Abstract
The budding yeast proteins Dma1 and Dma2 are members of the unique FHA-RING domain protein family and are linked to mitotic regulation and septin organization by ill-defined mechanisms. We show that Dma2 has ubiquitin ligase activity, and that septins Shs1 and Cdc11 are likely direct in vivo targets. We further propose that human RNF8, rather than Chfr, is the mammalian Dma homolog. As in yeast, RNF8 localizes to the centrosomes and cell division sites and promotes ubiquitylation of the septin SEPT7, whose depletion increases cell division anomalies. Together, these findings reveal evolutionary and functional conservation of Dma proteins, and suggest that RNF8 maintains genome stability through independent, yet analogous, nuclear and cytoplasmic ubiquitylation activities.
Keywords: Chfr, cytokinesis, Dma1, Dma2, E3 ubiquitin ligase, midbody, RNF8, septins, septin-ring, ubiquitin
Introduction
Schizosaccharomyces pombe Dma1 protein possesses FHA and RING finger domains that are, respectively, known to mediate phosphothreonine-dependent protein-protein interactions and E3 ubiquitin ligase activity.1 Dma1 acts in the spindle assembly checkpoint, and its FHA domain targets Dma1 to the spindle pole body (SPB), where it blocks recruitment of the polo-like kinase (Plo1)—most likely through Sid4 ubiquitylation2—when spindle function is compromised.3 Dma1 also localizes in an FHA-dependent manner to the site of cell division, but the functional relevance of this is unknown.3Saccharomyces cerevisiae possesses two redundant paralogs, Dma1 and Dma2, sharing the same domain organization as S. pombe Dma1. Although S. cerevisiae Dma1 and Dma2 are not required for the spindle checkpoint, they control mitotic exit and also control proper organization of septins (see below) by an unknown mechanism.4 Mammalian cells contain two orthologous Dma paralogs named Chfr and RNF8. The former was originally shown to control mitotic entry,5,6 while RNF8 controls DNA-damage responses through histone ubiquitylation (Jackson and Durocher, in press). Although RNF8 is implicated in regulating mitotic exit and mitotic arrest following nocodazole treatment,7,8 direct evolutionarily conserved targets for Dma/RNF8 ubiquitylation activity during mitosis are unknown.
Septins are conserved GTP binding proteins that assemble into filaments.9 Notably, S. cerevisiae septins first assemble at the incipient bud site. When buds emerge (G1/S), they reorganize to form a ring structure that eventually splits into two rings at cytokinesis.10 Rings then disassemble and the septin-assembly cycle starts again in G1. Yeast septins are implicated in bud-site selection, the control of bud growth, spindle positioning and cytokinesis.11 The filaments formed by septins serve as scaffolds that recruit other proteins, including myosin (Myo1) and the septation protein Hof1.12,13 During cytokinesis, the split septin rings act as a barrier to compartmentalize the cortex around the cell cleavage site.9 Analogously, human septins localize to each side of the cell-cleavage site and are important for cytokinesis,9,14 yet the mechanisms regulating septin activities remain obscure.
Here, we present evidence that Dma/RNF8—rather than Chfr—can ubiquitylate septins in vivo in an evolutionarily conserved manner. We also show that the Dma/RNF8 FHA domain is important for recruiting such proteins to sites of cell division, and that the RING domain mediates ubiquitylation needed for septin organization and function.
Results
Dma2 is an E3 ubiquitin ligase that colocalizes with septins during mitosis
The existence of RING finger domains in S. cerevisiae Dma1 and Dma2 suggested that they might be E3 ubiquitin ligases. By yeast two-hybrid analysis, we found that Dma2 interacted well with the ubiquitin E2 ligase Ubc5 and also with Ubc13 but not with the E2s Ubc7, Mms2 or Rad6 (Fig. S1A). Furthermore, we purified a glutathione-S-transferase (GST)-Dma2 fusion and tested it for auto-ubiquitylation. Western blot analysis with anti-GST and anti-ubiquitin (Ub) antibodies revealed the generation of ubiquitylated, higher molecular weight forms of GST-Dma2 when it was incubated with an E1, the E2 Ubch5, ubiquitin and ATP (Fig. S1B, lane 6). A Dma2 RING point mutant (Dma2C451A) lacked such activity (Fig. S1B, lane 7). In agreement with previous findings,15 these data established that Dma2 can serve as a RING-dependent E3 enzyme.
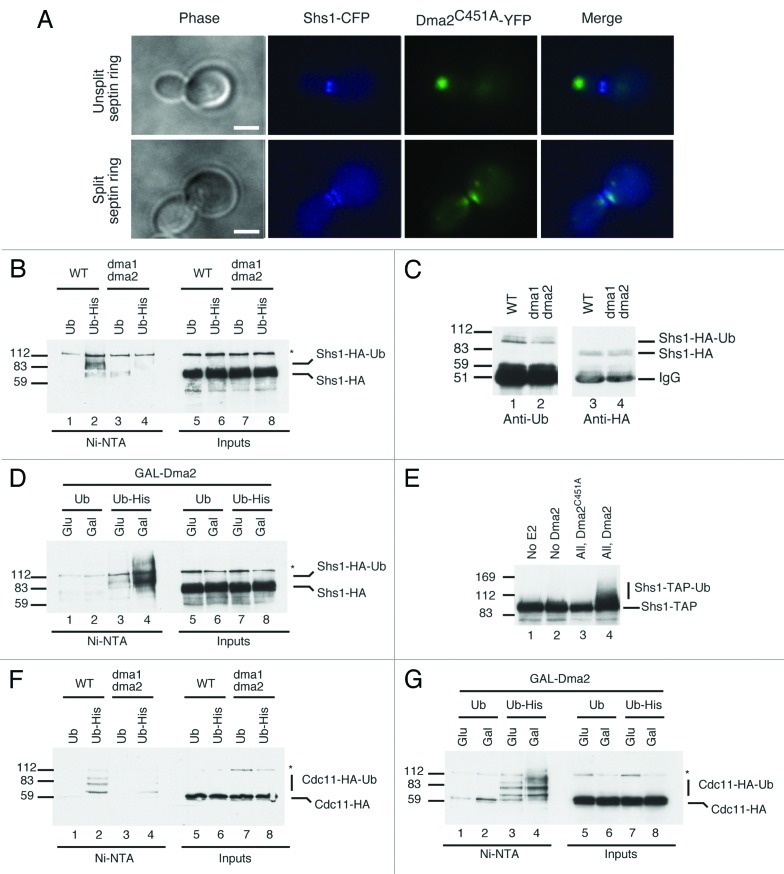
Overexpression of Dma2, or deletion of Dma2 and Dma1, interferes with septin organization and function,4 and Dma proteins control septin ring deposition and maintenance.16 Whether Dma proteins and septins physically associate in vivo, however, is unknown. Using a strain expressing endogenously tagged septin Shs1-CFP (Cyan-fluorescent protein) transformed with a 2-μ plasmid encoding Dma2-YFP (yellow fluorescent protein), we observed little or no colocalization of Shs1 and Dma2. Since Dma2 and Shs1 association might be transient, we speculated that it might be stabilized by inactivating Dma2 catalytic function. Indeed, we observed two distinct localization patterns for the Dma2C451A-YFP protein. For small- and large-budded cells with an unsplit septin ring, the bulk (~86%) of the Dma2C451A-YFP signal localized at the growing bud tip (Fig. 1A). In stark contrast, for cells where the septin hourglass structure had split into two defined rings, in the majority of cases (~95%), Dma2C451A-YFP localized at the mother-bud neck, where the septin rings are located (Fig. 1A). These data indicated that a catalytically functional RING domain is not required for localization of Dma2 at the bud neck, and, during the late stage of mitosis, Dma2 has the right localization to potentially interact with and ubiquitylate septins.
