Abstract
Although the sterile 20 (Ste20) serine/threonine protein kinase was originally identified as a component of the S. cerevisiae mating pathway, it has homologs in higher eukaryotes and is part of a larger family of Ste20-like kinases. Ste20-like kinases are involved in multiple cellular processes, such as cell growth, morphogenesis, apoptosis and immune response. Carrying out such a diverse array of biological functions requires numerous regulatory inputs and outputs in the form of protein-protein interactions and post-translational modifications. Hence, a thorough knowledge of Ste20-like kinase binding partners and phosphorylation sites will be essential for understanding the various roles of these kinases. Our recent study revealed that Schizosaccharomyces pombe Nak1 (a conserved member of the GC-kinase sub-family of Ste20-like kinases) is in a complex with the leucine-rich repeat-containing protein Sog2. Here, we show a novel and unexpected interaction between the Nak1-Sog2 kinase complex and Casein kinase 2 (Cka1, Ckb1 and Ckb2) using tandem-affinity purification followed by mass spectrometric analysis. In addition, we identify unique phosphosites on Nak1, Sog2 and the catalytic subunit of casein kinase 2, Cka1. Given the conserved nature of these kinases, we expect this work will shed light on the functions of these proteins both in yeast and higher eukaryotes.
Keywords: fission yeast, casein kinase 2, Ste20 kinase, polar growth
Introduction
Ste20 kinase in S. cerevisae functions as a MAP4K in the mating pheromone-response MAP kinase pathway.1 There are 28 kinases in this family in mammals, with some members functioning similarly to Ste20 in MAP kinase signaling cascades.2,3 The Ste20 family of protein kinases is further subdivided into p21-activated kinases (PAKs) and germinal center kinases (GCKs) based on their structural differences. Ste20 in budding yeast, Pak1/Shk kinases in fission yeast and corresponding PAK family homologs in mammals are regulated by the Rho/Cdc42 family of small GTPase.4,5 GCKs in mammals include MST1/2 and MST3/4, which act upstream of two NDR kinase signaling pathways (Hippo/LATS1/2 and NDR1/2), which have distinct roles in control of cell growth and apoptosis.6,7 In S. pombe, the GCK family members (Nak1, Sid1 and Ppk11) also function in distinct NDR kinase signaling pathways. Nak1 and Ppk11 kinases function in the MOR (morphogenesis Orb6 network) signaling pathway that regulates interphase polarized growth and cell separation at the end of cytokinesis through activation of the NDR1/2-related kinase Orb6.8-13 In contrast, Sid1 activates the LATS1/2-related kinase Sid2 during late mitosis to trigger cytokinesis through constriction of the actomyosin ring. Work from S. pombe showed that crosstalk between these signaling pathways is important for proper regulation of cell growth and division.10,14 Specifically, there appears to be mutual antagonism between the SIN and MOR pathways, whereby the SIN inhibits MOR signaling through phosphorylation of the Nak1 kinase.10,14 Nak1 is in a complex with the leucine-rich repeat-containing protein, Sog2, which might also be a target of SIN inhibition.15 Therefore, studying Nak1 phosphorylation and interacting proteins has provided insights into crosstalk between conserved NDR-kinase signaling networks.
Interestingly, the genetic screen for cell polarity mutants, which identified Nak1 and other MOR pathway components, also identified mutants in casein kinase 2.30 However, how casein kinase 2 fits in to the MOR signaling pathway has remained mysterious.
