Abstract
Background
The ancestries of genes form gene trees which do not necessarily have the same topology as the species tree due to incomplete lineage sorting. Available algorithms determining the probability of a gene tree given a species tree require exponential computational runtime.
Results
In this paper, we provide a polynomial time algorithm to calculate the probability of a ranked gene tree topology for a given species tree, where a ranked tree topology is a tree topology with the internal vertices being ordered. The probability of a gene tree topology can thus be calculated in polynomial time if the number of orderings of the internal vertices is a polynomial number. However, the complexity of calculating the probability of a gene tree topology with an exponential number of rankings for a given species tree remains unknown.
Conclusions
Polynomial algorithms for calculating ranked gene tree probabilities may become useful in developing methodology to infer species trees based on a collection of gene trees, leading to a more accurate reconstruction of ancestral species relationships.
Keywords: Incomplete lineage sorting, Coalescent history, Anomalous gene tree, Dynamic programming
Background
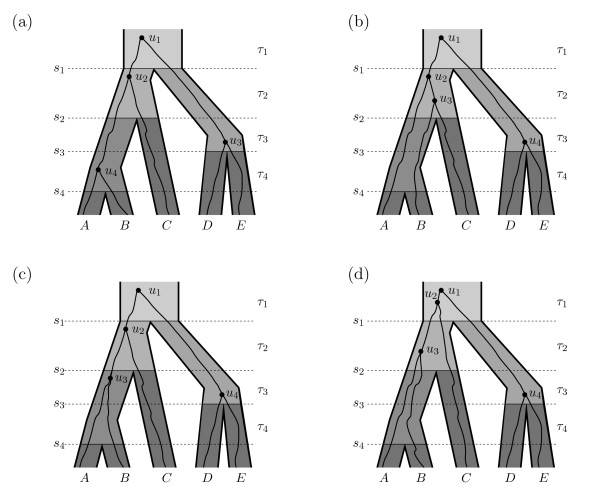
Phylogenetic reconstruction methods aim to infer the species phylogeny which gave rise to a group of extant species. Typically, this species phylogeny is obtained based on genetic data from representative individuals of each extant species. The ancestries of genes at different loci form gene trees which do not necessarily have the same topology as the species tree. Gene tree topologies and species tree topologies might be different due to such phenomena as incomplete lineage sorting, gene duplication, recombination within gene loci, and horizontal gene transfer [1]. In this paper, we focus on incomplete lineage sorting as the mechanism for incongruence of gene tree and species tree topologies, in which two gene lineages do not coalesce in the most recent population ancestral to the individuals from which the genes were sampled. As an example, the lineages sampled from species A and B in Figure 1b do not coalesce until the population ancestral to species A, B, and C, thus allowing the B and C lineages in the gene tree to have a more recent common ancestor than lineages A and B.
Figure 1.
In (a)–(d) the ranked species tree topology is(((A,B)4,C)2,(D,E)3)1. (a) The ranked gene tree matches the ranked species tree. (b) The (ranked or unranked) gene tree does not match the species tree, and there is an incomplete lineage sorting event (a deep coalescence) because the lineages from species A and B fail to coalesce more recently than s2. (c) The gene tree and species tree have the same unranked topology but have different ranked topologies, as D and E coalesce in the gene tree more recently than A and B, while A and B is the most recent divergence in the species tree. The gene tree in (c) has ranked topology (((A,B)3,C)2,(D,E)4)1. In (c), there are no incomplete lineage sorting events (no deep coalescences); however, there is an extra lineage at time s3 which leads to the gene tree and species tree having different rankings. In (c), all coalescences occur in the most recent possible interval consistent with the ranked gene tree, and we have ℓ1 = 2,ℓ2 = 3,ℓ3 = 5,ℓ4 = 5, and g1 = 2, g2 = 3, g3 = 5, g4 = 5. (d) A gene tree with the same ranked topology as the gene tree in (c) but with coalescences occurring in different intervals.
