Abstract
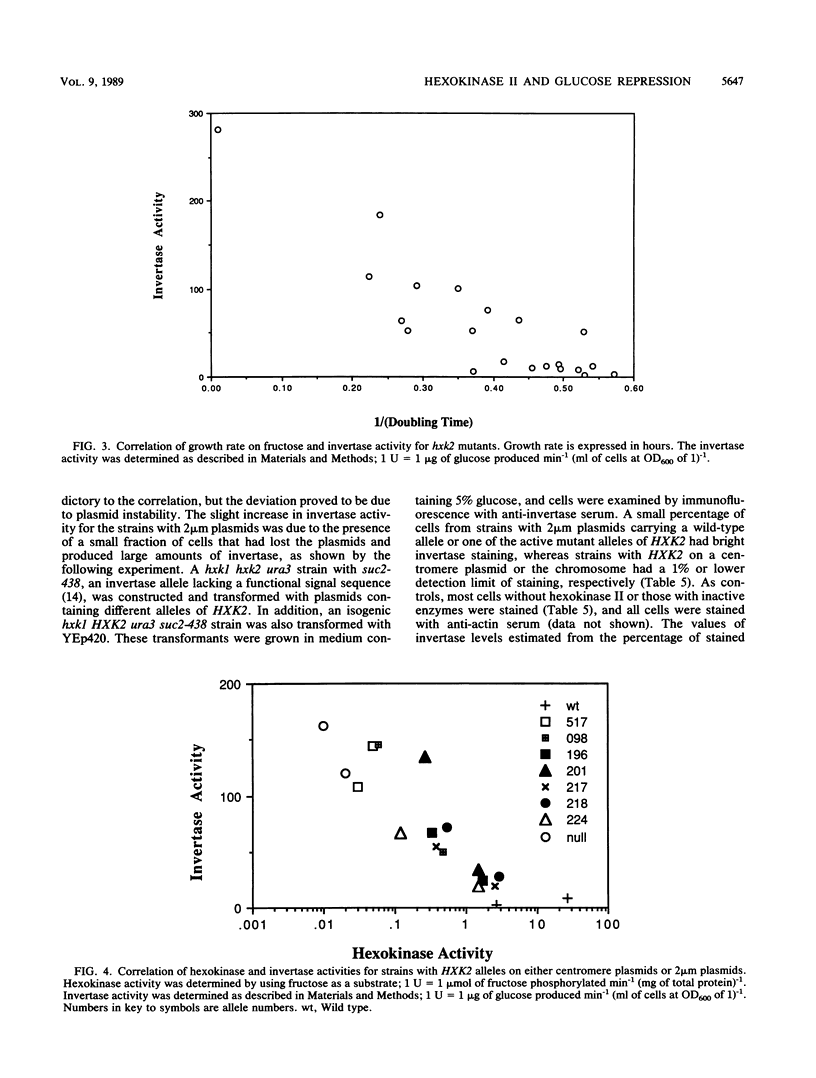
Saccharomyces cerevisiae mutants containing different point mutations in the HXK2 gene were used to study the relationship between phosphorylation by hexokinase II and glucose repression in yeast cells. Mutants showing different levels of hexokinase activity were examined for the degree of glucose repression as indicated by the levels of invertase activity. The levels of hexokinase activity and invertase activity showed a strong inverse correlation, with a few exceptions attributable to very unstable hexokinase II proteins. The in vivo hexokinase II activity was determined by measuring growth rates, using fructose as a carbon source. This in vivo hexokinase II activity was similarly inversely correlated with invertase activity. Several hxk2 alleles were transferred to multicopy plasmids to study the effects of increasing the amounts of mutant proteins. The cells that contained the multicopy plasmids exhibited less invertase and more hexokinase activity, further strengthening the correlation. These results strongly support the hypothesis that the phosphorylation activity of hexokinase II is correlated with glucose repression.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Broach J. R. Construction of high copy yeast vectors using 2-microns circle sequences. Methods Enzymol. 1983;101:307–325. doi: 10.1016/0076-6879(83)01024-1. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Organization of the SUC gene family in Saccharomyces. Mol Cell Biol. 1983 Mar;3(3):351–359. doi: 10.1128/mcb.3.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Celenza J. L., Eng F. J. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Mol Cell Biol. 1985 Nov;5(11):2894–2902. doi: 10.1128/mcb.5.11.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DelaFuente G. Specific inactivation of yeast hexokinase induced by xylose in the presence of a phosphoryl donor substrate. Eur J Biochem. 1970 Oct;16(2):240–243. doi: 10.1111/j.1432-1033.1970.tb01077.x. [DOI] [PubMed] [Google Scholar]
- Entian K. D. A carbon catabolite repression mutant of Saccharomyces cerevisiae with elevated hexokinase activity: evidence for regulatory control of hexokinase PII synthesis. Mol Gen Genet. 1981;184(2):278–282. doi: 10.1007/BF00272917. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Fröhlich K. U. Saccharomyces cerevisiae mutants provide evidence of hexokinase PII as a bifunctional enzyme with catalytic and regulatory domains for triggering carbon catabolite repression. J Bacteriol. 1984 Apr;158(1):29–35. doi: 10.1128/jb.158.1.29-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D. Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in carbon catabolite repression in yeast. Mol Gen Genet. 1980;178(3):633–637. doi: 10.1007/BF00337871. [DOI] [PubMed] [Google Scholar]