Results and Discussion
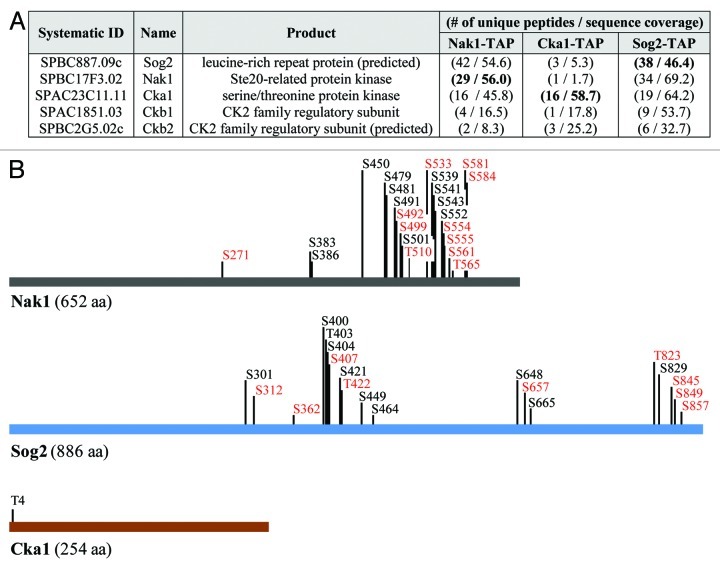
In order to identify proteins that physically interact with the Nak1 kinase, we constructed functional tandem affinity purification (TAP)-tagged Nak1 (Nak1-TAP) according to our protocol described at http://mendel.imp.ac.at/Pombe_tagging/.16 We used a TAP protocol to purify Nak1-TAP, and co-purifying proteins were identified by mass spectrometry. Among proteins with at least two unique peptides, we identified leucine-rich repeat protein Sog2, an uncharacterized protein SPBC2G5.02c, as well as catalytic (Cka1) and regulatory subunits (Ckb1) of casein kinase 2 (Fig. 1A). To confirm the specificity of these interactions, we also performed reciprocal purifications using functional TAP-tagged Sog2 (Sog2-TAP) and Cka1 (Cka1-TAP). Indeed, we found that Nak1, Cka1, Ckb1 and SPBC2G5.02c co-purified with Sog2-TAP and Nak1, Sog2, Ckb1 and SPBC2G5.02c co-purified with Cka1-TAP (Fig. 1A). Our finding that Nak1 co-purifies with Sog2 is consistent with our recent study15 and the observation from budding yeast that the Sog2 ortholog is known to interact with the Nak1 ortholog Kic1.17 More intriguing is our observation that Nak1 co-purifies with casein kinase 2 (CK2) (Fig. 1A). CK2 is an evolutionarily conserved serine/threonine protein kinase that regulates many cellular processes, including cell signaling and proliferation, DNA repair, apoptosis and senescence.18-22 Extensive evidence of its involvement in various cancers as well as neurodegenerative diseases had led to its emergence as a promising drug target.23 However, crosstalk between protein kinases from the Ste20 family and CK2 kinases is poorly understood.
Figure 1. Identification and characterization of proteins co-purifying with Nak1-TAP, Sog2-TAP and Cka1-TAP. (A) List of proteins identified by mass spectrometry co-purifying with S. pombe Nak1-TAP, Sog2-TAP and Cka1-TAP. Proteins were purified from cycling S. pombe cells expressing Nak1-TAP (JG15615), Sog2-TAP (JG16552) or Cka1-TAP (JG15429). Only proteins identified with at least two peptides are included. For a full list of identified proteins see the Table S1. Proteins found in other unrelated purifications (common contaminants) are omitted from this table. Number of unique peptides and sequence coverage are indicated. (B) Summary of phosphorylation sites identified by mass spectrometry. Protein purifications were performed as described in (A). Phosphorylation sites identified on Nak1, Cka1 and Sog2 in this study are indicated. New phosphorylation sites identified are indicated in bold. Sites with the R-X-X-S consensus motif are indicated in red. Asterisks indicate sites with the CK2 consensus motif S/T-X-X-D/E.
So far, the only evidence linking these two kinase families in humans appears to be from Chaar et al., who observed that human CK2 can phosphorylate the Ste20-like kinase family member SLK in vitro.24 It has also been shown that CK2 subunits in S. cerevisiae interact with either Ste20 and/or with Bem1, a cell polarity establishment protein that acts as a scaffold for Ste20.25,26 Few other studies in yeast and humans show interaction of CK2 subunits with Ste20-associated proteins. For instance, budding yeast Cka1 subunit associates with Ste20 target Ste11 (MAP3K).27 The CSK2B subunit of human CK2 contains the binding domain for p21 (CDKN1A), which is required for p21-mediated CK2 inhibition.28 In S. pombe, CK2 subunits Cka1 and Ckb1 interact with Tea1, a microtubule-associated polarity protein that is regulated through phosphorylation by the Pak1/Shk1 kinase.29 Together, it suggests that CK2 affects Ste20-mediated signaling to regulate processes such as polarized growth. Moreover, mutations in both nak1 and cka1 genes in S. pombe result in a similar phenotype—spherical cells, which are indicative of a loss of bipolar growth.30
We also observed that Cka1-GFP localizes to the nucleolus, cell tips, septa and the spindle pole bodies (Fig. 2A and B). While Nak1 localizes to the SPB only during early mitosis, Cka1-GFP appears to remain at the SPBs at all times. Therefore, it partially co-localizes with Nak1 at the SPB. Similar to Nak1,14 Cka1 also localizes to the cell tips and the septum (Fig. 2A). This reinforces the possibility that crosstalk between Nak1 and Cka1 kinases is important for the establishment of polar growth, and it will be interesting to investigate this in the future.