Given a fixed species tree, and assuming the gene tree evolved under the multi-species coalescent [1], the most probable gene tree topology can have a different topology from that of the species tree. Such a gene tree topology is called an anomalous gene tree. In fact, for every species tree topology with at least 5 leaves, we can choose edge lengths in the species tree topology such that anomalous gene trees exist [2]. This implies that the gene tree topology appearing most often when considering different genes might not agree with the species tree topology, thus we cannot use a simple majority-heuristic to infer the species tree from a collection of gene trees. Instead we need statistical tools rather than majority rule heuristics for inferring the species tree based on gene trees.
Current methods for inferring species trees from gene trees in this setting can be divided into topology-based and genealogy-based methods, in which the input for a reconstruction algorithm accepts either gene tree topologies or genealogies, i.e., gene trees with branch lengths (coalescence times). Topology-based methods include Minimize Deep Coalescence (MDC) [3,4], STAR [5], STELLS [6], rooted triple consensus [7] and other consensus and supertree methods [8,9]. Genealogy-based methods include Bayesian and likelihood methods such as BEST, *BEAST, and STEM [10-12] and clustering and distance-based methods [5,13-15]. Possible pros and cons of the two approaches are that topology-based methods can be computationally faster and less sensitive to errors in estimating gene trees (and gene tree branch lengths) from sequence data [16], while methods that use coalescence times, particularly using Bayesian modelling, can be the most accurate when model assumptions are correct [17].
Another possibility that has been so far unexplored in methods for inferring species trees from gene trees is to use ranked gene trees, in which the temporal order of the nodes of the gene tree (the coalescence times) is used, but not the continuous-valued branch lengths. This approach might therefore be intermediate between purely topology-based methods and genealogy-based methods. By preserving more of the temporal information in the gene tree nodes, the hope is to develop methods that are more powerful than purely topology-based methods and that are still computationally efficient and robust to errors in estimating gene trees and gene tree branch lengths from sequence data.
In [18], a first step toward developing methods that use ranked gene trees for inferring species trees was taken by providing formulae to calculate the probability of a ranked gene tree given a species tree. The previous work, however, was based on an exponential enumeration of what were called ranked coalescent histories and did not provide an algorithm for computing some of the key terms in the probability of individual ranked histories. In this paper, we improve this previous (computationally inefficient) approach, by providing a method for computing probabilities of ranked gene trees given species trees which is polynomial in the number of leaves using a dynamic programming approach.
Methods for computing probabilities of ranked gene trees efficiently may also be of interest in the context of computing probabilities of unranked gene trees, particularly because no polynomial time algorithm has been found for calculating the probability of a gene tree topology given a species tree under the multispecies coalescent [6,19-21]. The probability of an unranked gene tree topology can be obtained by summing over all ranked gene tree topologies with the same topology. Thus, for unranked gene trees with particular shapes where the number of rankings increases in polynomial time, using ranked gene trees can potentially increase the speed of computing probabilities of unranked gene trees as well. We note that a completely unbalanced gene tree has only one ranking, while the number of rankings can be exponential in the number of leaves when gene trees become more balanced. Thus, our approach for calculating unranked gene tree probabilities will be most useful for less balanced ranked gene trees.
The bulk of the paper consists of the derivation of the polynomial time method for computing ranked gene tree probabilities. The algorithm is summarized in section ‘An algorithm’. This is followed by a discussion of applications to computing probabilities of unranked gene tree topologies and to inferring ranked species trees under maximum likelihood and a modification to the MDC criterion.
Calculating the probability of a ranked gene tree topology
In the following, we will derive the probability of a ranked gene tree topology given a species tree, . Equations (1, 2, 3, 4, 8, 10) allow the calculation of in time O(n5). The model giving rise to the gene tree is the multi-species coalescent with constant population sizes [1]. Each species consists of a population of constant size where lineages merge according to the coalescent. Thus, lineages from two different species may coalesce any time previous to the split of the two species.
We begin with some notation, which is also summarized in Table 1. Let time be 0 today and increasing going into the past. Let be a species tree with n species, and thus n − 1 speciation events (denoted by 1,…,n − 1) occurring at times s1 >⋯> sn−1. Denote the interval between speciation event i − 1 and speciation event i by τi, see Figure 1.
