The use of within-array replicate spots for assessing differential expression in microarrays experiments. Bioinf 21: 2067–2075. [DOI] [PubMed] [Google Scholar]
- 49. Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 85: 289–300. [Google Scholar]
- 50. Robinson MD, Grigull J, Mohammad N, Hughes TR (2002) FunSpec: a web-based cluster interpreter for yeast. BMC Bioinf 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of nutrient imbalances on the cell population of S. cerevisiae EC1118 cultures during alcoholic fermentation at 24°C according to CO2 production. The synthetic medium contained 71 mg/L (SM71), 142 mg/L (SM142) or 425 mg/L (SM425) assimilable nitrogen, and 5% or 100% lipid factors (LF 5% or LF100%). The graphs are the result of smoothing of measurement series (at least 3 repetitions) using the software R.
(TIFF)
Effect of lipid factors on the rate of CO2 production of S. cerevisiae EC1118 cultures during alcoholic fermentation at 24°C. The synthetic medium contained 142 mg/L assimilable nitrogen (SM142) and 5% lipid factor (LF5%), with or without additional oleic acid or ergosterol (content as in LF 100%). The graphs are the result of smoothing of measurement series (at least 3 repetitions) using the software R.
(TIFF)
Effect of nitrogen sources on the rate of CO2 production of S. cerevisiae EC1118 cells during alcoholic fermentation at 24°C. The synthetic medium contained 71 mg/L assimilable nitrogen (SM71) and 5% lipid factors (LF5%), with or without additional arginine, glutamine, glutamate, histidine or proline (content as in SM142). The graphs are the result of smoothing of measurement series (at least 3 repetitions) using the software R.
(TIFF)
Effect of nutrient imbalances on the rate of CO2 production by cultures of S. cerevisiae EC1118 carrying the HSP12-GFP fusion during alcoholic fermentation at 24°C. The synthetic medium contained 71 mg/L (SM71) or 142 mg/L (SM142) assimilable nitrogen, and 5% or 100% lipid factors (LF5% or LF100%). The graphs are the result of smoothing of measurement series (at least 3 repetitions) using the software R.
(TIFF)
Effect of nutrient imbalances on the viability of S. cerevisiae EC1118 carrying the HSP12-GFP fusion during alcoholic fermentation at 24°C. The synthetic medium contained 71 mg/L (SM71) or 142 mg/L (SM142) assimilable nitrogen, and 5% lipid factors (LF5%). The graphs are the result of smoothing of measurement series (at least 3 repetitions) using the software R.
(TIFF)
Effect of nutrient imbalances on the amino acid content of S. cerevisiae EC1118 cells (µmol assimilable nitrogen/L).
(DOC)
Genes that were significantly down-regulated in the 59A SCH9 -deleted mutant.
(DOC)
Genes that were significantly up-regulated in the 59A SCH9 -deleted mutant.
(DOC)