Abstract
Id2 is a helix-loop-helix (HLH) transcription factor essential for normal development and its expression is dysregulated in many human neurological conditions. Although it is speculated that elevated Id2 levels contribute to the pathogenesis of these disorders, it is unknown whether dysregulated Id2 expression is sufficient to perturb normal brain development or function. Here, we show that mice with elevated Id2 expression during embryonic stages develop microcephaly, and that females in particular are prone to generalized tonic-clonic seizures. Analyses of Id2 transgenic brains indicate that Id2 activity is highly cell context specific: elevated Id2 expression in naive NSCs in early neuroepithelium induces apoptosis and loss of NSCs and intermediate progenitors. Activation of Id2 in maturing neuroepithelium results in less severe phenotypes and is accompanied by elevation of G1 Cyclin expression and p53 target gene expression. In contrast, activation of Id2 in committed intermediate progenitors has no significant phenotype. Functional analysis with Id2 over-expressing and Id2-null NSCs shows that Id2 negatively regulates NSC self-renewal in vivo, in contrast to previous cell culture experiments. Deletion of p53 function from Id2-transgenic brains rescues apoptosis and results in increased incidence of brain tumors. Furthermore, Id2 over-expression normalizes the increased self-renewal of p53-null NSCs, suggesting that Id2 activates and modulates the p53 pathway in NSCs. Together, these data suggest that elevated Id2 expression in embryonic brains can cause deregulated NSC self-renewal, differentiation and survival that manifest in multiple neurological outcomes in mature brains, including microcephaly, seizures, and brain tumors.
Keywords: neural stem cells, Id2, brain tumor, glioma, medulloblastoma, self-renewal, apoptosis, seizure, CyclinG1, Rett syndrome
INTRODUCTION
The mammalian brain develops from tightly controlled expansion and maturation of neuroepithelial stem cells in the rostral tip of the neural tube. The early neuroepithelium is composed of neural stem cells (NSCs) that rapidly divide symmetrically while maturing neuroepithelial stem cells divide asymmetrically to generate a daughter cell that remains as a NSC and another daughter cell that becomes an intermediate (basal) progenitor cell or a mature neuron1, 2. For normal brain development, proliferation and differentiation of NSCs and intermediate progenitors must be strictly orchestrated with the activation of differentiation genes, cell cycle withdrawal, and cellular migration. Many transcription factors have been shown to be critical for these processes, including bHLH genes Hes1, Hes5, Neurogenin1/2, Mash1 and Id1, 2, 3, and 43-6. In addition, altered expression of these genes, particularly the Id (inhibitor of differentiation) genes, has been associated with various neurological disorders in human, suggesting that they may be involved in pathogenesis7, 8.
Id genes encode helix-loop-helix proteins that are highly expressed in the developing nervous system. There are four Id gene family members (Id1-4) in mammals. Unlike basic-helix-loop-helix (bHLH) proteins that directly bind to DNA, ID proteins lack DNA binding domain and function by sequestering bHLH proteins in inactive heterodimeric complexes 8-10. Many bHLH proteins are involved in cell fate specification and differentiation; hence, it is thought that by inhibiting bHLH proteins, ID proteins block differentiation. In addition, ID proteins also actively promote proliferation by interacting with components of the cell cycle machinery. For example, ID2 promotes G1-S transition in neural cells by directly binding to the RB (Retinoblastoma) protein and inhibiting its checkpoint function11, 12. Id genes are expressed in the developing mammalian brain in overlapping patterns. Consistent with its function in blocking differentiation and promoting proliferation, Id genes are highly expressed in the VZ (ventricular zone) and SVZ (subventricular zone), proliferative zones containing NSCs and intermediate progenitors13, 14. In addition, Id2 is also expressed in postmitotic neurons, particularly in layer 5 neurons in somatosensory cortex 14, 15, consistent with its proposed role in regulation of neurite outgrowth16, 17.
Id genes are also implicated in stem cell regulation in vitro and in vivo. For example, Id genes can replace serum or BMP in mouse ES cell cultures to promote self-renewal and pluripotency18. In addition, Id1 marks and promotes self-renewal of adult NSCs in the SVZ19, and glioma initiating cells (glioma stem cells) in human GBMs20. Consistently, while Id1 and Id3 single null mice develop grossly normal brains, Id1/3 double mutant brains show precocious differentiation and severe depletion of neural precursors6, suggesting a functional redundancy between Id1 and Id3 in embryonic NSCs. In contrast, Id4 single null mice develop microcephalic brains due to compromised expansion of early neuroepithelial stem cells5, and ectopic expression of Id4 confers stem cell-like properties to Ink4/ARF−/− astrocytes in culture21. Together, these studies suggest a positive role for Id genes in stem cell maintenance and proliferation.
Other studies, however, paint a more complex picture of Id gene function. In some cell types, Id genes promote apoptosis and differentiation rather than proliferation, and function as tumor suppressors. For example, Id2 −/− mice develop intestinal tumors, suggesting that Id2 functions as a tumor suppressor in the intestinal epithelium22. In SW480 and MIA PaCa-2 cells, Id2 inhibits proliferation, and mutant p53 proteins promote cellular proliferation in part by suppressing Id2 expression in these cells23. In addition, Id genes can also promote apoptosis in a cell context-dependent manner. For example, Id1 promotes proliferation of mammary cells at low-density but induces apoptosis at high-density24. Furthermore, ectopic expression of Id2 in myeloid cells induces apoptosis through a mechanism independent of HLH protein interaction25. Hence, whether Id genes promote proliferation, differentiation or apoptosis appears to depend on the cell type and the cellular microenvironment, underscoring the need to decipher Id gene function in relevant cell contexts in vivo.
To determine the causal role of Id2 dysregulation in pathogenesis of neural conditions, we analyzed the consequences of altered Id2 expression levels in different populations of proliferating cells in the developing cortex. We report that elevated Id2 expression negatively regulates NSC self-renewal and survival in vivo, ultimately leading to microcephalic brains that are susceptible to seizures and brain tumor formation.
MATERIALS AND METHODS
Mice
All mouse work was performed according to the protocols approved by The Jackson Laboratory ACUC. All phenotypic analyses were performed by comparing at least three pairs of littermate control and transgenic embryos. FID2 mouse was generated in the Israel laboratory26. FVB-Tg(GFAP-cre)25Mes/J, B6.Cg-Tg(Nes-cre)1Kln/J, 129P2(Cg)-Foxg1<tm1(cre)Skm>/J, and Tg(Neurog1-cre)1Jejo/J were obtained from the Jackson Laboratory Repository. For embryo harvests, noon of the day when vaginal plug was observed was considered E0.5 days.
Immunohistochemistry
Embryo brains were fixed in fresh 4% paraformaldehyde overnight, and then prepared for OCT or paraffin sections. Coronal sections from matching levels were compared between control and transgenic littermates with Hematoxylin and Eosin-Y and standard immunohistochemistry protocol. Please see Supplementary Materials and Methods for details of antibodies used in the study.
Electroconvulsive threshold test and EEG
FID2;Nestin-Cre and control littermates on C57BL/6J background were tested for minimal forebrain clonic seizures - the mildest generalized seizure endpoint - following electrical stimulation, as previously described27. Young (5-6 weeks old) control and transgenic mice were tested once at a stimulus level estimated to be in the first quartile of response for wildtype. Because it is well established that females generally have a lower electroconvulsive threshold than males27, the sexes were tested separately at stimulus levels estimated to be in the first quartile response, respectively (6.5 mA for female and 8.0 mA for male). The pulse frequency, width and duration were fixed at 299 Hz, 1.6 ms and 0.2 s, respectively. Differential responses between transgenic and non-transgenic were recorded and analyzed using a one-tailed 2 × 2 contingency χ2 test for each sex. EEG electrode implant surgery and analysis was performed as previously28. Please see Supplementary Materials and Methods for more details.
