Abstract
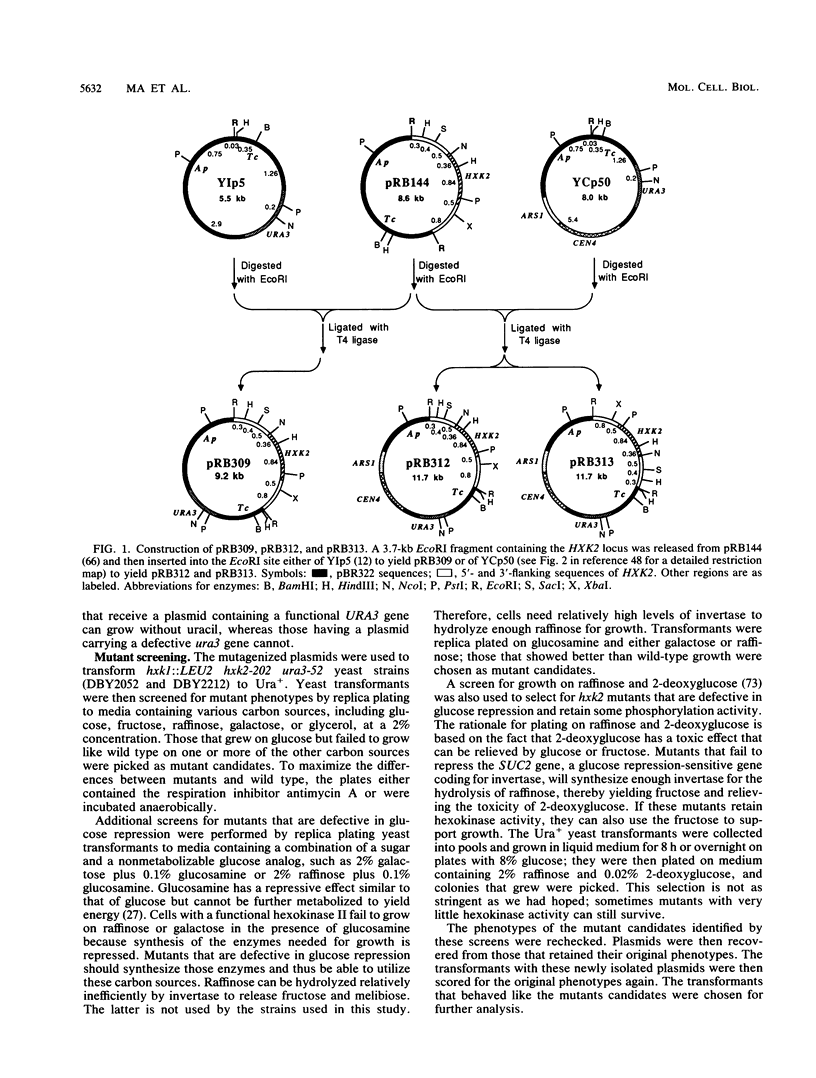
Several hundred new mutations in the gene (HXK2) encoding hexokinase II of Saccharomyces cerevisiae were isolated, and a subset of them was mapped, resulting in a fine-structure genetic map. Among the mutations that were sequenced, 35 were independent missense mutations. The mutations were obtained by mutagenesis of cloned HXK2 DNA carried on a low-copy-number plasmid vector and screened for a number of different phenotypes in yeast strains bearing chromosomal hxk1 and hxk2 null mutations. Some of these mutants were characterized both in vivo and in vitro; they displayed a wide spectrum of residual hexokinase activities, as indicated by three assays: in vitro enzyme activity, ability to grow on glucose and fructose, and ability to repress invertase production when growing on glucose. Of those that failed to support growth on fructose, only a small minority made normal-size, stable, and inactive protein. Analysis of the amino acid changes in these mutants in light of the crystallographically determined three-dimensional structure of hexokinase II suggests important roles in structure or catalysis for six amino acid residues, only two of which are near the active site.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T., Sun D. P., Nye J. A., Muchmore D. C., Matthews B. W. Temperature-sensitive mutations of bacteriophage T4 lysozyme occur at sites with low mobility and low solvent accessibility in the folded protein. Biochemistry. 1987 Jun 30;26(13):3754–3758. doi: 10.1021/bi00387a002. [DOI] [PubMed] [Google Scholar]
- Alber T., Sun D. P., Wilson K., Wozniak J. A., Cook S. P., Matthews B. W. Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature. 1987 Nov 5;330(6143):41–46. doi: 10.1038/330041a0. [DOI] [PubMed] [Google Scholar]
- Anderson C. M., McDonald R. C., Steitz T. A. Sequencing a protein by x-ray crystallography. I. Interpretation of yeast hexokinase B at 2.5 A resolution by model building. J Mol Biol. 1978 Jul 25;123(1):1–13. doi: 10.1016/0022-2836(78)90373-x. [DOI] [PubMed] [Google Scholar]
- Anderson C. M., Stenkamp R. E., McDonald R. C., Steitz T. A. A refined model of the sugar binding site of yeast hexokinase B. J Mol Biol. 1978 Aug 5;123(2):207–219. doi: 10.1016/0022-2836(78)90321-2. [DOI] [PubMed] [Google Scholar]
- Anderson C. M., Stenkamp R. E., Steitz T. A. Sequencing a protein by x-ray crystallography. II. Refinement of yeast hexokinase B co-ordinates and sequence at 2.1 A resolution. J Mol Biol. 1978 Jul 25;123(1):15–33. doi: 10.1016/0022-2836(78)90374-1. [DOI] [PubMed] [Google Scholar]