- Entian K. D., Hilberg F., Opitz H., Mecke D. Cloning of hexokinase structural genes from Saccharomyces cerevisiae mutants with regulatory mutations responsible for glucose repression. Mol Cell Biol. 1985 Nov;5(11):3035–3040. doi: 10.1128/mcb.5.11.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K. D., Mecke D. Genetic evidence for a role of hexokinase isozyme PII in carbon catabolite repression in Saccharomyces cerevisiae. J Biol Chem. 1982 Jan 25;257(2):870–874. [PubMed] [Google Scholar]
- Fernández R., Herrero P., Moreno F. Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by D-xylose. J Gen Microbiol. 1985 Oct;131(10):2705–2709. doi: 10.1099/00221287-131-10-2705. [DOI] [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C. A., Preuss D., Grisafi P., Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987 Jan 16;235(4786):312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- Kuo C. L., Campbell J. L. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8-1 mutation. Mol Cell Biol. 1983 Oct;3(10):1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lobo Z., Maitra P. K. Genetics of yeast hexokinase. Genetics. 1977 Aug;86(4):727–744. doi: 10.1093/genetics/86.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo Z., Maitra P. K. Physiological role of glucose-phosphorylating enzymes in Saccharomyces cerevisiae. Arch Biochem Biophys. 1977 Aug;182(2):639–645. doi: 10.1016/0003-9861(77)90544-6. [DOI] [PubMed] [Google Scholar]
- Ma H., Bloom L. M., Zhu Z. M., Walsh C. T., Botstein D. Isolation and characterization of mutations in the HXK2 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1989 Dec;9(12):5630–5642. doi: 10.1128/mcb.9.12.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Botstein D. Effects of null mutations in the hexokinase genes of Saccharomyces cerevisiae on catabolite repression. Mol Cell Biol. 1986 Nov;6(11):4046–4052. doi: 10.1128/mcb.6.11.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Kunes S., Schatz P. J., Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58(2-3):201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Yoshimatsu T., Oshima Y. Recessive mutations conferring resistance to carbon catabolite repression of galactokinase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1983 Mar;153(3):1405–1414. doi: 10.1128/jb.153.3.1405-1414.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes L. C., Pudles J. Studies on the active site of yeast hexokinase. Specific phosphorylation of a serine residue induced by D-xylose and ATPMg. Eur J Biochem. 1976 May 17;65(1):41–47. doi: 10.1111/j.1432-1033.1976.tb10387.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann F. K., Scheel I. Mutants of Saccharomyces cerevisiae resistant to carbon catabolite repression. Mol Gen Genet. 1977 Jul 7;154(1):75–82. doi: 10.1007/BF00265579. [DOI] [PubMed] [Google Scholar]