Realtime RT-PCR analysis
Total RNAs from E12.5 and E15.5 littermate cortices were extracted using Trizol reagent (Invitrogen). RNA samples were first treated with DNase1 (Ambion) and converted to cDNA using the iScript cDNA synthesis Kit (Bio-Rad). Realtime RT-PCR was performed using iQ SYBR Green Supermix (Bio-Rad) in iQ5 realtime PCR machine (Bio-Rad). Samples were normalized to internal 18s values. Realtime RT-PCR primer sequences are listed in Supplementary Materials and Methods.
Secondary neurosphere formation assay
E15.5 transgenic and littermate control cortices were dissected and dissociated into single cells to establish fresh neurosphere cultures. At passages 1 and 2, neurospheres were dissociated into single cells and were plated at a density of 1 cell/1ul in neural stem cell culture media (DMEM/F12 with B-27, 10ng/ml basic FGF, 20ng/ml EGF, and Penicillin/Streptomycin). After 7days, secondary neurosphere numbers were counted. All experiments were performed with at least three independent biological replicates and on at least two consecutive passages for technical replicates.
Cell cycle analysis
E12.5 cortices from control and FID2;Nestin-cre littermate embryos were microdissected and immediately dissociated and fixed in 70% ethanol. RNA was digested with RNAse (100μg/ml) and DNA was labeled with Propidium Iodide (50μg/ml) for 30 minutes. Samples were scanned on FACS Calibur to measure DNA contents. Cell cycle profiles were generated using the Flowjo software.
Statistical analysis
In all quantitative experiments, only littermate control and transgenic embryos were directly compared and samples from at least two independent litters were compared. Statistical significance was calculated using GraphPad Prism software, using unpaired t-test (except for Kaplan Meier survival curve) and error bars represent SEM unless noted otherwise.
RESULTS
Elevated Id2 expression induces apoptosis and depletes early NSCs in vivo
To determine the consequence of misregulated Id2 expression on brain development, we used a floxed allele of Id2 (FID2)26 to induce human ID2-EGFP fusion protein expression in different populations of cells using Cre-mediated recombination. To assess the function of Id2 in NSC proliferation and differentiation, we first used Foxg1-cre to activate FID2 expression in the early neuroepithelium (Fig. 1A). Foxg1-Cre is expressed in the developing forebrain at E9.5 29, corresponding to the time when neuroepithelial stem cells rapidly expand laterally through symmetric stem:stem divisions and prior to onset of neurogenesis in the cortex2.
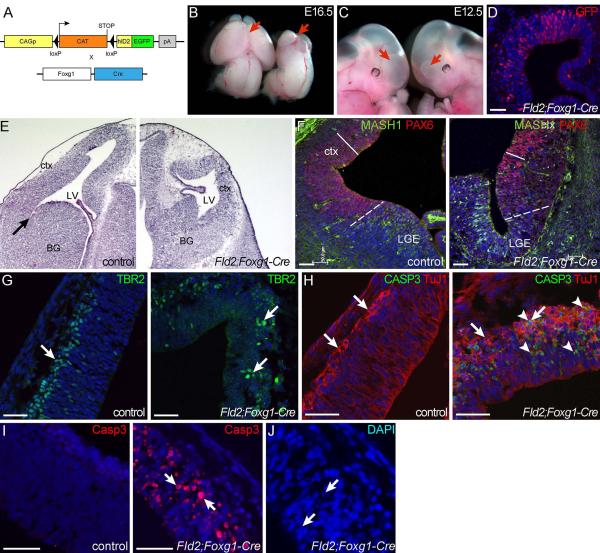
Figure 1. Elevation of Id2 expression in naive, pre-neurogenic neuroepithelium results in severe hypoplasia.
(A) A schematic of transgenic crosses to generate FID2;Foxg1-cre embryos. (B,C) Images of control and FID2;Foxg1-Cre littermate embryos at E16.5 (B) and E12.5 (C). Red arrows indicate forebrain regions. FID2;Foxg1-Cre embryos are on the right. (D) Antibody staining against GFP shows expression of the ID2-GFP fusion protein in the transgenic brain. (E) H&E staining of coronal sections through E12.5 control and FID2;Foxg1-Cre littermates shows severely reduced forebrain size in the transgenic embryo. Arrow marks the pallial-subpallial sulcus. F) Regional markers for dorsal (PAX6, red) and ventral (MASH1. green) forebrain show that the ventral forebrain show most of the residual epithelium is dorsal in FID2;Foxg1-Cre embryos. Dash lines indicate pallial-subpallial boundary, and solid bars indicate the thickness of the PAX6+ NSC zone. (G) Marker analysis for cortical progenitors (TBR2) shows no gross change in numbers. (H,I) Antibody staining for differentiating neurons (ß-III-Tubulin/TuJ1) and apoptotic cells (cleaved caspase 3) at E12.5 (H) and E10.5 (I). Arrows point to TuJ1+ neurons and arrowheads point to cleaved caspase3+ apoptotic cells. (J) DAPI staining in transgenic brain at E10.5 shows pyknotic and karyorrhexic nuclei. Scale bars= 50μm.
At mid-gestation, FID2;Foxg1-Cre forebrains were dramatically smaller than those of control littermates (arrows in Fig. 1B). Note that mid- and hindbrain regions of FID2;Foxg1-Cre embryos are normal in size, indicating that hypoplasia is observed only in the region where Id2 expression is elevated. This phenotype was obvious as early as E12.5 (mutant on the right, Fig. 1C), and robust transgene expression is detected at E12.5 with an antibody against the GFP fusion protein (Fig. 1D). H&E staining on coronal sections of control and FID2;Foxg1-Cre at E12.5 showed severely reduced lateral ventricle size due to compromised lateral expansion of the forebrain neuroepithelium (Fig. 1E). To determine the regional identify of the residual neuroepithelium in FID2;Foxg1-Cre forebrain, we analyzed expression patterns of dorsal and ventral forebrain VZ markers30 (PAX6 and MASH1, respectively). Most of the neuroepithlelium in the transgenic brain was PAX6+ and only the most ventral region was MASH1+ (Fig. 1F), suggesting that the residual tissue is mostly cortical even though both ventral and dorsal tissues were properly specified.
To assess the underlying cause of compromised brain growth in the transgenic forebrain, we analyzed proliferation, differentiation and survival of NSCs and intermediate progenitors in the residual FID2;Foxg1-cre cortex. Most neuroepithelial cells expressed NSC marker PAX6+, as expected, and the thickness of the PAX6+ neuroepithelium is reduced in transgenic brain (Fig. 1F). In the developing cortex of wildtype embryos, intermediate progenitors express a T-box transcription factor, TBR2, and are observed in the VZ and SVZ31. TBR2+ cells were present in disorganized clusters in FID2;Foxg1-cre cortices (arrows in Fig. 1G). Analysis of differentiating neurons (TuJ1/ß-III-tubulin) showed similar disorganization in radial columns and thickening of the postmitotic zone in FID2;Foxg1-Cre cortices (Fig. 1H), suggesting increased neurogenesis.