Table 1.
Notation used in the paper
| Symbol | Meaning |
|---|---|
|
|
Species tree with real-valued divergence times |
|
|
Ranked gene tree (real-valued coalescence times not specified) |
|
n |
The number of leaves of and |
|
si |
Speciation times, with s1 >⋯> sn−1, let s0 = ∞ |
|
τi |
Intervals between speciation times, τi = [si,si−1) |
|
ℓi |
The number of gene tree lineages at time si |
|
mi |
The number of coalescence events in interval τi |
|
|
The ranked gene tree observed from time 0 to time si |
|
gi |
The minimum number of gene tree lineages at time si |
|
yi,z |
Population z in interval τi in beaded tree |
|
ui |
Internal node (coalescence) with rank i in the gene tree, u1 is most ancient, un−1 is the most recent |
|
ki,j,z |
The number of lineages available for coalescence in population yi,z just after the jth coalescence (considered forward in time) in interval τi; ki,0,z is the number of lineages “exiting” at time si−1 |
|
δ(y),δ(u) |
The set of leaves descended from a node of the species tree or gene tree, respectively |
| lca(u) |
For a node u of the gene tree, the node y of the species tree with largest rank such that δ(u) ⊂ δ(y) |
|
τ(y) |
For a node y with rank i on the species tree, we denote τ(y) = τi (the interval immediately above y) |
|
λi,j |
The overall coalescence rate in interval τi immediately preceding (backwards in time) the jth coalescence |
|
Hk |
Number of sequences of coalescences above the root of the species tree starting with k lineages |
| fi | The joint density of coalescence times in interval τi |
Let be a ranked gene tree topology. It is convenient to use the same labels for the leaves of and of . This is a slight abuse of notation, as leaf A of refers to a population (or species), and A of refers to a gene sampled from population A. We denote the nodes of (which are coalescence events) by u1,…,un−1, where node uj has rank j, and where higher rank indicates a more recent coalescence. A ranked tree topology can be notated similarly to Newick notation, putting the rank as a subscript for each node, see also Figure 1.
Let be part of a ranked gene tree evolving on a species tree between time si and time 0 (i.e. the present). consists of ℓi gene tree lineages at speciation time si and the coalescent history of in time interval (0,si) is consistent with the ranked gene tree . Let gi be the minimum number of lineages required in the ranked gene tree at time si such that can be embedded into the species tree . Note that n ≥ ℓi ≥ gi > i. Next we provide a dynamic programming approach for calculating the probability of a ranked gene tree given a species tree. An efficient way to determine the required quantities g1,…,gn−1 is provided in Section ‘Calculation of gi and ki,j,z’.
Essentially, in our approach, we traverse the intervals between speciation events going back in time, τn−1,…,τ2 (formalized in Theorem 2), and calculate the probability of the appropriate coalescent events occurring in interval τi based on how many coalescent events happened in the later intervals τi+1,…,τn−1 (Theorem 3). Finally with Theorem 1, we account for the most ancestral time interval τ1.
Theorem 1
The probability of a ranked gene tree given a species tree is,
| (1) |
where
| (2) |
is the probability for the coalescences above the root appearing in the right order[22].
For precalculated (ℓ1 = 2,…,n) the complexity of calculating is thus O(n). Next, we will provide a recursive way to calculate for ℓ1 = 2,…,n in polynomial time, thus can be calculated in polynomial time.
Theorem 2
The probabilitycan be calculated for all i recursively (with li ≥ gi),
| (3) |
with
The complexity of calculatingfor ℓ1 = 2,…,n is O(n3), given we knowfor all i,ℓi,ℓi+1.