- Bennett W. S., Jr, Steitz T. A. Glucose-induced conformational change in yeast hexokinase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4848–4852. doi: 10.1073/pnas.75.10.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett W. S., Jr, Steitz T. A. Structure of a complex between yeast hexokinase A and glucose. I. Structure determination and refinement at 3.5 A resolution. J Mol Biol. 1980 Jun 25;140(2):183–209. doi: 10.1016/0022-2836(80)90102-3. [DOI] [PubMed] [Google Scholar]
- Bennett W. S., Jr, Steitz T. A. Structure of a complex between yeast hexokinase A and glucose. II. Detailed comparisons of conformation and active site configuration with the native hexokinase B monomer and dimer. J Mol Biol. 1980 Jun 25;140(2):211–230. doi: 10.1016/0022-2836(80)90103-5. [DOI] [PubMed] [Google Scholar]
- Borders C. L., Jr, Cipollo K. L., Jorkasky J. F., Neet K. E. Role of arginyl residues in yeast hexokinase PII. Biochemistry. 1978 Jun 27;17(13):2654–2658. doi: 10.1021/bi00606a031. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Broach J. R. Construction of high copy yeast vectors using 2-microns circle sequences. Methods Enzymol. 1983;101:307–325. doi: 10.1016/0076-6879(83)01024-1. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Cox E. C., Horner D. L. Dominant mutators in Escherichia coli. Genetics. 1982 Jan;100(1):7–18. doi: 10.1093/genetics/100.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnen G. E., Cox E. C. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J Bacteriol. 1974 Feb;117(2):477–487. doi: 10.1128/jb.117.2.477-487.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derechin M., Rustum Y. M., Barnard E. A. Dissociation of yeast hexokinase under the influence of substrates. Biochemistry. 1972 May 9;11(10):1793–1797. doi: 10.1021/bi00760a009. [DOI] [PubMed] [Google Scholar]
- Easterby J. S., Rosemeyer M. A. Purification and subunit interactions of yeast hexokinase. Eur J Biochem. 1972 Jul 13;28(2):241–252. doi: 10.1111/j.1432-1033.1972.tb01907.x. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Fröhlich K. U. Saccharomyces cerevisiae mutants provide evidence of hexokinase PII as a bifunctional enzyme with catalytic and regulatory domains for triggering carbon catabolite repression. J Bacteriol. 1984 Apr;158(1):29–35. doi: 10.1128/jb.158.1.29-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D. Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in carbon catabolite repression in yeast. Mol Gen Genet. 1980;178(3):633–637. doi: 10.1007/BF00337871. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Kopetzki E., Fröhlich K. U., Mecke D. Cloning of hexokinase isoenzyme PI from Saccharomyces cerevisiae: PI transformants confirm the unique role of hexokinase isoenzyme PII for glucose repression in yeasts. Mol Gen Genet. 1984;198(2):50–54. doi: 10.1007/BF00328699. [DOI] [PubMed] [Google Scholar]
- Fowler R. G., Degnen G. E., Cox E. C. Mutational specificity of a conditional Escherichia coli mutator, mutD5. Mol Gen Genet. 1974;133(3):179–191. doi: 10.1007/BF00267667. [DOI] [PubMed] [Google Scholar]