To determine whether cell death also contributes to reduced brain size, we analyzed FID2;Foxg1-Cre brains for apoptosis. Antibody staining for cleaved Caspase3 confirmed an increased number of apoptotic cells in the transgenic cortex (arrows in Fig. 1H). Most of the apoptotic cells were TuJ1/ß-III-tubulin+ differentiating neurons (Fig. 1H), suggesting that differentiation is incompatible with sustained high levels of Id2 expression. To determine whether undifferentiated neuroepithelial NSCs also die, we analyzed FID2;Foxg1-Cre brains at an earlier time point. At E10.5, 1 day after the Cre activation, we did not observe increased neurogenesis but did observe increased number of apoptotic cells in the transgenic brain, marked by cleaved Caspase3 staining (Fig. 1I) and the appearance of pyknotic and karyorrhexic nuclei (Fig. 1J). Together, these results show that early neuroepithelial stem cells fail to expand when Id2 expression is elevated due to increased apoptosis and premature differentiation of NSCs. These observations also explain the more severe hypoplasia observed in the ventral region of FID2;Foxg1-Cre brains since neurogenesis starts earlier in the ventral forebrain than in the dorsal region.
Id2 over-expression results in microcephaly and seizures
Because of the severity of the phenotype and embryonic lethality in FID2;Foxg1-Cre mice, it was not technically possible to further analyze molecular and cellular changes or determine functional consequences of Id2 over-expression in mature brains. Hence, we crossed FID2 mice to Nestin-Cre and GFAP-Cre transgenic mice (Fig. 2A). In these mice, the Cre recombinase expression is activated in both neural stem and progenitor cells in maturing neuroepithelium at E11.5 and E12.5, respectively. Overall phenotypes of FID2;GFAP-cre and FID2;Nestin-Cre embryos were very similar; hence, data from these strains are shown interchangeably below. Although the transgenic phenotype was 100% penetrant, we did observe differences in the severity of the phenotype in different individuals.
Figure 2. Id2 over-expression results in microcephaly, generalized tonic-clonic seizures and low seizure threshold.
(A) A schematic of transgenic crosses to generate FID2;Nestin-cre embryos. (B) RT-PCR analysis using primers from hID2 and GFP show the transgene expression in FID2;Nestin-cre cortex at E12.5. (C) Antibody staining against GFP shows robust expression (arrows) of the transgene at the protein level in FID2;Nestin-cre cortex at E12.5. (D) Control and littermate FID2-Nestin-cre brains in adult mice show significantly smaller transgenic brain. (E) Brain/body weight ratio comparison between control (n=7) and FID2;Nestin-cre (n=7) littermate females at p26 confirm microcephalic phenotype (p=0.0406). Paired t-test. (F) Onset of handling-associated generalized tonic-clonic seizures in Id2 transgenic female mice. (G) Video-EEG recording of a generalized tonic-clonic seizure observed in an Id2 transgenic female. The traces shown are the differential signal between each electrode pair (RF - right front electrode; LF - left front; RB –right back; LB – left back; positions relative to Bregma and midline). Numbered arrows denote the behavioral progression, as described in the text. Scale bar = 1 second. (H) A summary table of mice that experienced a minimal clonic seizure in response to a single subthreshold electroconvulsive stimulus. One-tailed p-values are shown from a Fisher Exact 2 × 2 contingency table test for each sex. Scale bar= 50μm
The hID2-GFP fusion transgene is robustly expressed at the RNA (Fig. 2B) and protein levels (Fig.2 C) in FID2;Nestin-cre brains at E12.5. FID2;Nestin-Cre double transgenic mice were viable; however, gross examination of postnatal brains indicated microcephaly in FID2;Nestin-cre mice (Fig. 2D, E). In addition, we observed frequent generalized tonic-clonic seizures in female FID2;Nestin-cre mice. Seizures occurred with an average age of onset of 3 months in females (5 of 12) but not in males (0 of 9) or any non-transgenic or Cre-minus control littermates (Fig. 2F). These events lasted for just over 30 seconds and consisted of partial seizure behaviors (whisker twitching, jaw clonus, forelimb clonus), and progressed to generalized behaviors (Straub tail, neck flexion, full loss of posture), followed by tonic-clonic movement in fore- and hindlimbs and ending in transient immobility. Several transgenic mice were examined by video-electroencephalography (EEG) in overnight recordings, and we were able to capture five spontaneous convulsive seizures in two transgenic mice (Fig. 2G, Supplementary Fig. 1, and not shown). The progression of the typical generalized tonic-clonic convulsion sequence was reflected in the EEG – beginning with an initial single spike with the mouse still (arrow 1 in Fig. 2G), then partial behaviors accompanied by high-amplitude faster spiking that spread from one region to another (arrows 2 and 3), then generalized behaviors accompanied by very high amplitude complex spiking (arrow 4), followed by complete stillness and post-ictal depression (arrow 5). We also examined electroconvulsive threshold in 5-6 week old Id2 transgenic mice using the electroconvulsive threshold test (Fig. 2H). Littermate control and transgenic male mice did not show significant differences in their electroconvulsive threshold (28.1% vs. 40%, p=0.367, Fig. 2H). However, female transgenic mice showed a significantly reduced electroconvulsive threshold when compared with non-transgenic or cre-minus control littermates (15.4% vs. 72.2%, p=0.0002). These data suggest that elevated Id2 levels in the developing brain is sufficient to lead to serious developmental abnormalities, manifesting in increased propensity for sex-specific spontaneous convulsions and low seizure threshold, and microcephaly. In addition, these results suggest that Id2 may contribute to sex-specific differentiation of the mammalian brain.
Id2 inhibits NSC self-renewal in vivo
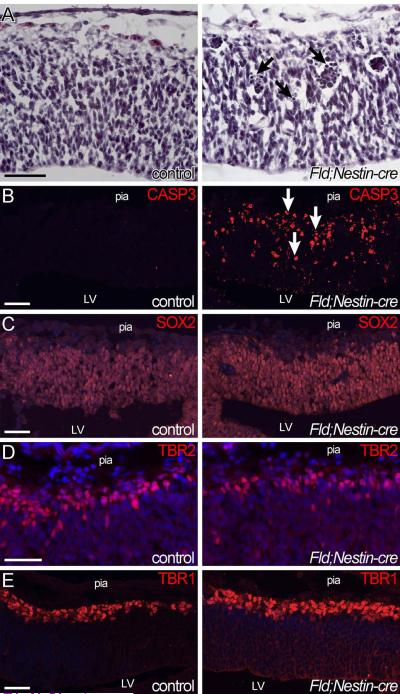
To understand the developmental origin of these behavioral abnormalities, we analyzed the brains of FID2;Nestin-Cre mice during embryogenesis. Nestin-cre is activated in the developing cortex at around E11.5; hence, we chose to analyze the brains from E12.5 onwards. At E12.5, the cortical thickness is similar between control and FID2;Nestin-cre transgenic littermates (Fig. 3A). However, pyknotic nuclei and clusters of darkly stained nuclei were present in Id2-overexpressing brains (arrows in Fig. 3A). Antibody staining with Cleaved Caspase3 clearly demonstrated dramatic increase in apoptosis in the transgenic brain, both in the proliferative zone and in the mantle region under the pia surface (Fig. 3B). Marker analyses showed equivalent density of SOX2+ NSCs in control and transgenic brains at E12.5 (Fig. 3C). Quantitation of TBR2+ intermediate progenitors showed a significant decrease in FID2;Nestin-cre brains (79%+/− 1.6% of control, p=0.0064, Fig. 3D). At the same time, the percentages of TBR1+ neurons tended to be higher in the transgenic brains, although the difference was not statistically significant (Fig. 3E). Together, these results suggest that an early effect of elevating Id2 expression level is increased apoptosis and loss of intermediate progenitors.