Proof
At the time of the most recent speciation event, sn−1, we have n lineages with probability 1, which is the initial value of the recursion. Calculating for i < n − 1 can be done in the following way,
Suppose is known. Given we calculated the probability for ℓi+1 = i + 2,…,n, then calculating for ℓi = i + 1,…,n requires calculations. Summing up over i = 1,…,n − 1 yields a complexity of . □
It remains to determine . Note that during the interval τi, we have i branches in the species tree. Let mi be the number of coalescent events in τi, so mi = ℓi − ℓi−1. Let the number of lineages on branch z just after the jth coalescent event (going forward in time) in τi be ki,j,z. Calculation of ki,j,z can be done efficiently as shown in Section ‘Calculation of gi and ki,j,z’.
Theorem 3
We have,
| (4) |
where and .
Proof
The density for the coalescence events in interval τi can be obtained by considering the waiting time to the “next” coalescent event (going backwards in time) as being due to competing exponentials in the different branches, where the coalescence rate within branch z is . Thus, the waiting time until the next coalescent event has rate .
We denote the time between the jth and (j + 1)st coalescent event as vj, where v0 is the time between si−1 and the first (least recent) coalescent event in τi and with being the time between si and coalescent event mi.
The density for the coalescent events in the interval τi is [18],
It remains to integrate over v, for which we distinguish between case (i) λi,0 = 0, and case (ii) λi,0 > 0.
Case (i): If λi,0 = 0 (which occurs if ℓi−1 = i, i.e., all lineages within each population coalesce), then we rewrite fi as,
| (5) |
Using the fact that the integral of the numerator of Equation (5) is a hypoexponential distribution based on the sum of mi exponential random variables [23] (with density functions , j = 1,…,mi), the probability of the coalescent events in the interval is the cumulative distribution function of the hypoexponential distribution evaluated at . Thus, with λi,j < λi,j+1,
| (6) |
where the second line follows because −λi,j = λi,0 − λi,j.
Case (ii): If λi,0 > 0, then we rewrite fi as,
| (7) |
For integrating fi, we use the fact that the integral of the numerator in Equation (7) is the convolution of mi + 1 exponential random variables with parameters , which is the hypoexponential distribution. Now, since λi,j < λi,j+1, we observe, using the probability density function of the hypoexponential distribution,
which is the same expression as for the λi,0 = 0 case (6). Note that for case (i) we made use of the cumulative distribution function of the hypoexponential distribution, while for case (ii) we made use of the density function of the hypoexponential distribution. Both cases yield the same final expression for , which establishes the proof. □
Corollary 4
The probabilitiesfor all possible i, mi and ℓi(recall that mi = ℓi − ℓi−1) are calculated in O(n5), given all λi,j.
Proof
For a fixed i, mi and ℓi, we require calculations to evaluate . We need to determine for all possible i, mi and ℓi. First, we observe that i ≤ ℓi−1 ≤ n, and thus for a fixed ℓi, we have, 0 ≤ mi ≤ ℓi − i. Second, i < ℓi ≤ n. And third, 2 ≤ i ≤ n − 1. Thus, the number of calculations needed to calculate for all possible i, mi and ℓi is,
□
Corollary 5
The quantities λi,j can be calculated for all possible i, mi, ℓi and j in O(n5), given all ki,j,z.
Proof
For a fixed i, mi, ℓi and j, we require O(i) calculations to evaluate λi,j. As j = 0,…,mi, with the same arguments as in Corollary 4, we obtain,
□
We note that the terms are analogous to the functions gi,j defined in [24],[25], which give the probability that i lineages coalesce into j within time t in a single population and are used extensively in computing probabilities related to unranked gene trees [6],[19],[26,27]. In particular, if only one population, say z∗, has coalescence events, then we have r
a product of gi,j functions with the denominator counting the number of sequences in which mi coalescences could have occurred. The terms allow for the coalescences to occur in separate populations, however, and are constrained by the ranking of the gene tree. For example, in interval τ3 of Figure 1c, there are two coalescences which occur in different populations. If the ranking of the gene tree were not important, the branches could be considered independent, and the probability of this event would be g2,1(s2 − s3)g2,1(s2 − s3). However, the gene tree ranking constrains the coalescence of A and B to be less recent than that of D and E, so the probability for events in this interval is, r
We illustrate that we get the same result from Theorem 3: there are two coalescence events in interval τ3, so we use j = 0,1,2, and calculate
Thus, Equation (4) from Theorem 3 evaluates to
Remark 6
The probability of a gene tree topology is the sum of the probabilities of each ranked gene tree with the given topology. A given tree topology hasrankings, whereciis the number of descendant leaves of interior vertex i. A proof can be found in[28]. For a completely balanced tree on n = 2k leaves, the number of rankings grows faster than polynomial: the numerator can be approximated by,
and the denominator can be approximated by,
showing that the ratio grows faster than polynomial in n.