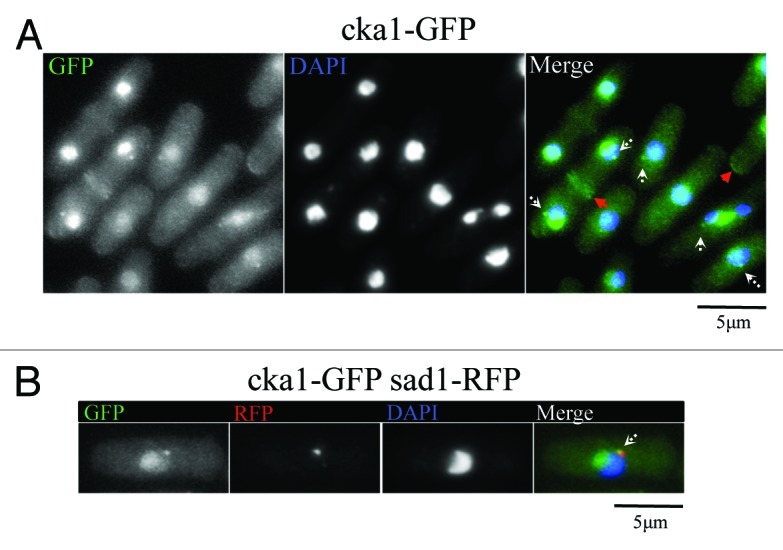
Figure 2. Cka1-GFP localizes similar to MOR pathway proteins like Nak1. (A) Fixed Cka1-GFP-expressing cells (YDM2969) were stained with DAPI. White arrows in the merge panel indicate localization of Cka1-GFP to dots that might correspond to the SPB. Red arrows indicate Cka1-GFP localization to cell septum and tips. (B) Cka1 co-localizes with the SPB marker Sad1. Fixed Cka1-GFP Sad1-RFP expressing cells (YDM3463) were stained with DAPI.
Protein kinase CK2 is a tetramer composed of two catalytically active (CK2 α isoforms) and two regulatory (CK2 β isoforms) subunits. In fission yeast, one catalytic subunit (Cka1) and one regulatory subunit (Ckb1) have been characterized.31 Our analysis showed that Cka1, in addition to Ckb1, co-purifies with an uncharacterized protein SPBC2G5.02c (Fig. 1A). Our sequence searches revealed that SPBC2G5.02c shows high sequence similarity to Ckb1 as well as to other CK2 β isoforms from various species (data not shown). This suggests that SPBC2G5.02c gene encodes for the second fission yeast CK2 β isoform, and we propose to call this gene ckb2 (CK2 beta isoform 2).