Figure 3. Id2 expression increases apoptosis and reduces the number of proliferating cells.
(A) H&E staining of E12.5 control and FID2;Nestin-cre littermate cortices. Arrows point to pyknotic nuclei and clusters of nuclei corresponding to dying cells (arrows). (B) Cleaved Caspase 3 staining shows dramatically increased number of apoptotic cells (arrows) in the transgenic cortex, both in the VZ and the mantle. (C) SOX2 staining shows that most cells in the VZ are NSCs in both control and transgenic cortices at this age. (D) TBR2 staining shows ~20% reduction in the number of intermediate progenitors in the transgenic cortex (p=0.0064). TBR2+ and DAPI+ nuclei were counted from same boxed area along the apical surface from 3 set of independent littermate (total # of images counted: wt: n=15, tg :n=16). To account for slight differences in developmental stage and rostrocaudal position of analyzed sections among different litters, percentages of TBR2+ cells were first normalized to littermate controls. (E) TBR1 staining for differentiating neurons do not show a significant difference at E12.5 (p=0.8781). Scale bar= 50μm
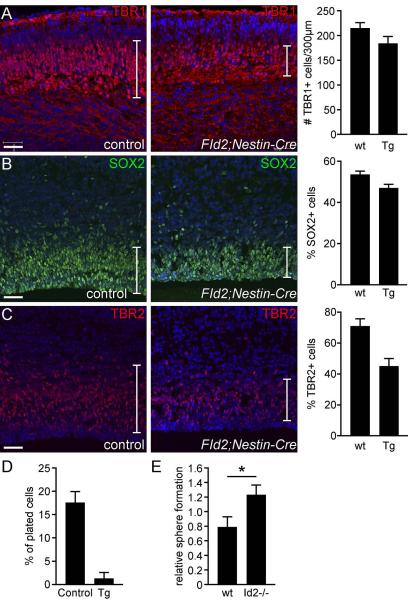
To further analyze the effect of Id2-over expression at the peak of neurogenesis, we analyzed FID2;Nestin-cre brains at E15.5. Immunofluorescence analyses with markers for differentiating neurons (TBR1) revealed a consistent decrease in the number of neurons in FID2;Nestin-Cre transgenic brains (Fig. 4A, p=0.0851). At E15.5, both SOX2+ NSCs and TBR2+ intermediate progenitor numbers were significantly decreased in transgenic brains (Fig. 4B,C, SOX2 p= 0.0179, TBR2 p=0.0141), suggesting that elevated Id2 level depletes both NSCs and progenitors.
Figure 4. Id2 expression in NSCs inhibits self-renewal.
(A) Antibody staining against markers for early-born cortical neurons (TBR1 in red) shows consistently reduced number of TBR1+ cells in the transgenic cortex at E15.5 (p-value= 0.0851). (B.C) Numbers of SOX2+ NSCs and TBR2+ progenitors are significantly reduced in the transgenic cortices at E15.5 (p=0.0179 for SOX2, and p=0.0141 for TBR2). (D) Secondary neurosphere formation assay at passage1 in vitro shows a significantly reduced number of NSCs in FID2;GFAP-Cre E15.5 cortical cultures compared to littermate controls (n>7). (E) Self-renewal assay with Id2−/− and control littermate cortices at passage1 shows significant increase in self-renewal in Id2 mutant cultures (p=0.0529). Scale bar= 50μm
These marker analyses yielded unexpected outcome, i.e., over-expression of Id2 reduces rather than increases cell numbers in the developing brain. In particular, Id2 elevation results in reduced numbers of proliferating cells. To functionally test whether NSC self-renewal is compromised in Id2-over expressing cells, we isolated NSCs from E15.5 control and transgenic littermate cortices and cultured them in vitro. Freshly established neurospheres (p0) were dissociated into single cells and plated at a clonal density at passage 1and 2 and numbers of secondary spheres were counted 7 days later. We observed a consistent and dramatic reduction in the number of secondary neurospheres that form in transgenic cultures (Fig. 4D). This confirmed the marker analysis in vivo and suggested that Id2 suppresses NSC self-renewal.
To test whether negative regulation of NSC self-renewal is a normal function of Id2, we isolated neurospheres from E15.5 Id2−/− and control littermate cortices and analyzed them for NSC self-renewal in vitro. We observed a subtle but statistically significant increase in neurosphere formation in Id2−/− cultures (Fig. 4E, p= 0.0529), complementing our findings in Id2-GOF brains. Together, these gain- and loss-of-function analyses show that Id2 negatively regulates NSC self-renewal in the developing mouse cortex.
Id2−/− mice develop grossly normal brains at birth, most likely due to functional compensation by other Id genes (not shown). However, a recent study reported that Id2−/− adults have reduced numbers of olfactory interneurons, due to increased gliogenesis at the expense of neurogenesis in the rostral migratory stream32. To test whether Id2 expression is sufficient to induce gliogenesis in vivo, we analyzed FID2;Nestin-cre brains at E15.5 with markers for gliogenesis. Antibody staining with GFAP and OLIG2 did not show any differences between the control and FID2;Nestin-cre brains (Supplementary Fig. 2), suggesting that Id2 is not sufficient to promote premature gliogenesis during embryogenesis.
NSCs and intermediate progenitors respond differently to elevated Id2 expression
While the overall phenotypes of FID2;Foxg1-cre and FID2;Nestin-cre were consistent, we were struck by the difference in phenotypic severity. To test whether this reflects cell-context dependence of Id2 function, we over-expressed Id2 in intermediate progenitor cells, using the Neurogenin1-Cre driver33, 34 (Supplementary Fig. 3A). Neurogenin1-Cre mice express the Cre recombinase in both intermediate progenitors and differentiating neurons in the cortex (arrowheads in Supplementary Fig. 3B). FID2;Neuro-cre transgenic mice were viable and had normal brain size (Supplementary Fig. 3C). In stark contrast to Id2 expression in FID2;Foxg1-Cre brains (Fig. 1), there are no observable morphological changes in FID2;Neuro-Cre brains compared to their littermates (Supplementary Fig. 3D). Marker analyses for NSCs, progenitors, and neurons showed no significant difference between the control and transgenic littermates (Supplementary Fig. 3E,F,G). In addition, there was no increased apoptosis in the FID2;Neuro-cre cortices (Supplementary Fig. 3H), suggesting that ectopic Id2 expression in committed intermediate progenitors does not induce apoptosis. Together, these data indicate that the two proliferating cell types in the developing brain, NSCs and intermediate progenitors, display differential sensitivity to misregulated Id2 expression.
Id2 over-expression results in enhanced G1-S transition but blocked G2/M transition
To better understand the cause of reduced proliferating cell numbers in FID2;Nestin-cre cortices, we tested whether cell cycle progression is compromised in Id2-over expressing cells. We analyzed cells that are in different phases of the cell cycle using two different approaches at E12.5 (one day after the onset of the transgene expression). At this age, there are very few neurons in the cortex (Fig. 3E) and the majority of the cells are NSCs and intermediate progenitors (Fig. 3C,D). To label cells in the S-phase of the cell cycle, pregnant dams were injected with BrdU for 1 hour prior to harvest. Due to interkinetic nuclear movement, S-phase cells (BrdU+) are positioned away from the apical surface while phospho-Histone 3-positive M-phase nuclei are positioned along the apical surface in the neuroepithelium 35. Immunofluorescence analysis with antibodies again BrdU show equivalent percentages of BrdU+ cells (79.53+/−6.51% of counted cells in control and 79.123+/−14.38% in transgenic); however, the percent of PH3+ cells in the transgenic cortex was consistently decreased (10.22+/−1.71% of counted cells in control and 8.487+/−1.24% in transgenic cortices, p-value= 0.3354, Fig. 5A). The ratio of PH3/BrdU cells was decreased in the transgenic cortices suggesting a compromised cell cycle progression (0.127+/−0.007 in control and 0.106+/−0.005 in transgenic, p= 0.0804).