Calculation of gi and ki,j,z
Calculation of gi
If and have the same ranked topology, then gi = i + 1. In general, to compute gi, we let lca (uj) be the least common ancestor node on the species tree for a node uj on the ranked gene tree – i.e., the node with the largest rank on the species tree which is ancestral to all species represented in uj. For a node y on the species tree, let τ(y) be the interval immediately above y. For example, in Figure 1c, τ(lca(u4)) = τ3 where u4 is the gene tree node with rank 4 — the node ancestral to D and E only. In order to compute gi, we count the number of gene tree nodes which may occur closer to the present than si. These are precisely all gene tree nodes uj where lca (uj) is in any of the intervals τi+1,…,τn−1. Since at the present, n lineages are able to coalesce, we can express gi as,
| (8) |
where τj < τi iff j < i, and where I(·) is an indicator function taking the value 1 if the condition holds and otherwise 0. Assuming each lca() operation is O(1) [29,30], preprocessing allows all lca terms to be computed in O(n) time. Thus, calculating g1,…,gn−1 can be done, based on Equation 8, in O(n3).
Calculation of ki,j,z
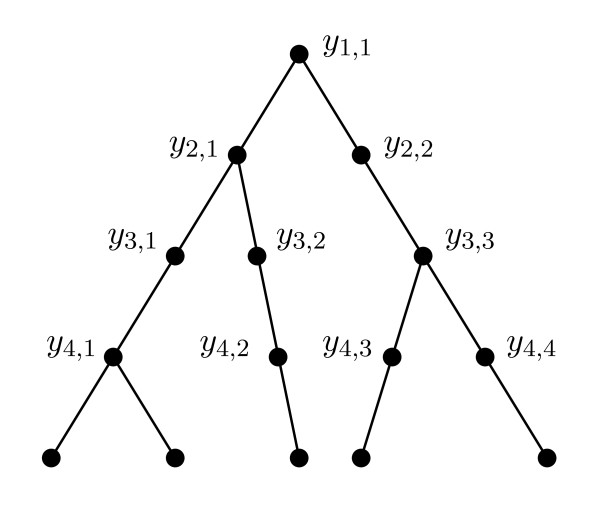
We let yi,j be the jth population (read left to right) in interval τi (equivalently, the jth branch or jth node subtending the branch). In order to label every population before and after a speciation time si uniquely, extra nodes can be added to the species tree to form a beaded species tree (Figure 2), so that there are i nodes at time si, . For each i ∈ {1,…,n−1}, there is one node of outdegree 2, and i − 1 nodes of outdegree 1. Thus, population yi,j corresponds to a branch (equivalently, a node) in the beaded species tree. We denote the outdegree of a node y by outdeg(y).
Figure 2.
The beaded version of the species tree topology in Figure1a–d.
In the remainder of this section, we compute the values ki,j,z, i.e. the number of lineages on branch yi,z of the beaded species tree during the interval immediately after the jth coalescence event (going forward in time), with ki,0,z being the number of lineages “exiting” the branch at time si−1. For example, in Figure 1b, we have
The value of ki,j,z depends on the number of lineages entering branch i, ℓi, as well as the number of lineages exiting the branch, and not just on the number of coalescence events in the interval. For example, in Figure 1c, k2,0,1 = 1 and k2,1,1 = 2, while in Figure 1d, k2,0,1 = 2 and k2,1,1 = 3, although the two gene trees have the same ranked topology and m2 = 1 for both cases.