Figure 1. Effects of Dma on septin proteins. (A) Cells expressing endogenous-tagged Shs1-CFP and YFP-Dma2C451A were grown to mid-log, fixed and analyzed. Unsplit septin ring (top) and split septin ring (bottom) are presented (scale bar is 5 μm). (B) WT and dma1∆ dma2∆ cells expressing Shs1-HA and overexpressing Ub or His-Ub were purified and blotted with anti-HA. Protein markers (kDa) are on the left. (C) WT and dma1∆ dma2∆ cells expressing Shs1-HA were purified and blotted with anti-Ub (left); the blot was stripped and reprobed with anti-HA (right). (D) WT cells expressing GAL-Dma2, Shs1-HA and overexpressing Ub or His-Ub were analyzed as in (B). (E) Shs1-TAP was purified from yeast extracts and resuspended in 10 μl of ubiquitylation buffer containing E1 and E2 enzymes, ubiquitin, ATP and recombinant Dma2 or Dma2C451A was added or omitted, as indicated. Samples were analyzed by blotting with anti-TAP antibody. (F) WT and dma1∆ dma2∆ cells expressing Cdc11-HA and overexpressing Ub or His-Ub were analyzed as in (B). (G) WT cells expressing GAL-Dma2, Cdc11-HA and overexpressing Ub or His-Ub were analyzed as in (D).
Dma1 and Dma2 modulate ubiquitylation of septins Shs1 and Cdc11 in vivo
We expressed tagged Shs1 (Shs1-HA) in a (wild-type) WT strain and in a strain deleted for both DMA1 and DMA2 (to avoid redundancies4). The strains used were copper-inducible for either ubiquitin (Ub) or hexa-histidine-tagged ubiquitin (His-Ub). At mid-log phase, ubiquitin or His-Ub was induced, ubiquitylated proteins were purified, and bound materials were analyzed by blotting with an Shs1-HA antibody. Modified Shs1 was readily detectable from WT strains expressing His-Ub, clearly representing ubiquitylated forms of Shs1 (Fig. 1B, compare lanes 1 and 2). In these analyses, we routinely detected two closely migrating modified bands and believe that the lower migrating species represents monoubiquitylated Shs1, while the upper band could either be a diubiquitylated form or Shs1 with additional modification(s). Notably, Shs1 is phosphorylated and sumoylated in vivo,17 yet mutating the established Shs1 sumoylation sites, lysines 426 and 437, into arginine essentially abolished sumoylated Shs1 derivatives (Fig. S1C, left) but maintained ubiquitylated Shs1 (Fig. S1C, right). These data therefore suggested that Shs1 sumoylation and ubiquitylation occur on different lysine residues. Notably, the levels of ubiquitylated Shs1 were reduced in dma1∆ dma2∆ cells (Fig. 1B). In line with these findings, immunoprecipitation of Shs1-HA followed by western analysis with an anti-Ub antibody indicated that DMA1 and DMA2 inactivation led to lower levels of ubiquitylated Shs1 (Fig. 1C). This phenotype is unlikely to be due to a defective cell cycle progression, because dma1∆ dma2∆ cells have a comparable cell cycle distribution profile to WT cells (Fig. S1D). Collectively, these data indicated that Dma1 and Dma2 control Shs1 ubiquitylation, and suggested that they might directly target Shs1 in vivo.
If Shs1 was a direct target of Dma1 and Dma2, then overexpression of these ubiquitin ligases might increase Shs1 ubiquitylation. We therefore engineered strains expressing copper-inducible Ub or His-Ub to permit the controlled overexpression of Dma2 from a galactose-inducible promoter. After purification and western blotting, extracts prepared from cells overexpressing Dma2 contained markedly increased levels of ubiquitylated Shs1 (Fig. 1D, lane 4), suggesting that Shs1 could be a direct substrate of Dma2. Indeed, we found that recombinant WT Dma2 but not the RING mutant Dma2C451A could ubiquitylate purified TAP-tagged Shs1 in the presence of Ubch5, ubiquitin and ATP (Fig. 1E). We thus conclude that the septin Shs1 is likely to be a direct substrate of Dma1 and Dma2.
Using the above assay, we checked if other septins might be ubiquitylated in a Dma2-dependent manner. We were unable to detect ubiquitylated Cdc10-HA (data not shown), but found that the septin Cdc11-HA was modified by ubiquitin in vivo (Fig. 1F, lane 2; like Shs1, Cdc11 was probably monoubiquitylated at more than one site). The fact that deletion of DMA1 and DMA2 decreased the intensity of these ubiquitylated species (Fig. 1F, lane 4), and that their levels were increased by Dma2 overexpression (Fig. 1G) indicated that Cdc11 also likely serves as an in vivo substrate for Dma1 and Dma2. Accordingly, a non-biased screen for dosage compensation of temperature-sensitive (ts) alleles of lethal yeast deletions18 showed that Cdc11-ts is rescued by septin proteins (Cdc3, Cdc10 and Cdc11) but also by the expression of the ubiquitin gene Ubi4 or Dma1 (Fig. S1E).