Figure 5. Id2 activates G1 cyclins in the developing cortex.
(A) Antibody staining against BrdU (S-phase) and phosphorylated Histone 3 (PH3, M-phase). Quantification of M-phase cells in the VZ (apical PH3+ cells) shows a slight but not statistically significant (p=0.3354) decrease in FID2;Nestin-Cre brains at E12.5. The percentages of BrdU+ cells are equivalent between control and transgenic cortices. The ratio of PH3+/BrdU+ cells is reduced in the transgenic cortex (p=0.0804). (B) Cell cycle analysis using Propidium Iodide staining of acutely dissociated control and FID2;Nestin-cre cortices at E12.5 shows statistically significant increase in percentage of cells in S-phase (p=0.0269) and a significant decrease in G0/1 (p=0.028) in FID2;Nestin-Cre cortices compared to littermate controls. The percentage of M-phase cells is reduced but does not reach statistical significance in this assay (p=0.4167). (C) RT-PCR analysis of G1 Cyclins shows consistent increase in FID2;Nestin-cre brains at E12.5, compared to littermate controls. Scale bar= 50μm
To quantify the proportion of cells in different phases of the cell cycle using a different approach, we micro-dissected cortices of control and FID2;Nestin-Cre littermates at E12.5 and acutely dissociated them into single cells. These were fixed and stained with Propidium Iodide (PI) to label their DNA contents. FACS analyses of PI labeled cells showed that FID2;Nestin-cre cortices have significantly decreased percentages of cells in G0/G1 (control: 80.67+/−1.36% and transgenic: 76.33+/−1.76%, p=0.028) and an increased percentage of cells in S-phase (control:13.4+/−1.97% and FID2;Nestin-Cre: 18.47+/− 1.65, p=0.027) of the cell cycle (Fig. 5B). The percentage of M-phase cells in the transgenic brains was also reduced. Together, these results suggest that elevation of Id2 expression in the developing neuroepithelium may cause aberrant cell cycle progression.
Id2 activates G1 Cyclin expression and the p53 pathway in vivo
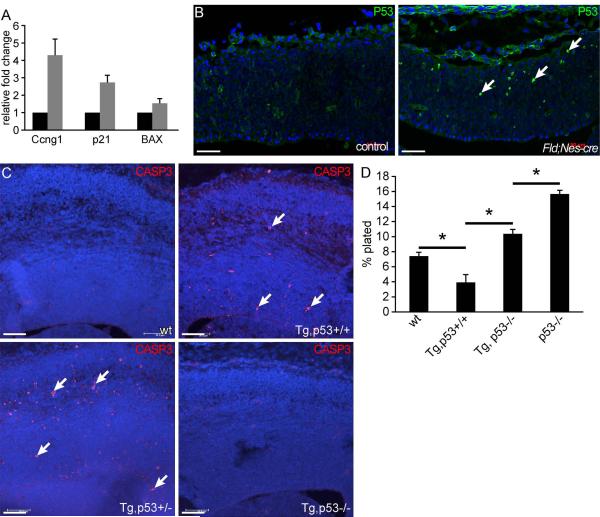
To gain better molecular understanding of these phenotypes, we examined the transcriptomes of control and FID2;Nestin-cre forebrains at E12.5. No statistically significant gene expression differences were detected (likely due to variable severity of the transgenic phenotype from embryo to embryo), although many genes showed different expression levels between control and FID2;Nestin-cre littermate brains (not shown). Transcriptome analyses of FID2;GFAP-cre cortices at E13.5 identified 17 genes that showed significant expression level differences between control and transgenic cortices (q<0.05 and > +/−1.4 fold change). Among the up-regulated genes were cell cycle regulators (CyclinD1 and p21(Cdkn1a)), CyclinG1, and p53 target genes, Pdrg1 and p21 (Supplementary Table 1). Activation of CyclinG1 and p53-target genes suggested that Id2-over expression might induce DNA damage response in neuroepithelial cells that activates the p53 pathway.
We first validated elevated expression levels of CyclinD1, CyclinG1, p21, and Pdrg1 in independent sets of control and FID2;Nestin-Cre and FID2;GFAP-cre brains using quantitative realtime RT-PCR (Fig. 5C and 6A, and Supplementary Fig. 4). Elevated CyclinD1 expression is consistent with enhanced G1-S transition we observed in FID2;Nestin-cre cortices at E12.5 (Fig. 5B). Consistently, other G1 cyclins, Cyclin D2 and E1, were also up-regulated (Fig. 5C). On the other hand, increased p21 and CyclinG1 levels (Fig. 6A) suggests activation of the DNA damage check point, which is consistent with reduced number of M-phase cells we observed in FID2;Nestin-Cre cortices at E12.5 (Fig. 5A,B). Activation of the DNA damage checkpoint also suggests that the p53 pathway may be activated in the transgenic brain. To test this idea, we analyzed the expression pattern of the p53 protein by immunohistochemistry. The p53 protein level is low in normal cells and is not detectable in the control cortex, but it was up-regulated in scattered cells in the FID2;Nestin-Cre and FID2;GFAP-cre cortices at E12.5 (Fig. 6B, and Supplementary Fig. 5). We also tested whether other p53 target genes involved in apoptosis were elevated, and observed that Bax level was also elevated in FID2;Nestin-cre brains (Fig. 6A). Together, these results suggest that increased apoptosis in Id2-GOF brains is mediated through the p53 pathway.
Figure 6. Id2 activates and modulates the p53 pathway.
(A) Realtime RT-PCR validation of selected genes from the microarray analysis that are known targets of p53 comparing control and FID2;Nestin-cre littermates at E12.5. (B) Elevated p53 protein expression in the FID2;Nestin-Cre cortex at E12.5. Arrows point to P53high cells in the transgenic cortex. (C) Cleaved Caspase 3 staining in wildtype, FID2;GFAP-cre, FID2;GFAP-cre;p53+/−, and FID2;GFAP-cre;p53−/− cortices at E15.5. Arrows point to apoptotic, cleaved Casp3+ cells. (D) Secondary neurosphere formation assay using wildtype, FID2;GFAP-cre, FID2;GFAP-cre;p53−/−, and p53−/− cells at low passages. * indicates p<0.006, error bars represent STDEV. Scale bar= 50μm
To genetically test this hypothesis, we crossed p53−/− mice to FID2;GFAP-Cre mice to generate FID2;GFAP-Cre;p53−/− embryos. As shown in Fig. 6C, loss of p53 rescued the apoptosis phenotype in FID2;GFAP-Cre brains. To test whether blocking apoptosis is sufficient to rescue reduced NSC self-renewal in FID2;GFAP-cre cortices, we isolated NSCs from wildtype, FID2;GFAP-cre, p53−/− and FID2;GFAP-cre;p53−/− cortices and tested them for secondary sphere formation at low passages (p2 and p3). p53−/− NSC cultures contained approximately twice more NSCs than littermate p53+/+ NSC cultures (Fig. 6D), as previously reported 36-38. Loss of p53 rescued the reduced self-renewal ability of FID2;GFAP-cre NSCs, restoring their level to that of wildtype cells (Fig. 6D). Interestingly, FID2;GFAP-cre;p53−/− NSCs showed significantly lower self-renewal ability than p53−/− NSCs (p=0.0003, Fig. 6D), suggesting that Id2 suppresses self-renewal of NSCs downstream of p53 and independent of NSC survival. Together, these data suggest that elevated Id2 expression activates and modulates the p53 pathway.