To determine the terms ki,j,z we note that the number of coalescences that have occurred more recently than interval τi is n − ℓi. In a given interval τi, we let z(1) and z(2) be the left and right children, respectively, of population z of outdegree 2, and let z(1) = z(2) be the only child of a node z of outdegree 1.
The number of lineages available to coalesce in population z of interval τi is
| (9) |
where the z(j) are the daughter populations (one or two) of z. Further, kn,0,z = 0 for all z. Since the beaded species tree has n2/2 nodes, precalculating outdeg(yi,z) requires O(n2). For 0 ≤ j < mi, we have
| (10) |
Consequently, determining a particular ki,j,z is O(1). Thus determining ki,j,z for all possible i, mi and ℓi is (see also Corollary 4),
Note that taking the sum over all z is not necessary, as in all but one branch the ki,j,z equals the ki,j+1,z.
An algorithm
In summary, we derived an algorithm with runtime O(n5) for calculating the probability of a ranked gene tree given a species tree on n tips:
1. Calculate g1,…gn−1 using Equation (8).
2. Calculate ki,j,z (for i,j = 1,…,n;z = 1…i), using Equations (9) and (10).
3. Calculate (for i,j = 1,…,n).
4. Calculate (for i = 2,…,n; ℓi−1 = gi−1,…,n; ℓi = gi,…,n), using Theorem 3.
5. Calculate using Theorem 2.
6. Calculate using Theorem 1.
Conclusions
In this paper, we provide a polynomial-time algorithm (O(n5) where n is the number of species) to calculate the probability of a ranked gene tree topology given a species tree, summarized in Section ‘An algorithm’. We now discuss applying these results to computing probabilities of unranked gene tree topologies and to inferring ranked species trees.
Computing probabilities of unranked gene tree topologies
Previous work on computing probabilities of unranked gene tree topologies used the concept of coalescent histories, which specify the branches in the species tree in which each node of the gene tree occurs. An unranked gene tree probability can then be computed by enumerating all coalescent histories and computing the probability of each. The number of coalescent histories grows at least exponentially when the (unranked) gene tree matches the species tree, making this approach computationally intensive. Coalescent histories can be enumerated either recursively (e.g., in PHYLONET [31] or [20]) or nonrecursively (COAL [19]).
A much faster approach using dynamic programming similar to that used in this paper is implemented in STELLS [6], which conditions on the ancestral configuration in each branch rather than the number of lineages. Here an ancestral configuration keeps track not only of the number of lineages in a branch in the species tree, but also the particular nodes of the gene tree. Different ancestral configurations can potentially have the same number of lineages within a population. Enumerating ancestral configurations turns out to have exponential running time for arbitrarily shaped trees, but the number of ancestral configurations is still much smaller than the number of coalescent histories. When computing probabilities of ranked gene tree topologies, however, the ranking specifies the sequence of coalescence events, leading to a unique ancestral configuration given the number of lineages in a time interval. This fortuitously enables probabilities of ranked gene tree topologies to be computed in polynomial time.
We note that although the number of rankings for a gene tree is not polynomial in the number of leaves in general, the number of rankings can be small for certain tree shapes. For example, if the gene tree has a caterpillar shape, in which each internal node has a leaf as a descendant, then there is only one ranking, and thus computing the ranked and unranked gene tree are equivalent. For a pseudo-caterpillar, a tree made by replacing the subtree with four leaves of a caterpillar with a balanced tree on four leaves [20], there are only two rankings possible, and for a bicaterpillar[20], for which the left subtree is a caterpillar with nL leaves and the right subtree is a caterpillar with n − nL leaves, there are rankings. Thus computing unranked gene tree probabilities by summing ranked gene tree probabilities can be done in polynomial time for some tree shapes. We note that for the approach used by STELLS, some tree shapes can also be computed in polynomial time, including the cases we mentioned with a polynomial number of rankings (caterpillar and pseudo-caterpillar). An open question is whether there are any classes of unranked gene trees which have a polynomial number of rankings but an exponential number of ancestral configurations, or vice versa.