- Fraenkel D. G. Mutants in glucose metabolism. Annu Rev Biochem. 1986;55:317–337. doi: 10.1146/annurev.bi.55.070186.001533. [DOI] [PubMed] [Google Scholar]
- Fröhlich K. U., Entian K. D., Mecke D. Cloning and restriction analysis of the hexokinase PII gene of the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1984;194(1-2):144–148. doi: 10.1007/BF00383509. [DOI] [PubMed] [Google Scholar]
- Fröhlich K. U., Entian K. D., Mecke D. The primary structure of the yeast hexokinase PII gene (HXK2) which is responsible for glucose repression. Gene. 1985;36(1-2):105–111. doi: 10.1016/0378-1119(85)90074-5. [DOI] [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Grütter M. G., Gray T. M., Weaver L. H., Wilson T. A., Matthews B. W. Structural studies of mutants of the lysozyme of bacteriophage T4. The temperature-sensitive mutant protein Thr157----Ile. J Mol Biol. 1987 Sep 20;197(2):315–329. doi: 10.1016/0022-2836(87)90126-4. [DOI] [PubMed] [Google Scholar]
- Grütter M. G., Hawkes R. B., Matthews B. W. Molecular basis of thermostability in the lysozyme from bacteriophage T4. Nature. 1979 Feb 22;277(5698):667–669. doi: 10.1038/277667a0. [DOI] [PubMed] [Google Scholar]
- Haltiner M., Kempe T., Tjian R. A novel strategy for constructing clustered point mutations. Nucleic Acids Res. 1985 Feb 11;13(3):1015–1025. doi: 10.1093/nar/13.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M. H., Nelson H. C., Sauer R. T. Mutations in lambda repressor's amino-terminal domain: implications for protein stability and DNA binding. Proc Natl Acad Sci U S A. 1983 May;80(9):2676–2680. doi: 10.1073/pnas.80.9.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M. H., Sturtevant J. M., Sauer R. T. Effect of single amino acid replacements on the thermal stability of the NH2-terminal domain of phage lambda repressor. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5685–5689. doi: 10.1073/pnas.81.18.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopetzki E., Entian K. D., Mecke D. Complete nucleotide sequence of the hexokinase PI gene (HXK1) of Saccharomyces cerevisiae. Gene. 1985;39(1):95–101. doi: 10.1016/0378-1119(85)90113-1. [DOI] [PubMed] [Google Scholar]
- Korneluk R. G., Quan F., Gravel R. A. Rapid and reliable dideoxy sequencing of double-stranded DNA. Gene. 1985;40(2-3):317–323. doi: 10.1016/0378-1119(85)90055-1. [DOI] [PubMed] [Google Scholar]
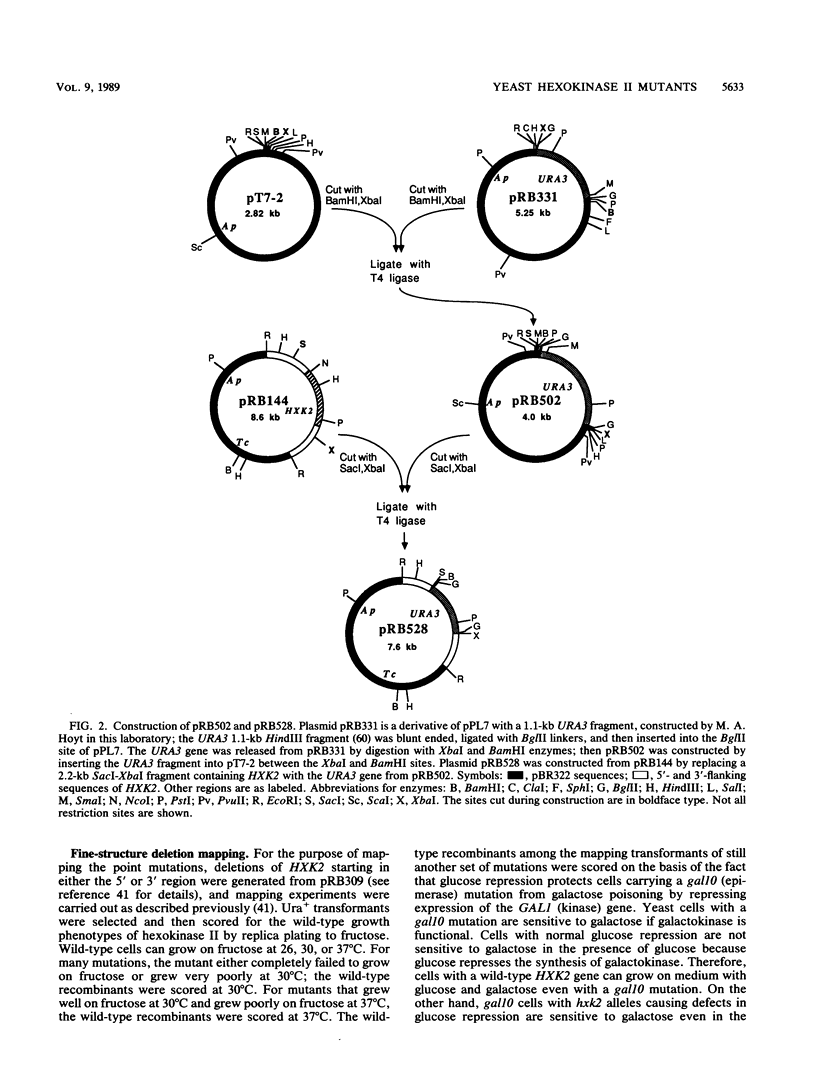