Septin organization requires the Dma2 RING and FHA domains
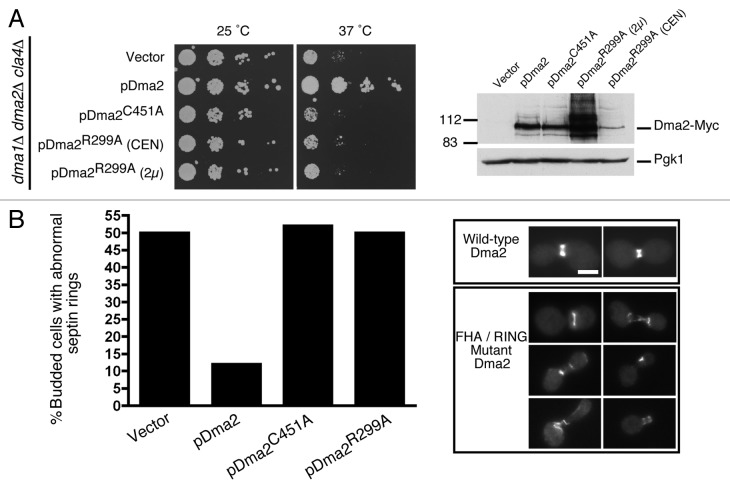
For viability, cells lacking Dma1 and Dma2 require Cla4,4,16 a protein kinase required for correct septin ring assembly.11 Consequently, dma1∆ dma2∆ mutants expressing a ts CLA4 allele, cla4-75, are inviable at 37°C, presumably due to severe septin organization defects caused by the combined loss of Dma and Cla4 activities.4,16 We therefore took a dma1∆ dma2∆ cla4∆ mutant strain expressing the cla4-75 allele from a plasmid and transformed it with empty vector or with constructs encoding WT Dma2 (pDma2-Myc), FHA mutant Dma1 (pDma2R299A-Myc) or RING mutant Dma1 (pDma2C451A-Myc). Interestingly, incubation of these strains at 25°C or 37°C (the restrictive temperature) revealed that cells expressing the FHA- or RING- mutants remained temperature-sensitive, while expression of WT Dma2 rescued the thermo-sensitivity phenotype of the dma1∆ dma2∆ cla4-75 strain (Fig. 2A, left panels). This indicates that these domains are required for Dma2 activity, where viability is likely to reflect septin function. Western blot analysis indicated that the Dma2C451A-Myc mutant expressed at levels similar to WT, but expression of the Dma2R299A-Myc mutant was slightly lower (Fig. 2A, right panel). However, even when overexpressed from a high copy-number 2 μ-based plasmid, the Dma2R299A-Myc mutant failed to rescue the temperature sensitivity of the dma1∆ dma2∆ cla4-75 strain. Collectively, these results suggested that both the Dma2 FHA and RING domains contribute to normal septin function.
Figure 2. Dma2 ubiquitin ligase activity is required for viability of cla4 mutants and for normal septin organization. (A) A dma1∆ dma2∆ cla4∆ mutant strain (cla4-75 allele) was transformed with an empty vector or with a plasmid encoding pDma2WT, pDma2C451A or pDma2R299A. Dma2 proteins were Myc-tagged while Dma2R299A was expressed from a low- (CEN) or high- (2 μ) copy plasmid. Cells were incubated at 25°C or 37°C for 3 d (left). Extracts from (A) were analyzed by sequential blotting with anti-Myc (right) then anti-Pgk1 as loading control. (B) dma1∆ dma2∆ mutants transformed as in (A) were transformed with a plasmid expressing GFP-Cdc3. Cells were grown at 25°C, shifted to 37°C for 3 h, and percentages of budded cells showing abnormal septin ring morphology or position were scored by microscopy (left; n = 200). Examples of cells scored as having abnormal septin rings are shown on the right. Scale bar is 5 μm.
We next tested the importance of the Dma2 FHA and RING domains for septin positioning and organization. Thus, dma1∆ dma2∆ cla4-75 cells expressing septin Cdc3-GFP from a plasmid were transformed with control, pDma2-Myc, pDma2R299A-Myc or pDma2C451A-Myc plasmids. After growth at 25°C followed by a shift to 37°C for 3 h, cells were fixed and visualized by microscopy. As described previously,4 we found a high incidence of mis-positioned septin rings in dma1∆ dma2∆ cla4-75 mutants (Fig. 2B). In addition, we observed a high number of cells with disorganized septin filaments. Importantly, while this phenotype was rescued by reintroducing WT Dma2, cells expressing the Dma2 FHA or RING mutants showed septin positioning and organization defects comparable to dma1∆ dma2∆ cells transformed with empty vector (Fig. 2B). These results therefore indicated that the Dma2 FHA and RING domains are required for efficient septin positioning and organization.
Human RNF8, not Chfr, colocalizes with septins in an FHA-dependent manner
Akin to S. cerevisiae Dma1 and Dma2, human RNF8 and Chfr contain both FHA and RING domains. Notably, however, sequence analyses have suggested that Chfr and RNF8 diverged in a vertebrate genome duplication event independent of the budding yeast duplication.1 Furthermore, our phylogenetic analyses indicated that RNF8 homologs cluster together with Dma proteins, while Chfr homologs appear to be vertebrate-specific (Fig. S2). Perhaps the main molecular distinction is that Chfr family members possess a poly-cysteine rich region at their C terminus,5 while RNF8/Dma proteins are devoid of this but, instead, possess a predicted coiled-coil region between the FHA and RING domains. During interphase, RNF8 and Chfr localize to the nucleus and are involved in the DNA damage response.19-22 Chfr participates in a cell cycle checkpoint response that acts at the end of interphase to delay entry into mitosis when cells suffer microtubule damage.5,6 Notably, during mitosis and until reassembly of the nuclear membrane, RNF8 and Chfr may diffuse in the cytoplasm and could thus function independently from their roles in interphase. Due to its domain organization and links to mitotic events, it has been assumed that Chfr, not RNF8, is the Dma functional counterpart.3,4 Since both S. pombe Dma13 and S. cerevisiae Dma2 (Fig. 1C) localize to the site of cell division, we sought to determine whether RNf8 and/or Chfr adopt a similar localization. To examine and simultaneously compare their subcellular localizations, we microinjected a synchronized population of HeLa cells, 10 h after release, with equal amounts of two plasmids: YFP-Chfr and CFP-RNF8, and analyzed mitotic progression. We found that while the most pronounced accumulation of CFP-RNF8 was at the midbody, a transient structure that marks the site of cell abscission at the end of cytokinesis, YFP-Chfr never localized to such sites but instead accumulated in daughter cells at the time that the cell cleavage furrow had fully ingressed (Fig. 3A; Movie 1). A closer analysis of RNF8 localization also indicated that it associates with centrosomes in mid-mitosis (Fig. 3B; Movie 2). Thus, of the two proteins, the localization pattern of RNF8 is most similar to that of the yeast Dma proteins, suggesting that Chfr has possibly evolved to perform alternative mitotic functions.