Inferring species trees from ranked gene trees
Our fast calculation of the probability of ranked gene tree topologies can be used to determine the maximum likelihood species tree from a collection of known gene trees. Assume we have observed N ranked gene trees (i.e., N loci). Now the maximum likelihood species tree (with branch lengths on internal branches) is
where
| (11) |
is a multinomial likelihood. Here can be determined with our polynomial-time algorithm, we let denote the ith ranked topology, and ni is the number of times ranked topology i is observed, with . Note in particular that the ranked topology of might differ from the most frequent ranked gene tree topology [18].
Our derivation of the ranked gene tree probability also suggests a way to infer a ranked species tree topology from ranked gene tree topologies with a similar flavor as the MDC criterion. In MDC, for an input gene tree and candidate species tree, the number of extra lineages (lineages which necessarily fail to coalesce due to topological differences between gene and species trees) on each edge of the species tree is counted. For MDC, whether the edge of the species tree is long or short does not affect the deep coalescence cost. In working with ranked gene trees, however, we can keep track of the minimum number of extra lineages within each time interval τi. The total number of extra lineages in this sense is
| (12) |
Minimizing (12) as a criterion for the ranked species tree will tend to penalize long edges of the species tree which have multiple lineages persisting through multiple species divergence events. As an example, in Figure 1b, the gene tree has a MDC cost of 1 since there are two lineages exiting the population immediately ancestral to A and B; however the cost according (12) is 2 because there are two edges on the beaded version of the species tree (Figure 2) that each have an extra lineage. In Figure 1c, the gene tree has a MDC cost of 0 for the species tree since it has the matching unranked topology; however, the number of extra lineages from equation (12) is 1. We note that in Figure 1c, interval τ3, incomplete lineage sorting (and deep coalescence) have not occurred as these concepts are normally used. To capture the idea that coalescence has nevertheless occurred in a more ancient time interval than allowed, we might refer to the coalescence of A and B in Figure 1c as an “ancient lineage sorting” event (rather than incomplete lineage sorting event) or an ancient coalescence rather than a deep coalescence. We could therefore refer to minimizing equation (12) as the Minimize Ancient Coalescence (MAC) criterion, which would provide an interesting comparison to the usual topology-based MDC criterion.
In practice, a method of inferring a species tree from ranked gene trees would require estimating the ranked gene trees. This would require clock-like gene trees, or trees with times estimated for nodes, which can also be inferred under relaxed clock models in BEAST [32]. To account for the uncertainty in the gene trees, the counts for different ranked gene trees could be weighted by their posterior probabilities obtained from Bayesian estimation of the gene trees [33]. Thus, in equation (11), we would let nik be the posterior probability of ranked topology i at locus k, and use as the estimated number of times that ranked topology i was observed. Similarly, for equation (12), the coalescence cost at a locus could be distributed over multiple topologies weighted by their posterior probabilities.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Both authors contributed equally to all parts of this work. Both authors read and approved the final manuscript.
Contributor Information
Tanja Stadler, Email: tanja.stadler@env.ethz.ch.
James H Degnan, Email: j.degnan@math.canterbury.ac.nz.
Acknowledgements
We thank David Bryant for suggesting the dynamic programming approach to this problem and two anonymous referees for valuable comments, particularly on calculating gi and ki,j,z. JHD was funded by the New Zealand Marsden fund and by a Sabbatical Fellowship at the National Institute for Mathematical and Biological Synthesis, an Institute sponsored by the National Science Foundation, the U.S. Department of Homeland Security, and the U.S. Department of Agriculture through NSF Award #EF-0832858, with additional support from The University of Tennessee, Knoxville. TS was funded by the Swiss National Science Foundation.