Next, we mapped the phosphorylation sites on proteins identified in our purifications. Nak1 was phosphorylated on 22 residues, Sog2 on 19 residues and Cka1 on a single residue (Fig. 1B). Previous studies found 15 residues phosphorylated on Nak1 (S373, S376, S383, S386, S407, T446, S448, S450, S479, S481, S539, S541, S543, S549, S552) and 11 residues phosphorylated on Sog2 (S301, T392, T395, S398, S400, T403, S404, S464, S648, S829, S839).15,32,33 In this study, we show 11 and nine new phosphosites on Nak1 and Sog2, respectively (Fig. 1B, indicated in bold). Sites with an RXXS consensus motif are targeted by NDR kinases. We previously showed that the RXXS sites on Nak1 are phosphorylated by the SIN pathway NDR kinase, Sid2. All Sid2 phosphorylation sites identified by this study were mutated previously (ref. 15 and unpublished data; Fig. 1B, shown in red). Analysis of the mutants showed that Sid2 phosphorylation of Nak1 causes removal of Nak1 from the spindle pole bodies, which prevents premature activation of the SIN in early mitosis. Moreover, Sid2 phosphorylation of Nak1 blocks MOR signaling by preventing interaction of Nak1 with the scaffold protein Mor2.15 The presence of RXXS site phosphorylation on the Nak1 binding partner Sog2 suggests that it might also be phosphorylated by the SIN kinase Sid2. To completely understand the role of phosphorylation in the regulation of Nak1, Sog2 and Cka1, it will be important to identify relevant protein kinases and counteracting phosphatases, analyze possible periodicity during the cell cycle and study functional consequences of mutating these phosphorylation sites. We suggest that sites with the phospho-motif pSP may be targets of the Cdk1 kinase.34 Furthermore, both Nak1 and Sog2 contain the consensus phosphorylation motif S/T-X-X-D/E, which is recognized by CK2.31,35 Some of these sites have been identified by our analysis (Fig. 1B, indicated by asterisks). This indicates a possibility that Nak1 and Sog2 may be potential CK2 substrates. However, further mutational studies will be required to confirm this. Because most of the phosphorylation sites on Nak1 cluster in the C-terminal non-kinase half of the protein, we expect that this region is a key regulatory module targeted by multiple kinases.
Taken together, our results presented in this study provide a thorough biochemical analysis of Nak1-interacting partners and their post-translational modifications. We confirmed that Nak1 co-purifies with Sog2. Significantly, Nak1 shows interaction with the CK2 complex. We therefore established a physical interaction between members of two major kinase families, namely Ste20-related kinases and the Casein kinase 2 family. Further exploration should reveal a better understanding of the functional relationship between them.
Materials and Methods
Strains
S. pombe strains expressing either Nak1-TAP (JG15615, h− nak1-TAP::KanMX), Cka1-TAP (JG15429, h− cka1-TAP::KanMX) or Sog2-TAP (JG16552, h− sog2-TAP::KanMX) were grown in complete yeast extract medium (YE + 5S).36-38 TAP-tagging was confirmed by PCR and immunoblotting.16 The TAP epitope was detected using PAP antibodies (rabbit antiperoxidase antibody linked to peroxidase, Dako) at 1:50,000 dilution in PBS-T.
Protein purification
Six-liter cultures of strains expressing TAP-tagged proteins were grown to mid-log phase (OD ~0.8) and collected by centrifugation. Yeast cell powder was made from frozen pellet using SPEX SamplePrep 6870 Freezer/Mill. Proteins were extracted using IPP150 buffer (50 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% NP-40, complete protease and phosphatase inhibitors, 1 mM PMSF). All washing steps were performed in Poly-Prep columns by gravity flow (Bio-Rad Laboratories, Ges.m.b.H). Five hundred µl of IgG Sepharose™ 6 Fast Flow beads (GE Healthcare, cat.# 17-0969-01) was washed with IPP150 buffer, mixed with protein extract and rotated for 2 h at 4°C. Beads were washed with 20 volumes of IPP150 buffer and with 5 volumes of TEV cleavage buffer (TCB, 10 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% NP-40, 0.5 mM EDTA, 1 mM DTT). Cleavage step was performed in 2 ml of TCB buffer supplemented with 400 units of AcTEV™ protease (Invitrogen, cat.# 12575-015) for 2 h at 16°C. Two ml of eluate was supplemented with 6 µl of 1 M CaCl2 and mixed with 6 ml of Calmodulin binding buffer 1 (CBB1, 10 mM Tris pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% NP-40, 1 mM imidazole, 1 mM Mg-Acetate, 2 mM CaCl2, 10 mM β-mercaptoethanol). One hudred and fifty µl of Calmodulin Sepharose™ 4B beads (GE Healthcare, cat.# 17-0529-01) was washed with CBB1 buffer, added to a mixture of eluate and CBB1 buffer and incubated for 2 h at 4°C. The beads were washed with 10 volumes of CBB1 and 5 volumes of Calmodulin binding buffer 2 (CBB2, 10 mM Tris pH 8.0, 150 mM NaCl, 1 mM Mg-Acetate, 2 mM CaCl2, 1 mM β-mercaptoethanol). The proteins were step-eluted using bead volume of elution buffer (EB, 10 mM Tris pH 8.0, 150 mM NaCl, 1 mM Mg-acetate, 2 mM EGTA, 1 mM β-mercaptoethanol). Eluted proteins were separated on SDS-PAGE and checked by silver staining. Eluates from peak fraction were submitted for LC-MS/MS analysis.16,39,40
Enzymatic digest, LC-MS/MS analysis and data analysis
The pH of the sample was adjusted to 8.5. Disulfide bonds were reduced with DTT (1:10 of the estimated amount of protein) for 30 min at 56°C and subsequently alkylated with iodoacetamide (1:2 of the estimated amount of protein) for 20 min at RT protected from light. DTT (1:20 of the estimated amount of protein) was added to consume excess iodoacetamide and proteins were digested either with trypsin (recombinant, proteomics grade, Roche; 1:25 of the estimated amount of protein) at 37°C overnight. Digestion was stopped by addition of trifluoroacetic acid to pH 2.