- Kunes S., Ma H., Overbye K., Fox M. S., Botstein D. Fine structure recombinational analysis of cloned genes using yeast transformation. Genetics. 1987 Jan;115(1):73–81. doi: 10.1093/genetics/115.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. L., Campbell J. L. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8-1 mutation. Mol Cell Biol. 1983 Oct;3(10):1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lobo Z., Maitra P. K. Genetics of yeast hexokinase. Genetics. 1977 Aug;86(4):727–744. doi: 10.1093/genetics/86.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo Z., Maitra P. K. Physiological role of glucose-phosphorylating enzymes in Saccharomyces cerevisiae. Arch Biochem Biophys. 1977 Aug;182(2):639–645. doi: 10.1016/0003-9861(77)90544-6. [DOI] [PubMed] [Google Scholar]
- Ma H., Bloom L. M., Walsh C. T., Botstein D. The residual enzymatic phosphorylation activity of hexokinase II mutants is correlated with glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5643–5649. doi: 10.1128/mcb.9.12.5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Botstein D. Effects of null mutations in the hexokinase genes of Saccharomyces cerevisiae on catabolite repression. Mol Cell Biol. 1986 Nov;6(11):4046–4052. doi: 10.1128/mcb.6.11.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58(2-3):201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Maitra P. K. Glucokinase from yeast. Methods Enzymol. 1975;42:25–30. doi: 10.1016/0076-6879(75)42087-0. [DOI] [PubMed] [Google Scholar]
- Maitra P. K., Lobo Z. Genetics of yeast glucokinase. Genetics. 1983 Nov;105(3):501–515. doi: 10.1093/genetics/105.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald R. C., Steitz T. A., Engelman D. M. Yeast hexokinase in solution exhibits a large conformational change upon binding glucose or glucose 6-phosphate. Biochemistry. 1979 Jan 23;18(2):338–342. doi: 10.1021/bi00569a017. [DOI] [PubMed] [Google Scholar]
- Muratsubaki H., Katsume T. Distribution of hexokinase isoenzymes depending on a carbon source in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1979 Feb 28;86(4):1030–1036. doi: 10.1016/0006-291x(79)90220-1. [DOI] [PubMed] [Google Scholar]
- Neff N. F., Thomas J. H., Grisafi P., Botstein D. Isolation of the beta-tubulin gene from yeast and demonstration of its essential function in vivo. Cell. 1983 May;33(1):211–219. doi: 10.1016/0092-8674(83)90350-1. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Sauer R. T. Interaction of mutant lambda repressors with operator and non-operator DNA. J Mol Biol. 1986 Nov 5;192(1):27–38. doi: 10.1016/0022-2836(86)90461-4. [DOI] [PubMed] [Google Scholar]
- Otieno S., Bhargava A. K., Serelis D., Barnard E. A. Evidence for a single essential thiol in the yeast hexokinase molecule. Biochemistry. 1977 Sep 20;16(19):4249–4255. doi: 10.1021/bi00638a019. [DOI] [PubMed] [Google Scholar]
- Philips M., Pho D. B., Pradel L. A. An essential arginyl residue in yeast hexokinase. Biochim Biophys Acta. 1979 Feb 9;566(2):296–304. doi: 10.1016/0005-2744(79)90033-0. [DOI] [PubMed] [Google Scholar]
- Pho D. B., Roustan C., Tot A. N., Pradel L. A. Evidence for an essential glutamyl residue in yeast hexokinase. Biochemistry. 1977 Oct 4;16(20):4533–4537. doi: 10.1021/bi00639a031. [DOI] [PubMed] [Google Scholar]