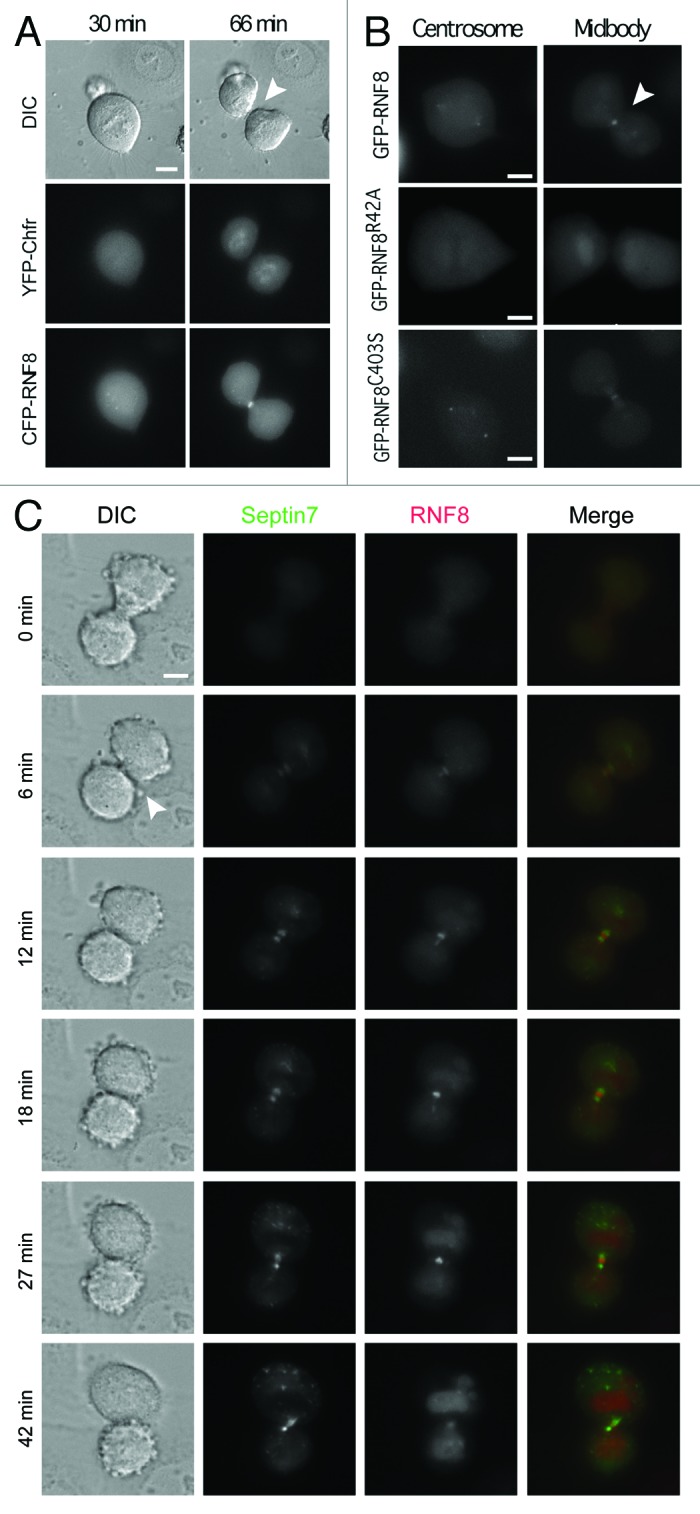
Figure 3. RNF8 localizes to the midbody and centrosomes in an FHA-dependent manner. (A) G1/S synchronized HeLa cells were released for 10 h, injected with equimolar YFP-Chfr and CFP-RNF8 plasmids. Cells entering mitosis were identified by DIC microscopy and then monitored by time-lapse microscopy. (B) HeLa cells were synchronized as in (A), injected with pGFP-RNF8, GFP-RNF8R42A or GFP-RNF8C403S (C). As in (B), but cells were co-injected with YFP-RNF8 and CFP-SEPT7. Scale bar is 5 μm. Arrowheads indicate the midbody. Experiments were repeated multiple times, and a representative movie for each is shown in supplemental data as Movies 1–5.
To determine which domains of RNF8 were important for the observed localization pattern, we injected HeLa cells with plasmids expressing WT or mutant GFP-RNF8. While WT RNF8 localized to both the midbody and centrosomes, these associations were abolished by mutating a FHA domain residue that is predicted to be important for phospho-protein binding activity (GFP-RNF8R42A, Fig. 3B; Movie 3). By contrast, the GFP-RNF8C403S protein, a mutation that abrogates RNF8 ubiquitin ligase activity,23 had a localization pattern similar to the WT protein (Fig. 3B; Movie 4). These data therefore revealed that the FHA domain but not an intact RING is required for the correct subcellular localization pattern of human RNF8, a situation strikingly analogous to that of Dma proteins of both fission yeast3 and budding yeast (Fig. 1A).