Digests were separated on an UltiMate 3000 RSLC nano LC system (Dionex, Thermo Fisher Scientific). Peptides were loaded on a trapping column (PepMap C18, 5 µm particle size, 300 μm i.d. × 5 mm, Dionex, Thermo Fisher Scientific) equilibrated with 0.1% TFA and separated on an analytical column Acclaim PepMap RSLC C18 (50 cm × 75 μm × 2 μm, 100 Å, Dionex, Thermo Fisher Scientific) applying a 30 min resp. 60 min linear gradient from 2% up to 40% acetonitrile. The HPLC was directly coupled to a Q Exactive mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray ionization source (Proxeon, Thermo Fisher Scientific). The electrospray voltage was set to 1,900 V. The mass spectrometer was operated in the data-dependent mode: one full scan (m/z: 350–2,000, resolution 70,000) with lock mass enabled was followed by maximal 10 MS/MS scans. The lock mass was set at the signal of polydimethylcyclosiloxane at m/z 445.120025. The 10 most intense ions were fragmented by higher energy collisional dissociation (HCD) with normalized collision energy of 30. Fragment spectra were acquired with a resolution of 17,500. The ion target value for full MS was set to 1,000,000 for MS/MS to 100,000. Fragmented ions were excluded from further selection for 20 sec.
Peptide identification was performed using the SEQUEST algorithm in the Proteome Discoverer 1.3.0.339 software package (Thermo Fisher Scientific). Spectra were searched against the Sanger S. pombe database (04/23/2012). Following search parameters were used: peptide tolerance was set to 10 ppm, MS/MS fragment tolerance to 50 mmu, trypsin was selected as protease and two missed cleavages were allowed, carbamidomethylcysteine was set as static modification, oxidation of methionine and phosphorylation of serine, threonine and tyrosine as a variable modification. The results were filtered at the XCorr values to an FDR of 1% on the peptide level.
Supplementary Material
Acknowledgments
This work was supported by Austrian Science Fund grants P23609, P21437 and F3403, HFSP grant RGY0069/2010 and by the Slovak Research and Development Agency contract No. APVV-0111-12 and APVV-0334-12. L.C. was supported by the (European Community’s) Seventh Framework Programme (FP7/2007–2013) under grant agreement number PERG07-GA-2010-268167. J.G. was supported by the (European Community’s) Seventh Framework Programme (FP7/2007–2013) under grant agreement number PCIG11-GA-2012-322300. D.M. and S.G. were supported by the National Institutes of Health grant GM058406-14 to D. McCollum.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/24095
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/24095
References
- 1.Boyce KJ, Andrianopoulos A. Ste20-related kinases: effectors of signaling and morphogenesis in fungi. Trends Microbiol. 2011;19:400–10. doi: 10.1016/j.tim.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Delpire E. The mammalian family of sterile 20p-like protein kinases. Pflugers Arch. 2009;458:953–67. doi: 10.1007/s00424-009-0674-y. [DOI] [PubMed] [Google Scholar]
- 3.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–30. doi: 10.1016/S0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 4.Chan PM, Manser E. PAKs in human disease. Prog Mol Biol Transl Sci. 2012;106:171–87. doi: 10.1016/B978-0-12-396456-4.00011-0. [DOI] [PubMed] [Google Scholar]
- 5.Ong CC, Jubb AM, Zhou W, Haverty PM, Harris AL, Belvin M, et al. p21-activated kinase 1: PAK’ed with potential. Oncotarget. 2011;2:491–6. doi: 10.18632/oncotarget.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cornils H, Kohler RS, Hergovich A, Hemmings BA. Downstream of human NDR kinases: impacting on c-myc and p21 protein stability to control cell cycle progression. Cell Cycle. 2011;10:1897–904. doi: 10.4161/cc.10.12.15826. [DOI] [PubMed] [Google Scholar]