Id2 over-expression increases brain tumor incidence in p53−/− mice
Id2 is over-expressed in many human cancers, including brain tumors 8, 39-41. Not surprisingly, Id2 is not sufficient to transform neural cells on its own since we did not observe brain tumors in any of the FID2;Nestin-cre or FID2;GFAP-cre mice. However, since p53 is mutated in >50% of human gliomas (TCGA)42 and p53 activity is necessary to remove aberrant cells through apoptosis in Id2-GOF brains, we tested whether p53 loss combined with Id2 over-expression is sufficient to transform neural cells. We aged control and FID2;GFAP-Cre;p53−/− mice and observed them for signs of brain tumor. All FID2;GFAP-Cre;p53−/− mice died by 181 days of age (Fig. 7A). Interestingly, 40% of FID2;GFAP-Cre;p53−/− mice developed obvious neurologic symptoms of brain tumor while none of the p53−/− littermate mice developed these symptoms. Upon harvest and analysis, we confirmed that non-transgenic p53−/− mice did not have brain tumors but died from lymphomas and sarcomas, as reported previously43. In contrast, 40% (2 of 5) of the FID2;GFAP-Cre;p53−/− mice developed malignant astrocytoma and medulloblastoma (Fig. 7B,C,D,E). Analysis of these tumors by a neuropathologist indicated that the glioma in the forebrain had typical histological features of malignant astrocytoma and showed high levels of GFAP and PCNA expression (Supplementary Fig. 6). In another mouse, a tumor on the cerebellum was composed of small, EGL progenitor-like cells (Fig. 7D,E). This tumor was highly vascularized and proliferative (Fig. 7D, E, F), and many tumor cells expressed a stem cell marker OLIG2 (Fig. 7H). Very few cells within the tumor expressed GFAP (Fig. 7I), consistent with it being a medulloblastoma. Together, these observations show that elevated Id2 expression promotes brain tumor formation when combined with cooperating mutations in tumor suppressor genes such as p53.
Figure 7. Spontaneous brain tumor formation in FID2;GFAP-Cre;p53−/− mice.
(A) Kaplan-Meier survival curve for FID2;GFAP-cre;p53−/−, FID2;GFAP-Cre;p53+/−, FID2;GFAP-Cre;p53+/+, and non-transgenic p53−/− mice. (B) H&E staining of astrocytoma that formed in the hippocampus of a FID2;GFAP-cre;p53−/− mouse. (C) Higher magnification of boxed area in (B). (D) H&E staining of a medulloblastoma that formed in a FID2;GFAP-cre;p53−/− mouse. (E) A higher magnification view of tumor in (D). (F) PCNA, (G) ID2, (H) OLIG2, and (I) GFAP staining in the medulloblastoma. Scale bar= 50μm
DISCUSSION
Id2 expression is upregulated in many pathological human conditions. For example, all four Id genes are elevated significantly in the cortices of patients with Rett Syndrome7, an autism disorder characterized by microcephaly, seizures, autistic behavior, and other developmental regression and delays. Furthermore, mice with mutated Mecp2, the orthologue of the most commonly mutated gene in Rett patients, also have elevated levels of Id gene expression in the cortex7, suggesting that Id gene dysregulation may be a contributing factor to the pathology. In addition, Id genes are highly expressed in many human cancers, including gliomas and neuroblastomas40. Despite these suggestions, however, it is unknown whether elevated Id2 expression is sufficient to disrupt normal brain development and cause neurological symptoms associated with Retts or brain tumors. Results presented here demonstrate for the first time that deregulated Id2 expression can be causal in inducing some of the pathological features in these human conditions.
To elucidate the role of Id2 expression in normal brain development and the etiology of these disorders, we analyzed Id2 gain- and loss-of-function mutant mouse brains. First, we show that elevated Id2 expression is sufficient to cause aberrant brain development that recapitulates some of the features associated with Retts and brain tumors. Second, we provide evidence that Id2 activates and modulates the p53 pathway to regulate NSC self-renewal and survival. Third, we show that Id2 function is highly cell context specific, even within two different populations of proliferating cells in the developing brain. Finally, contrary to most previous studies performed in cell lines in vitro, our in vivo analysis shows that over-expression of Id2 reduces, rather increases, neural cell numbers and generates microcephalic brains.
Our results show that Id2 elevation is sufficient to disrupt normal brain development where the severity of the phenotype inversely correlated with the maturity of the target cell type. In particular, apoptosis was more apparent in younger embryos than in older embryos, suggesting that either specific cellular state of neuroepithelial cells are more sensitive to Id2 dose or that cells that survive the initial change in Id2 level learn to adapt or molecularly compensate to prevent activation of the apoptosis program. Cell-type specific activation of the Id2-GFP transgene by Foxg1-cre and Neurogenin1-cre clearly demonstrate that cellular state is a critical determinant of Id2 function. It should be noted that though even the cells that survive do not mature normally since FID2;Nestin-cre transgenic adults develop seizures. In addition, these cells are primed for cellular transformation since the incidence rate of brain tumors in FID2;GFAP-Cre;p53−/− mice is much higher than that in p53−/− mice (5%)43. Hence, this study provides the first in vivo evidence that Id2 over-expression could be an initiating event in brain tumor formation. It is well documented that activation of many oncogenes induces apoptosis or senescence 44-46, and induction of apoptosis by Id2 in neuroepithelial cells is consistent with the idea that Id2 has oncogenic activity.
Our loss- and gain-of-function analyses show that elevated Id2 expression suppresses NSC self-renewal/ proliferation and induces apoptosis. This idea is somewhat contrary to the prevalent theory that Id genes function to block differentiation and promote proliferation. Consistent with previous in vitro studies, we do observe elevated expression of G1 cyclins, including CyclinD1, in Id2-GOF brains. However, we also observe concurrent increase in CyclinG1 and other p53 target gene expression, which suggests activation of a DNA damage response and G2/M check point. The reduced percentage of PH3+ M-phase cells and increased apoptosis in Id2-GOF brains support the model that elevated Id2 expression may promote G1-S transition but that these cells incur DNA damage that blocks cell cycle progression and activate p53-mediated apoptosis. On the other hand, elevation of CyclinD1 may also explain transient increase in neurogenesis observed in the early neuroepithelium of FID2;Foxg1-cre brains (Fig. 1H). Others have reported that CyclinD1 expression can promote differentiation of neural stem/progenitor cells. For example, forced expression of CyclinD1 in chick spinal cord increased neurogenesis without affecting cell cycle progression47. Finally, it should be noted that our observation that Id2 suppresses proliferation and enhances differentiation in vivo is incongruous with previous in vitro studies that showed increased proliferation of neural precursors when Id2 is activated in vitro26, 48. Whether these apparent inconsistencies in our observations arise from use of neural precursors at different developmental stages (postnatal vs. embryonic cortices) or in vivo vs. in vitro manipulations remains to be resolved in the future. Together with other studies, our results suggest that Id2 over-expression may promote or inhibit differentiation and proliferation depending on the cellular context, providing a potential explanation for the reported oncogenic and tumor suppressive functions of Id2 in different cell types.