RNF8 regulates septin ubiquitylation and physiology in human cells
Having shown that S. cerevisiae Dma2 is an E3 ubiquitin ligase affecting septin Shs1 and Cdc11 ubiquitylation, and since RNF8 possesses ubiquitin ligase activity in vitro24 and in vivo,23 we wondered whether RNF8 might also target septins. Sequence comparisons indicated that SEPT7 is the closest human homolog of yeast Shs1 (data not shown). While it is not clear if these two proteins are indeed functional homologs,11 such sequence similarity prompted us to test whether SEPT7 colocalizes with and is ubiquitylated by RNF8. Using HeLa cells microinjected with YFP-RNF8 and CFP-SEPT7 plasmids followed by time-lapse microscopy, we established that RNF8 and SEPT7 simultaneously localized to the midbody and that the distribution of RNF8 appeared to considerably overlap with that of SEPT7 at this structure (Fig. 3C; Movie 5) in a manner comparable to the localization of the Rts1 protein that has been suggested to regulate Shs1 phosphorylation status at the bud neck.25
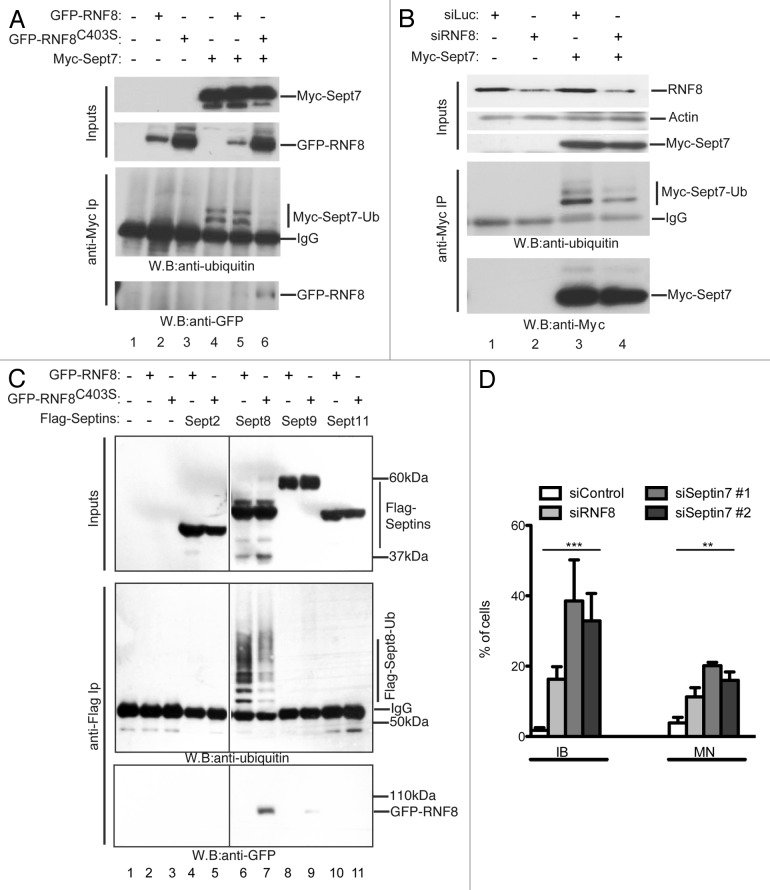
To test whether RNF8 can ubiquitylate SEPT7, a construct encoding Myc-tagged SEPT7 was co-transfected into human 293 cells with either a vector expressing GFP-RNF8 or a catalytically inactive RNF8 RING mutant (GFP-RNF8C403S). We then prepared cell lysates, performed immunoprecipitations with an anti-Myc antibody and analyzed samples by western blotting. Strikingly, upon Myc-SEPT7 expression, two bands were detected by the anti-Ub antibody, thereby indicating that SEPT7 is ubiquitylated in cells (Fig. 4A, lane 4). Since these two bands are sharp and discrete, we inferred that SEPT7 is not polyubiquitylated but is probably monoubiquitylated at one or more sites, or perhaps monubiquitylated or diubiquitylated at a single site. Moreover, while expression of GFP-RNF8 did not alter levels of ubiquitylated SEPT7 (Fig. 4A; lanes 4 and 5), expression of the RNF8 RING mutant considerably decreased the intensity of both ubiquitylated species (Fig. 4A, lanes 4 and 6). The most likely explanation for this is that overexpressed GFP-RNF8C403S acts as a dominant-negative mutant by competing with the WT, endogenously expressed RNF8, for targeting SEPT7. In support of this idea, we detected the GFP-RNF8C403S protein (and also the analogous WT protein at lower levels) in SEPT7 immunoprecipitates (Fig. 4A bottom).
Figure 4. RNF8 controls septin SEPT7 ubiquitylation. (A) 293 cells were co-transfected with GFP-RNF8 or GFP-RNF8C403S and Myc-SEPT7, as indicated. Cells were lysed 48 h later and immunoprecipitated with anti-Myc (anti-Myc IP). Samples were analyzed by blotting with anti-Ub or anti-GFP. (B) 293 cells were siRNA treated and 48h later were transfected with a plasmid encoding Myc-SEPT7 and grown for a further 48 h. Immunoprecipitates were prepared as in (A) then blotted with indicated antibodies (C). As (A) but instead of Myc-SEPT7, cells were co-transfected with empty plasmid or FLAG-tagged SEPT2, 4, 8 or 11. (D) Quantification of cellular anomalies in RNF8 depleted cells or in cells treated with two independent siRNAs targeting SEPT7. All experiments were independently repeated multiple times; unpaired t-test (**, p < 0.01; ***, p < 0.001).
As a complementary approach to investigate the impact of RNF8 on SEPT7 ubiquitylation, we treated 293 cells with short-interfering RNAs (siRNAs) targeting RNF8 or with a control siRNA (targeting firefly luciferase) and then transfected the cells with a plasmid encoding Myc-SEPT7. We then prepared cell lysates, immunoprecipitated Myc-SEPT7 and analyzed samples by western immunoblotting with an anti-Ub antibody. Although the siRNA treatment caused only partial RNF8 depletion, this considerably decreased the levels of ubiquitylated SEPT7 as compared with cells treated with control siRNA (Fig. 4B, lanes 3 and 4). To explore whether the effect on SEPT7 was specific and did not reflect an artifact generally affecting septins, we conducted the same experiment shown in Figure 4A but with Flag-tagged septins: SEPT2, SEPT8, SEPT9 or SEPT11 (tested by Nagata et al.26). This analysis showed that the RNF8-dependent ubiquitylation of septins is directed toward SEPT7 and possibly SEPT8 but not the other septins we tested (Fig. 4C). This effect therefore bears strong similarity to the ubiquitylation of Shs1 and Cdc11 by yeast Dma proteins.
Finally, we postulated that because RNF8 ubiquitylates SEPT7 in vivo, the loss of either should manifest comparable cellular anomalies. To test this idea, we depleted RNF8 or SEPT7 from human U2OS cells and analyzed these cells by immunofluorescence microscopy (Fig. S3). Thus, we observed that RNF8 or SEPT7 depletion produced similar phenotypes in terms of increased inter-cellular bridges (IB) and multi-nucleated (MN) cells, both of which can result from aberrant cytokinesis and cell abscission (Fig. 4D). These observations are supported by similar findings for depletion of other septin proteins,7,8,14,27 thereby reinforcing the idea that adequate cell abscission depends on septin ubiquitylation. Our results therefore established that RNF8 affects SEPT7 ubiquitylation and infer that interference with this process had deleterious effects during cellular abscission by affecting the equal distribution of the genome into the two daughter cells.