- 7.Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–64. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- 8.Goshima T, Kume K, Koyano T, Ohya Y, Toda T, Hirata D. Fission yeast germinal center (GC) kinase Ppk11 interacts with Pmo25 and plays an auxiliary role in concert with the morphogenesis Orb6 network (MOR) in cell morphogenesis. J Biol Chem. 2010;285:35196–205. doi: 10.1074/jbc.M110.176867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu G, Young D. Conserved Orb6 phosphorylation sites are essential for polarized cell growth in Schizosaccharomyces pombe. PLoS ONE. 2012;7:e37221. doi: 10.1371/journal.pone.0037221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray S, Kume K, Gupta S, Ge W, Balasubramanian M, Hirata D, et al. The mitosis-to-interphase transition is coordinated by cross talk between the SIN and MOR pathways in Schizosaccharomyces pombe. J Cell Biol. 2010;190:793–805. doi: 10.1083/jcb.201002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanai M, Kume K, Miyahara K, Sakai K, Nakamura K, Leonhard K, et al. Fission yeast MO25 protein is localized at SPB and septum and is essential for cell morphogenesis. EMBO J. 2005;24:3012–25. doi: 10.1038/sj.emboj.7600782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonhard K, Nurse P. Ste20/GCK kinase Nak1/Orb3 polarizes the actin cytoskeleton in fission yeast during the cell cycle. J Cell Sci. 2005;118:1033–44. doi: 10.1242/jcs.01690. [DOI] [PubMed] [Google Scholar]
- 13.Goyal A, Takaine M, Simanis V, Nakano K. Dividing the spoils of growth and the cell cycle: The fission yeast as a model for the study of cytokinesis. Cytoskeleton (Hoboken) 2011;68:69–88. doi: 10.1002/cm.20500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta S, McCollum D. Crosstalk between NDR kinase pathways coordinates cell cycle dependent actin rearrangements. Cell Div. 2011;6:19. doi: 10.1186/1747-1028-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta S, Mana-Capelli S, McLean JR, Chen CT, Ray S, Gould KL, et al. Identification of SIN Pathway Targets Reveals Mechanisms of Crosstalk between NDR Kinase Pathways. Curr Biol. 2013;23:333–8. doi: 10.1016/j.cub.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cipak L, Spirek M, Novatchkova M, Chen Z, Rumpf C, Lugmayr W, et al. An improved strategy for tandem affinity purification-tagging of Schizosaccharomyces pombe genes. Proteomics. 2009;9:4825–8. doi: 10.1002/pmic.200800948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson B, Kurischko C, Horecka J, Mody M, Nair P, Pratt L, et al. RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol Biol Cell. 2003;14:3782–803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominguez I, Sonenshein GE, Seldin DC. Protein kinase CK2 in health and disease: CK2 and its role in Wnt and NF-kappaB signaling: linking development and cancer. Cell Mol Life Sci. 2009;66:1850–7. doi: 10.1007/s00018-009-9153-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dodson GE, Limbo O, Nieto D, Russell P. Phosphorylation-regulated binding of Ctp1 to Nbs1 is critical for repair of DNA double-strand breaks. Cell Cycle. 2010;9:1516–22. doi: 10.4161/cc.9.8.11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moucadel V, Prudent R, Sautel CF, Teillet F, Barette C, Lafanechere L, et al. Antitumoral activity of allosteric inhibitors of protein kinase CK2. Oncotarget. 2011;2:997–1010. doi: 10.18632/oncotarget.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor KM, Kille P, Hogstrand C. Protein kinase CK2 opens the gate for zinc signaling. Cell Cycle. 2012;11:1863–4. doi: 10.4161/cc.20414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez DI, Gil C, Martinez A. Protein kinases CK1 and CK2 as new targets for neurodegenerative diseases. Med Res Rev. 2011;31:924–54. doi: 10.1002/med.20207. [DOI] [PubMed] [Google Scholar]