Several lines of evidence we present here indicate that Id2 interacts with a major tumor suppressor pathway, the p53 pathway, in neural cells. Previous studies have shown that Id2 is downstream of another tumor suppressor pathway, the RB pathway, and that Id2 modulates RB function in neural cells 49, 50. For example, Id2 deletion rescues the neural phenotype in Rb−/− embryos 11. Interestingly, germline Rb−/− embryos show increased apoptosis and misregulated cell cycle progression in the developing brain, similar to Id2-GOF embryos shown here; however, tissue-specific deletion of the Rb gene showed that increased apoptosis observed in Rb−/− cortex is due to non cell-autonomous defects51, 52. Hence, it is unlikely that Id2-induced apoptosis is mediated through inhibition of the RB pathway. On the other hand, we show that Id2 over-expression stabilizes the p53 protein, activates p53 target gene expression, and induces apoptosis. Importantly, we show genetically that deletion of p53 rescues the apoptosis and NSC self-renewal phenotype in Id2-GOF brains, providing strong evidence for Id2 interaction with the p53 pathway in vivo. Furthermore, we show that increased NSC self-renewal of p53−/− NSCs can be normalized by Id2-over expression, suggesting that Id2 also modulates the activity of p53 downstream targets that regulate NSC self-renewal. Importantly, this observation provides strong evidence that the suppression of NSC self-renewal by Id2 is not merely due to increased apoptosis in these cells.
In summary, we provide in vivo evidence that deregulated Id2 expression has dire consequences to a developing brain. We show that elevation of Id2 expression is sufficient to disrupt normal brain development, increasing susceptibility to develop seizures, especially in females. This sex-specific phenotype suggests that Id2 may play a role in regulating sex-specific differences in mammalian brains. Finally, our study clearly demonstrates the contextual effect of Id2 function, even within different populations of proliferating cells in the same tissue in vivo, a concept that needs further exploration in other stem cell systems and organs.
Supplementary Material
S Figure 1. Video-EEG recording of a control mouse. Age-matched control mouse showed no seizure activity. The traces shown are the differential signal between each electrode pair (RF - right front electrode; LF - left front; RB –right back; LB – left back; positions relative to Bregma and midline). Scale bar=1 second.
S Figure 2. Id2 is not sufficient to induce premature gliogenesis in FID2;Nestin-cre mice. Antibody staining against GFAP (green) and OLIG2 (red) show no ectopic expression in the transgenic cortex at E15.5.
S Figure 3. Id2 expression in committed neural progenitors does not change proliferation or survival. (A) A schematic for generating FID2;Neurogenin1-cre embryos. (B) Robust transgene expression, detected by antibody staining against GFP, in intermediate progenitors and differentiating neurons in the transgenic cortex at E15.5. (c) FID2;Neuro1-cre brains are not microcephalic at p20. (D) H&E staining of E15.5 control and FiD2;Neurogenin1-cre littermate cortices shows no observable morphological phenotype. (E) SOX2, (F) TBR2, and (G) TBR1 antibody staining also shows no significant differences between control and littermate transgenic embryos at E15.5. (H) Cleaved Caspase3 staining shows no change in expression, indicating that ectopic ID2 expression in progenitor cells do not induce apoptosis.
S Figure 4. Realtime RT-PCR analysis of FID2;GFAP-cre cortices at E13.5
Realtime RT-PCR validation of selected genes from the microarray analysis and other G1-S regulators and p53 targets (n=5).
S Figure 5. P53 is activated in FID2;GFAP-cre cortex at E15.5
Antibody staining against p53 shows stabilization of p53 the protein in the transgenic cortex
S Figure 6. Astrocytoma in FID2;GFAP-cre;p53−/− express markers of gliomas.
GFAP and PCNA staining confirm glioma formation in FID2;GFAP-cre;p53−/− brain shown in Figure 7B.
Acknowledgment
We thank Jesse Hammer and Stephen Sampson for assistance with manuscript preparation, Michaela Petit and Kasima Brown for technical assistance, Barbara Beyer for EEG electrode implantation and recording, Gene Expression services, Computational Sciences, and Imaging Services at JAX for their assistance with the project. We are grateful to Susan Ackerman and Robert Burgess for their comments on the manuscript. This work was supported by an NIH grant to WNF (NS031348) and TJL Cancer Center Support grant CA034196 to KY.
Footnotes
author contributions: Mingi Hong: collection of data, data analysis and interpretation
Hee Jung Park: collection of data, data analysis and interpretation
Rod Bronson: data analysis and interpretation
Mark Israel: provision of study material, manuscript writing
Wayne Frankel: collection of data, data analysis and interpretation, manuscript writing
Kyuson Yun: conception and design, financial support, collection of study material, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
References
- 1.Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 2.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 3.Kageyama R, Ohtsuka T, Hatakeyama J, et al. Roles of bHLH genes in neural stem cell differentiation. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Schuurmans C, Guillemot F. Molecular mechanisms underlying cell fate specification in the developing telencephalon. Curr Opin Neurobiol. 2002;12:26–34. doi: 10.1016/s0959-4388(02)00286-6. [DOI] [PubMed] [Google Scholar]
- 5.Yun K, Mantani A, Garel S, et al. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131:5441–5448. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- 6.Lyden D, Young AZ, Zagzag D, et al. Id1 and Id3 are required for neurogenesis, angiogenesis, and vascularization of tumor xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 7.Peddada S, Yasui DH, Lasalle JM. Inhibitors of differentiation (ID1, ID2, ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome. Hum Mol Genet. 2006;15:2003–2014. doi: 10.1093/hmg/ddl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 9.Benezra R, Davis RL, Lockshon D, et al. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 10.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 11.Lasorella A, Noseda M, Beyna M, et al. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 12.Lasorella A, Rothschild G, Yokota Y, et al. Id2 mediates tumor initiation, proliferation, and angiogenesis in Rb mutant mice. Mol Cell Biol. 2005;25:3563–3574. doi: 10.1128/MCB.25.9.3563-3574.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jen Y, Manova K, Benezra R. Each member of the Id gene family exhibits a unique expression pattern in mouse gastrulation and neurogenesis. Dev Dyn. 1997;208:92–106. doi: 10.1002/(SICI)1097-0177(199701)208:1<92::AID-AJA9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 14.Neuman T, Keen A, Zuber MX, et al. Neuronal Expression of regulatory helix-loop-helix factor Id2 gene in mouse. Developmental Biology. 1993;160:186–195. doi: 10.1006/dbio.1993.1297. [DOI] [PubMed] [Google Scholar]
- 15.Rubenstein JL, Anderson S, Shi L, et al. Genetic control of cortical regionalization and connectivity. Cereb Cortex. 1999;9:524–532. doi: 10.1093/cercor/9.6.524. [DOI] [PubMed] [Google Scholar]
- 16.Yu P, Zhang YP, Shields LB, et al. Inhibitor of DNA binding 2 promotes sensory axonal growth after SCI. Experimental neurology. 2011;231:38–44. doi: 10.1016/j.expneurol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Lasorella A, Stegmuller J, Guardavaccaro D, et al. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442:471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 18.Ying QL, Nichols J, Chambers I, et al. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 19.Nam HS, Benezra R. High levels of Id1 expression define B1 type adult neural stem cells. Cell stem Cell. 2009;5:515–526. doi: 10.1016/j.stem.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anido J, Saez-Borderias A, Gonzalez-Junca A, et al. TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 21.Jeon HM, Jin X, Lee JS, et al. Inhibitor of differentiation 4 drives brain tumor-initiating cell genesis through cyclin E and notch signaling. Genes Dev. 2008;22:2028–2033. doi: 10.1101/gad.1668708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell RG, Lasorella A, Dettin LE, et al. Id2 drives differentiation and suppresses tumor formation in the intestinal epithelium. Cancer Res. 2004;64:7220–7225. doi: 10.1158/0008-5472.CAN-04-2095. [DOI] [PubMed] [Google Scholar]