Discussion
Dma2 interacts with ubiquitin E2 enzymes and has robust E3 ubiquitin ligase activity in vitro, and prior work has indicated that dma1∆ dma2∆ mutants have septin organization defects.4 Our data suggest that this reflects a direct effect of Dma proteins on septins, as we have found that septins Shs1 and Cdc11 are direct targets of Dma1 and Dma2 ubiquitin ligase activity. This conclusion is based on the observations that deletion of DMA1 and DMA2 decreases the abundance of the ubiquitylated forms of Shs1 and Cdc11 present in the cell and that Dma2 overexpression has the opposite effect. Furthermore, we have established that Shs1 can be ubiquitylated by Dma2 in vitro. Notably, these effects appear to be specific to certain members of the septin family, because we were unable to detect ubiquitylation of Cdc10 in vivo. Although Chfr has been considered as the ortholog of S. pombe Dma1 and S. cerevisiae Dma1 and Dma2,3,4 this idea was not supported by our analyses of Chfr subcellular localization. Instead, our results indicate that RNF8 is likely to be the genuine mammalian ortholog of the yeast Dma proteins. Thus, we have found that RNF8 localizes to centrosomes and to the cell division site, and it promotes the ubiquitylation of specific members of the septin family (SEPT7 and possibly SEPT8 but not SEPT2, 4 and 11), two key characteristics shared with the yeast Dma proteins. Furthermore, as has been shown for S. pombe Dma1,3 we have established that the FHA domain of RNF8 controls its subcellular localization. A phylogenetic analysis of available protein sequences having an FHA followed by a RING finger domain indicates that these molecules belong to two distinct families: one containing Chfr, the other containing RNF8 and the yeast Dma proteins (Fig. S2). A distinctive feature of Chfr family members is that they possess a poly-cysteine rich region at their C terminus,5 while the Dma/RNF8 proteins are devoid of this but, instead, possess a predicted coiled-coil region between the FHA and the RING finger domains. The phylogenetic analysis also suggests that the Chfr family of proteins arose during vertebrate evolution, possibly adopting mitotic functions distinct from those of RNF8 (Fig. S2).
Cells lacking Dma1 and Dma2 require the protein kinase Cla4 for viability, presumably because of the severe septin organization defect caused by the combined loss of Dma and Cla4 activities.4 We have shown that this lethality cannot be complemented by the catalytically inactive Dma2C451A RING-finger mutant, a result strongly suggesting that septin ubiquitylation by Dma1 and Dma2 is important for the viability of Cla4-defective cells. Furthermore, we have established that the ubiquitin ligase activity of Dma1 and Dma2 is important for the correct positioning and organization of the septin ring in dma1∆ dma2∆ cla4-75 cells grown at 37°C, a result reinforcing the importance of septin ubiquitylation. Although it is not yet clear how ubiquitylation of septins might affect their functions, it is unlikely to promote their proteasomal degradation, as we found no evidence for polyubiquitylation of Shs1 or Cdc11. Furthermore, levels of Cdc11 are constant during the cell cycle,17 and we have not observed accumulation of septins in cells where Dma1/2 activity was altered (Fig. 2). Because Dma2 colocalizes with septins late during mitosis, this suggests that septin ubiquitylation could happen at the site of cell division during cytokinesis. Although this time marks the beginning of septin ring disassembly, we did not find dma1∆ dma2∆ mutants to be defective in this process, and, as such, we cannot rule out the possibility that septin ubiquitylation occurs during interphase.
In budding yeast, the Cla4 kinase is required for septin ring formation at the time of bud emergence (G1/S). The genetic interaction between DMA1/2 and CLA4 reported here and by Fraschini et al.4 is consistent with septins becoming ubiquitylated at, or before, bud emergence, with this modification facilitating septin ring assembly. Although deletion of DMA1 and DMA2 markedly reduced levels of ubiquitylated Shs1 and Cdc11, we still detected residual ubiquitylation of these proteins. This suggested that an additional ubiquitin ligase(s) might compensate for the lack of Dma1 and Dma2, and raised the possibility that a complete lack of septin ubiquitylation would cause a more severe phenotype than that observed in dma1∆ dma2∆ mutants. Identification and mutation of the ubiquitylated residues on septins will be required to address this possibility.
Our observations that RNF8 localizes at the midbody in a manner that overlaps with and is concurrent with SEPT7 localization—and akin to the localization of the Rts1 protein with respect to yeast Shs125—suggests that it might play a key role in regulating cytokinesis. Consistent with this, we have observed a modest but significant increase in the occurrence of multi-nucleated cells and cells with inter-cellular bridges when RNF8 or SEPT7 are depleted (Fig. 4), findings that are in agreement with other studies.7,8,14,27 Since septins are required for cytokinesis,11,28,29 it is possible that the cytokinesis defects we observed are caused by loss of septin ubiquitylation. This could be addressed by analyzing cells in which the ubiquitylation sites of SEPT7, and potentially of other mammalian septins, are mutated. However, drawing upon the established role of RNF8 in histone modification, it is possible that there are parallels between the ubiquitylation activities of RNF8 in the cytoplasm and in the nucleus. First, the recruitment of RNF8 seems to depend on its FHA domain, while the RING domain appears to be dispensable for this recruitment but essential for subsequent RNF8 E3 ubiquitin ligase activity. Moreover, we note that septins and nucleosomes form vaguely similar higher-order octameric structures.30 Together, this suggests that RNF8 could promote independent but analogous ubiquitylation activities in the cytoplasm and in the nucleus at different phases of the cell cycle to maintain both cellular and genomic integrity. Finally, it is tempting to speculate that septin ubiquitylation by Dma/RNF8 is not only needed to bring about effective cell division and genome stability but also provides opportunities for the cell to sense mitotic progression and perhaps modulate it in response to environmental cues. Further analyses of Dma/RNF8 should shed light on such intriguing possibilities.
Materials and Methods
Yeast strains and methods
Genotypes of strains are listed in Table S1. All strains were S288 derivatives except ySP3018 (W303 background). Standard methods were used throughout and gene deletions and epitope tagging were PCR-mediated.