- 23.Hanif IM, Hanif IM, Shazib MA, Ahmad KA, Pervaiz S. Casein Kinase II: an attractive target for anti-cancer drug design. Int J Biochem Cell Biol. 2010;42:1602–5. doi: 10.1016/j.biocel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Chaar Z, O’reilly P, Gelman I, Sabourin LA. v-Src-dependent down-regulation of the Ste20-like kinase SLK by casein kinase II. J Biol Chem. 2006;281:28193–9. doi: 10.1074/jbc.M605665200. [DOI] [PubMed] [Google Scholar]
- 25.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, et al. The genetic landscape of a cell. Science. 2010;327:425–31. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fasolo J, Sboner A, Sun MG, Yu H, Chen R, Sharon D, et al. Diverse protein kinase interactions identified by protein microarrays reveal novel connections between cellular processes. Genes Dev. 2011;25:767–78. doi: 10.1101/gad.1998811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 28.Götz C, Kartarius S, Scholtes P, Montenarh M. Binding domain for p21(WAF1) on the polypeptide chain of the protein kinase CK2 beta-subunit. Biochem Biophys Res Commun. 2000;268:882–5. doi: 10.1006/bbrc.2000.2230. [DOI] [PubMed] [Google Scholar]
- 29.Snaith HA, Thompson J, Yates JR, 3rd, Sawin KE. Characterization of Mug33 reveals complementary roles for actin cable-dependent transport and exocyst regulators in fission yeast exocytosis. J Cell Sci. 2011;124:2187–99. doi: 10.1242/jcs.084038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snell V, Nurse P. Genetic analysis of cell morphogenesis in fission yeast--a role for casein kinase II in the establishment of polarized growth. EMBO J. 1994;13:2066–74. doi: 10.1002/j.1460-2075.1994.tb06481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roussou I, Draetta G. The Schizosaccharomyces pombe casein kinase II alpha and beta subunits: evolutionary conservation and positive role of the beta subunit. Mol Cell Biol. 1994;14:576–86. doi: 10.1128/mcb.14.1.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, et al. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch A, Krug K, Pengelley S, Macek B, Hauf S. Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci Signal. 2011;4:rs6. doi: 10.1126/scisignal.2001588. [DOI] [PubMed] [Google Scholar]
- 34.Maller Jl, Gautier J, Langan TA, Lohka MJ, Shenoy S, Shalloway D, et al. Maturation-promoting factor and the regulation of the cell cycle. J Cell Sci Suppl. 1989;12:53–63. doi: 10.1242/jcs.1989.supplement_12.6. [DOI] [PubMed] [Google Scholar]
- 35.Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, et al. A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol. 1996;16:6486–93. doi: 10.1128/mcb.16.11.6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dudas A, Ahmad S, Gregan J. Sgo1 is required for co-segregation of sister chromatids during achiasmate meiosis I. Cell Cycle. 2011;10:951–5. doi: 10.4161/cc.10.6.15032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cipak L, Hyppa RW, Smith GR, Gregan J. ATP analog-sensitive Pat1 protein kinase for synchronous fission yeast meiosis at physiological temperature. Cell Cycle. 2012;11:1626–33. doi: 10.4161/cc.20052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cipak L, Zhang C, Kovacikova I, Rumpf C, Miadokova E, Shokat KM, et al. Generation of a set of conditional analog-sensitive alleles of essential protein kinases in the fission yeast Schizosaccharomyces pombe. Cell Cycle. 2011;10:3527–32. doi: 10.4161/cc.10.20.17792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rumpf C, Cipak L, Schleiffer A, Pidoux A, Mechtler K, Tolić-Nørrelykke IM, et al. Laser microsurgery provides evidence for merotelic kinetochore attachments in fission yeast cells lacking Pcs1 or Clr4. Cell Cycle. 2010;9:3997–4004. doi: 10.4161/cc.9.19.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rumpf C, Cipak L, Dudas A, Benko Z, Pozgajova M, Riedel CG, et al. Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle. 2010;9:2657–62. doi: 10.4161/cc.9.13.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.