- 23.Yan W, Liu G, Scoumanne A, et al. Suppression of inhibitor of differentiation 2, a target of mutant p53, is required for gain-of-function mutations. Cancer research. 2008;68:6789–6796. doi: 10.1158/0008-5472.CAN-08-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parrinello S, Lin CQ, Murata K, et al. Id-1, ITF-2, and Id-2 comprise a network of helix-loop-helix proteins that regulate mammary epithelial cell proliferation, differentiation, and apoptosis. The Journal of biological chemistry. 2001;276:39213–39219. doi: 10.1074/jbc.M104473200. [DOI] [PubMed] [Google Scholar]
- 25.Florio M, Hernandez MC, Yang H, et al. Id2 promotes apoptosis by a novel mechanism independent of dimerization to basic helix-loop-helix factors. Mol Cell Biol. 1998;18:5435–5444. doi: 10.1128/mcb.18.9.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paolella BR, Havrda MC, Mantani A, et al. p53 directly represses Id2 to inhibit the proliferation of neural progenitor cells. Stem Cells. 2011;29:1090–1101. doi: 10.1002/stem.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frankel WN, Taylor L, Beyer B, et al. Electroconvulsive thresholds of inbred mouse strains. Genomics. 2001;74:306–312. doi: 10.1006/geno.2001.6564. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Mahaffey CL, Berube N, et al. Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS genetics. 2007;3:e124. doi: 10.1371/journal.pgen.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hébert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Developmental Biology. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- 30.Yun K, Potter S, Rubenstein JL. Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development. 2001;128:193–205. doi: 10.1242/dev.128.2.193. [DOI] [PubMed] [Google Scholar]
- 31.Englund C, Fink A, Lau C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Havrda MC, Harris BT, Mantani A, et al. Id2 is required for specification of dopaminergic neurons during adult olfactory neurogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:14074–14086. doi: 10.1523/JNEUROSCI.3188-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quinones HI, Savage TK, Battiste J, et al. Neurogenin 1 (Neurog1) expression in the ventral neural tube is mediated by a distinct enhancer and preferentially marks ventral interneuron lineages. Developmental biology. 2010;340:283–292. doi: 10.1016/j.ydbio.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim EJ, Hori K, Wyckoff A, et al. Spatiotemporal fate map of neurogenin1 (Neurog1) lineages in the mouse central nervous system. The Journal of comparative neurology. 2011;519:1355–1370. doi: 10.1002/cne.22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sauer FC. Mitosis in the neural tube. The Journal of comparative neurology. 1935;62:377–405. [Google Scholar]
- 36.Meletis K, Wirta V, Hede SM, et al. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 37.Armesilla-Diaz A, Bragado P, Del Valle I, et al. p53 regulates the self-renewal and differentiation of neural precursors. Neuroscience. 2009;158:1378–1389. doi: 10.1016/j.neuroscience.2008.10.052. [DOI] [PubMed] [Google Scholar]
- 38.Stecca B, Ruiz i Altaba A. A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. The EMBO journal. 2009;28:663–676. doi: 10.1038/emboj.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andres-Barquin PJ, Hernandez MC, Hayes TE, et al. Id genes encoding inhibitors of transcription are expressed during in vitro astrocyte differentiation and in cell lines derived from astrocytic tumors. Cancer research. 1997;57:215–220. [PubMed] [Google Scholar]
- 40.Iavarone A, Lasorella A. Id proteins in neural cancer. Cancer Lett. 2004;204:189–196. doi: 10.1016/S0304-3835(03)00455-5. [DOI] [PubMed] [Google Scholar]
- 41.Mikami S, Hirose Y, Yoshida K, et al. Predominant expression of OLIG2 over ID2 in oligodendroglial tumors. Virchows Arch. 2007;450:575–584. doi: 10.1007/s00428-007-0394-7. [DOI] [PubMed] [Google Scholar]
- 42.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harvey M, McArthur MJ, Montgomery CA, Jr., et al. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nature genetics. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 44.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 45.Bartkova J, Horejsi Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 46.Bartkova J, Rezaei N, Liontos M, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 47.Lukaszewicz AI, Anderson DJ. Cyclin D1 promotes neurogenesis in the developing spinal cord in a cell cycle-independent manner. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11632–11637. doi: 10.1073/pnas.1106230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jung S, Park RH, Kim S, et al. Id proteins facilitate self-renewal and proliferation of neural stem cells. Stem cells and development. 2010;19:831–841. doi: 10.1089/scd.2009.0093. [DOI] [PubMed] [Google Scholar]
- 49.Toma JG, El-Bizri H, Barnabe-Heider F, et al. Evidence that helix-loop-helix proteins collaborate with retinoblastoma tumor suppressor protein to regulate cortical neurogenesis. J Neurosci. 2000;20:7648–7656. doi: 10.1523/JNEUROSCI.20-20-07648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iavarone A, Garg P, Lasorelle A, et al. The helix-loop-helix protein Id2 enhances cell proliferation and binds to the retinoblastoma protein. Genes and Development. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 51.de Bruin A, Wu L, Saavedra HI, et al. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:6546–6551. doi: 10.1073/pnas.1031853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacPherson D, Sage J, Crowley D, et al. Conditional mutation of Rb causes cell cycle defects without apoptosis in the central nervous system. Mol.Cell Bio. 2003;23:1044–1053. doi: 10.1128/MCB.23.3.1044-1053.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S Figure 1. Video-EEG recording of a control mouse. Age-matched control mouse showed no seizure activity. The traces shown are the differential signal between each electrode pair (RF - right front electrode; LF - left front; RB –right back; LB – left back; positions relative to Bregma and midline). Scale bar=1 second.
S Figure 2. Id2 is not sufficient to induce premature gliogenesis in FID2;Nestin-cre mice. Antibody staining against GFAP (green) and OLIG2 (red) show no ectopic expression in the transgenic cortex at E15.5.
S Figure 3. Id2 expression in committed neural progenitors does not change proliferation or survival. (A) A schematic for generating FID2;Neurogenin1-cre embryos. (B) Robust transgene expression, detected by antibody staining against GFP, in intermediate progenitors and differentiating neurons in the transgenic cortex at E15.5. (c) FID2;Neuro1-cre brains are not microcephalic at p20. (D) H&E staining of E15.5 control and FiD2;Neurogenin1-cre littermate cortices shows no observable morphological phenotype. (E) SOX2, (F) TBR2, and (G) TBR1 antibody staining also shows no significant differences between control and littermate transgenic embryos at E15.5. (H) Cleaved Caspase3 staining shows no change in expression, indicating that ectopic ID2 expression in progenitor cells do not induce apoptosis.
S Figure 4. Realtime RT-PCR analysis of FID2;GFAP-cre cortices at E13.5
Realtime RT-PCR validation of selected genes from the microarray analysis and other G1-S regulators and p53 targets (n=5).
S Figure 5. P53 is activated in FID2;GFAP-cre cortex at E15.5
Antibody staining against p53 shows stabilization of p53 the protein in the transgenic cortex
S Figure 6. Astrocytoma in FID2;GFAP-cre;p53−/− express markers of gliomas.
GFAP and PCNA staining confirm glioma formation in FID2;GFAP-cre;p53−/− brain shown in Figure 7B.