Plasmids
The DMA2 ORF plus 270 bp of upstream and 250 bp of downstream sequences was amplified by PCR with EcoRI and XhoI sites appended to the upstream and downstream primers respectively. This PCR product was cloned in pRS413 (HIS3 CEN6 ARS4) cut at the same sites to generate pDMA2. A 13× Myc epitope tag was added to Dma2 by the following procedure: the stop codon of DMA2 in pDMA2 was mutated to an AgeI site using the QuikChange site directed mutagenesis kit (Stratagene) and a PCR product encoding 13 Myc epitopes, generated with upstream and downstream primers having, respectively, AgeI and XhoI restriction sites appended, was cloned in pDMA2 cut at the same sites to generate pDMA2-Myc. The pDMA2C451A-Myc plasmid was generated by changing the nucleotides encoding Cys-451 of the RING to an Ala codon using the QuikChange site directed mutagenesis kit (Stratagene). To generate plasmids pGST-DMA2 and pGST-DMA2C451A, full-length DMA2 ORF (without the Myc tag) was amplified from pDMA2-Myc and pDMA2C451A-Myc plasmids, respectively, and the generated PCR products (engineered to have EcoRI sites at both ends) were cloned in pGEX-4T-3 (Amersham Biosciences) at the same site. Plasmid pDma2C451A-YFP was created by replacing the 13xMyc module of pDma2C451A-Myc with a PCR-amplified YFP sequence. The DMA2-YFP sequence was then subcloned in pRS426 (2 µ, URA3). Yeast two-hybrid plasmids were created by cloning the complete coding sequences of Ubc5 or Ubc7 in pGBKT7 (Clontech) or the complete coding sequence of Dma2 in pGADT7 (Clontech). Two-hybrid plasmids encoding fusions of Ubc13, Mms2 or Rad6 with the GAL DNA binding domain were a kind gift from Dr Stefan Jentsch and are described elsewhere.31 Plasmids permitting copper-inducible expression of ubiquitin or His-Ub (respectively, pUb175 and pUb221) were kind gifts from Dr Daniel Finley. The pCdc3-GFP plasmid was kindly provided by Dr Simonetta Piatti. Full-length RNF8 was PCR-amplified from a human thymus cDNA library and cloned in vectors peGFP and peCFP (Clontech) to create GFP-RNF8 and CFP-RNF8 respectively. RNF8 point mutants were generated with the QuikChange site directed mutagenesis Kit (Stratagene). The Myc-SEPT7 construct was kindly provided by Dr Koh-ichi Nagata and is described elsewhere.26
Cell culture and synchronization
For HeLa cell G1/S synchronization, the day after seeding, cells were blocked with thymidine (2.5 mM, Sigma) for 24 h, released for 12 h and then blocked with aphidicolin (5 mg/ml, Sigma) for 24 h before release into fresh medium.
Transfections and siRNA treatments
Transfections of plasmids and siRNAs were with Metafectene (Cambio) and Oligofectamine (Invitrogen), respectively. siRNAs were from Dharmacon and their sequences are described in supplemental data.
Detection of septin-ubiquitin conjugates
For experiments involving Ub-His expression, cells were grown in selective medium containing 2% glucose to early log phase. Ubiquitin expression was induced by addition of 250 μM CuSO4 for 3 h. For experiments involving Dma2 overexpression, cells were grown in selective medium containing 2% raffinose to early log-phase. Copper sulfate was added to cultures for 1 h, then 2% galactose was added and cells grown for an additional 3 h. Cells were washed in water, resuspended in lysis buffer (6 M guanidium-HCl, 50 mM sodium phosphate buffer pH 8, 10 mM TRIS-HCl pH 8, 300 mM NaCl, 5 mM N-ethymaleimide, 10 mM imidazole) equal to the cell pellet volume, and extracts were prepared with a One Shot cell disruptor (Constant Systems) at a pressure of 30 kPSI. Purification of ubiquitylated proteins was performed as previously described.32 Bound proteins were eluted by addition of 25 μl of 2× SDS loading buffer and analyzed by western blotting with an anti-HA antibody. For Figure 2A, cells were lysed in a buffer containing 8 M urea, 50 mM sodium phosphate pH 8, 10 mM TRIS-HCl pH 8, 300 mM NaCl, 5 mM N-ethymaleimide and 10 mM imidazole; the urea concentration was then diluted 10-fold and Shs1-HA was immunoprecipitated from 4.5 mg of protein extracts with 1 μg of anti-HA antibody. Human 293 cells transfected with Myc-SEPT7, Flag-SEPT2, Flag-SEPT8, Flag-SEPT9 or Flag-SEPT1126 were collected 48 h after transfection and washed with cold PBS. Cells were then lysed in a buffer containing 8 M urea, 100 mM NaH2PO4, 10 mM Tris pH 8 and 0.2% Triton-X supplemented with 10 mM β-mercaptoethanol and 20 mM N-ethylmaleimide. One mg of protein extract was diluted ten times in buffer-T [10 mM Tris pH 8, 1% (v/v) Tween-20, 10 mM β-mercaptoethanol, 20 mM N-ethylmaleimide], and Myc-SEPT7 was immunoprecipitated with anti-Myc antibody. Other septins were immunoprecipitated with anti-Flag (M2) antibody.
Microinjection and time-lapse imaging and analysis
For microinjection and during microscopy observation, cells were grown in a 35-mm diameter glass-bottomed dish (WillCo Wells). Medium was replaced by Leibovitz’s L-15 medium (Gibco BRL) with 10% serum and antibiotics. Plasmids were microinjected at 3 ng/ml in G2 cells with a semiautomatic microinjector (Eppendorf) on a Leica DMIRBE microscope. Images were captured every 3 min with a DeltaVision Spectris deconvolution microscope (Applied Biosystems) with a 60× lens. Further details are in supplemental data. A modified version of ImageJ software (NIH) was used for movies.
Yeast microscopy
Cells were fixed for 15 min in 3.7% formaldehyde, washed twice in PBS, sonicated briefly and visualized on an integrated DeltaVision system (Applied Precision) with a 100× (numerical aperture, 1.4) objective. Captured images were exported to Adobe Photoshop.