- Purich D. L., Fromm H. J., Rudolph F. B. The hexokinases: kinetic, physical, and regulatory properties. Adv Enzymol Relat Areas Mol Biol. 1973;39:249–326. doi: 10.1002/9780470122846.ch4. [DOI] [PubMed] [Google Scholar]
- Rose M., Botstein D. Structure and function of the yeast URA3 gene. Differentially regulated expression of hybrid beta-galactosidase from overlapping coding sequences in yeast. J Mol Biol. 1983 Nov 15;170(4):883–904. doi: 10.1016/s0022-2836(83)80193-4. [DOI] [PubMed] [Google Scholar]
- Rose M., Grisafi P., Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- Rose M., Winston F. Identification of a Ty insertion within the coding sequence of the S. cerevisiae URA3 gene. Mol Gen Genet. 1984;193(3):557–560. doi: 10.1007/BF00382100. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Grisafi P., Benkovic S. J., Botstein D. Gap misrepair mutagenesis: efficient site-directed induction of transition, transversion, and frameshift mutations in vitro. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1588–1592. doi: 10.1073/pnas.79.5.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachelek C., Stachelek J., Swan J., Botstein D., Konigsberg W. Identification, cloning and sequence determination of the genes specifying hexokinase A and B from yeast. Nucleic Acids Res. 1986 Jan 24;14(2):945–963. doi: 10.1093/nar/14.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Anderson W. F., Fletterick R. J., Anderson C. M. High resolution crystal structures of yeast hexokinase complexes with substrates, activators, and inhibitors. Evidence for an allosteric control site. J Biol Chem. 1977 Jul 10;252(13):4494–4500. [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TRAYSER K. A., COLOWICK S. P. Properties of crystalline hexokinase from yeast. I. Analyses for possible co-factors. Arch Biochem Biophys. 1961 Jul;94:156–160. doi: 10.1016/0003-9861(61)90023-6. [DOI] [PubMed] [Google Scholar]
- TRAYSER K. A., COLOWICK S. P. Properties of crystalline hexokinase from yeast. II. Studies on ATP-enzyme interaction. Arch Biochem Biophys. 1961 Jul;94:161–168. doi: 10.1016/0003-9861(61)90024-8. [DOI] [PubMed] [Google Scholar]
- TRAYSER K. A., COLOWICK S. P. Properties of crystalline hexokinase from yeast. III. Studies on glucose-enzyme interaction. Arch Biochem Biophys. 1961 Jul;94:169–176. doi: 10.1016/0003-9861(61)90025-x. [DOI] [PubMed] [Google Scholar]
- Viola R. E., Cleland W. W. Use of pH studies to elucidate the chemical mechanism of yeast hexokinase. Biochemistry. 1978 Oct 3;17(20):4111–4117. doi: 10.1021/bi00613a001. [DOI] [PubMed] [Google Scholar]
- Zimmermann F. K., Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol Gen Genet. 1977 Jul 7;154(1):75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]