Supplementary Material
Acknowledgments
We thank S. Piatti, D. Finley, S. Jentsch and K. Nagata for strains and plasmids. We also thank C. Chahwan for insightful suggestions, M. Scharff for support, J. Pines, K.Dry and A. Moldon for critical reading of the manuscript and K. Dry for editorial help. S.G. was supported by an HFSP fellowship. Research in the S.P.J. Lab is supported by grants from CRUK (C6/A11226), the European Research Council, the European Community’s Seventh Framework Program grant agreement no. HEALTH-F2-2010-259893 (DDResponse) and by core infrastructure funding from CRUK and the Wellcome Trust.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/23947
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/23947
References
- 1.Brooks L, 3rd, Heimsath EG, Jr., Loring GL, Brenner C. FHA-RING ubiquitin ligases in cell division cycle control. Cell Mol Life Sci. 2008;65:3458–66. doi: 10.1007/s00018-008-8220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson AE, Gould KL. Dma1 ubiquitinates the SIN scaffold, Sid4, to impede the mitotic localization of Plo1 kinase. EMBO J. 2011;30:341–54. doi: 10.1038/emboj.2010.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guertin DA, Venkatram S, Gould KL, McCollum D. Dma1 prevents mitotic exit and cytokinesis by inhibiting the septation initiation network (SIN) Dev Cell. 2002;3:779–90. doi: 10.1016/S1534-5807(02)00367-2. [DOI] [PubMed] [Google Scholar]
- 4.Fraschini R, Bilotta D, Lucchini G, Piatti S. Functional characterization of Dma1 and Dma2, the budding yeast homologues of Schizosaccharomyces pombe Dma1 and human Chfr. Mol Biol Cell. 2004;15:3796–810. doi: 10.1091/mbc.E04-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scolnick DM, Halazonetis TD. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature. 2000;406:430–5. doi: 10.1038/35019108. [DOI] [PubMed] [Google Scholar]
- 6.Matsusaka T, Pines J. Chfr acts with the p38 stress kinases to block entry to mitosis in mammalian cells. J Cell Biol. 2004;166:507–16. doi: 10.1083/jcb.200401139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plans V, Guerra-Rebollo M, Thomson TM. Regulation of mitotic exit by the RNF8 ubiquitin ligase. Oncogene. 2008;27:1355–65. doi: 10.1038/sj.onc.1210782. [DOI] [PubMed] [Google Scholar]
- 8.Tuttle RL, Bothos J, Summers MK, Luca FC, Halazonetis TD. Defective in mitotic arrest 1/ring finger 8 is a checkpoint protein that antagonizes the human mitotic exit network. Mol Cancer Res. 2007;5:1304–11. doi: 10.1158/1541-7786.MCR-07-0388. [DOI] [PubMed] [Google Scholar]
- 9.Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol. 2012;13:183–94. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- 10.Lippincott J, Shannon KB, Shou W, Deshaies RJ, Li R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J Cell Sci. 2001;114:1379–86. doi: 10.1242/jcs.114.7.1379. [DOI] [PubMed] [Google Scholar]
- 11.Versele M, Thorner J. Some assembly required: yeast septins provide the instruction manual. Trends Cell Biol. 2005;15:414–24. doi: 10.1016/j.tcb.2005.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bi E, Maddox P, Lew DJ, Salmon ED, McMillan JN, Yeh E, et al. Involvement of an actomyosin contractile ring in Saccharomyces cerevisiae cytokinesis. J Cell Biol. 1998;142:1301–12. doi: 10.1083/jcb.142.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998;143:1947–60. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim MS, Froese CD, Estey MP, Trimble WS. SEPT9 occupies the terminal positions in septin octamers and mediates polymerization-dependent functions in abscission. J Cell Biol. 2011;195:815–26. doi: 10.1083/jcb.201106131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loring GL, Christensen KC, Gerber SA, Brenner C. Yeast Chfr homologs retard cell cycle at G1 and G2/M via Ubc4 and Ubc13/Mms2-dependent ubiquitination. Cell Cycle. 2008;7:96–105. doi: 10.4161/cc.7.1.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merlini L, Fraschini R, Boettcher B, Barral Y, Lucchini G, Piatti S. Budding yeast dma proteins control septin dynamics and the spindle position checkpoint by promoting the recruitment of the Elm1 kinase to the bud neck. PLoS Genet. 2012;8:e1002670. doi: 10.1371/journal.pgen.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–94. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magtanong L, Ho CH, Barker SL, Jiao W, Baryshnikova A, Bahr S, et al. Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nat Biotechnol. 2011;29:505–11. doi: 10.1038/nbt.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Chen Y, Lu LY, Wu Y, Paulsen MT, Ljungman M, et al. Chfr and RNF8 synergistically regulate ATM activation. Nat Struct Mol Biol. 2011;18:761–8. doi: 10.1038/nsmb.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 21.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–40. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–14. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bekker-Jensen S, Mailand N. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 2011;585:2914–9. doi: 10.1016/j.febslet.2011.05.056. [DOI] [PubMed] [Google Scholar]
- 24.Ito K, Adachi S, Iwakami R, Yasuda H, Muto Y, Seki N, et al. N-Terminally extended human ubiquitin-conjugating enzymes (E2s) mediate the ubiquitination of RING-finger proteins, ARA54 and RNF8. Eur J Biochem. 2001;268:2725–32. doi: 10.1046/j.1432-1327.2001.02169.x. [DOI] [PubMed] [Google Scholar]
- 25.Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4:345–57. doi: 10.1016/S1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- 26.Nagata K, Asano T, Nozawa Y, Inagaki M. Biochemical and cell biological analyses of a mammalian septin complex, Sept7/9b/11. J Biol Chem. 2004;279:55895–904. doi: 10.1074/jbc.M406153200. [DOI] [PubMed] [Google Scholar]
- 27.Estey MP, Di Ciano-Oliveira C, Froese CD, Bejide MT, Trimble WS. Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J Cell Biol. 2010;191:741–9. doi: 10.1083/jcb.201006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita M, Kumar S, Mizoguchi A, Ide C, Kinoshita A, Haraguchi T, et al. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 1997;11:1535–47. doi: 10.1101/gad.11.12.1535. [DOI] [PubMed] [Google Scholar]
- 29.Surka MC, Tsang CW, Trimble WS. The mammalian septin MSF localizes with microtubules and is required for completion of cytokinesis. Mol Biol Cell. 2002;13:3532–45. doi: 10.1091/mbc.E02-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMurray MA, Thorner J. Septins: molecular partitioning and the generation of cellular asymmetry. Cell Div. 2009;4:18. doi: 10.1186/1747-1028-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–97. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaiser P, Flick K, Wittenberg C, Reed SI. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell. 2000;102:303–14. doi: 10.1016/S0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 33.Leverson JD, Joazeiro CA, Page AM, Huang Hk, Hieter P, Hunter T. The APC11 RING-H2 finger mediates E2-dependent ubiquitination. Mol Biol Cell. 2000;11:2315–25. doi: 10.1091/mbc.11.7.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.