References
- Degnan JH, Rosenberg NA. Gene tree discordance, phylogenetic inference, and the multispecies coalescent. Trends Ecol Evol. 2009;24:332–340. doi: 10.1016/j.tree.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg NA. Discordance of species trees with their most likely gene trees. PLoS Genet. 2006;2:762–768. doi: 10.1371/journal.pgen.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Knowles LL. Inferring phylogeny despite incomplete lineage sorting. Syst Biol. 2006;55:21–30. doi: 10.1080/10635150500354928. [DOI] [PubMed] [Google Scholar]
- Than C, Nakhleh L. Species tree inference by minimizing deep coalescences. PLoS Comput Biol. 2009;5:e1000501. doi: 10.1371/journal.pcbi.1000501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yu L, Pearl DK, Edwards SV. Estimating species phylogenies using coalescence times among sequences. Syst Biol. 2009;58:468–477. doi: 10.1093/sysbio/syp031. [DOI] [PubMed] [Google Scholar]
- Wu Y. Coalescent-based species tree inference from gene tree topologies under incomplete lineage sorting by maximum likelihood. Evolution. 2011,. [DOI] [PubMed]
- Ewing GB, Ebersberger I, Schmidt HA, von Haeseler A. Rooted triple consensus and anomalous gene trees. BMC Evol Biol. 2008;8:118. doi: 10.1186/1471-2148-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan JH, DeGiorgio M, Bryant D, Rosenberg NA. Properties of consensus methods for inferring species trees from gene trees. Syst Biol. 2009;58:35–54. doi: 10.1093/sysbio/syp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Degnan JH. Performance of matrix representation with parsimony for inferring species from gene trees. Stat Appl Genet Mol Biol. 2011;10:21. [Google Scholar]
- Heled J, Drummond AJ. Bayesian inference of species trees from multilocus data. Mol Biol Evol. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatko LS, Carstens BC, Knowles LL. STEM: Species tree estimation using maximum likelihood for gene trees under coalescence. Bioinformatics. 2009;25:971–973. doi: 10.1093/bioinformatics/btp079. [DOI] [PubMed] [Google Scholar]
- Liu L, Pearl DK. Species trees from gene trees: Reconstructing bayesian posterior distributions of a species phylogeny using estimated gene tree distributions. Syst Biol. 2007;56:504–514. doi: 10.1080/10635150701429982. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L. Estimating species trees from unrooted gene trees. Syst Biol. 2011;60:661–667. doi: 10.1093/sysbio/syr027. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Pearl DK. Maximum tree: a consistent estimator of the species tree. J Math Biol. 2010;60:95–106. doi: 10.1007/s00285-009-0260-0. [DOI] [PubMed] [Google Scholar]
- Mossel E, Roch S. Incomplete lineage sorting: consistent phylogeny estimation from multiple loci. IEEE/ACM Trans Comp Biol Bioinf. 2010;7:166–171. doi: 10.1109/TCBB.2008.66. [DOI] [PubMed] [Google Scholar]
- Huang H, He Q, Kubatko LS, Knowles LL. Sources of error for species-tree estimation: Impact of mutational and coalescent effects on accuracy and implications for choosing among different methods. Syst Biol. 2009;59:573–583. doi: 10.1093/sysbio/syq047. [DOI] [PubMed] [Google Scholar]
- Liu L, Yu L, Kubatko LS, Pearl DK, Edwards SV. Coalescent methods for estimating phylogenetic trees. Mol Phylogenet Evol. 2009;53:320–328. doi: 10.1016/j.ympev.2009.05.033. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Rosenberg N, Stadler T. The probability distribution of ranked gene trees on a species tree. Math Biosci. 2012;235:45–55. doi: 10.1016/j.mbs.2011.10.006. [DOI] [PubMed] [Google Scholar]
- Degnan JH, Salter LA. Gene tree distributions under the coalescent process. Evolution. 2005;59:24–37. [PubMed] [Google Scholar]
- Rosenberg NA. Counting coalescent histories. J Comput Biol. 2007;14:360–377. doi: 10.1089/cmb.2006.0109. [DOI] [PubMed] [Google Scholar]
- Than C, Ruths D, Innan H, Nakhleh L. Confounding factors in HGT detection: statistical error, coalescent effects, and multiple solutions. J Comput Biol. 2007;14:517–535. doi: 10.1089/cmb.2007.A010. [DOI] [PubMed] [Google Scholar]
- Edwards AWF. Estimation of the branch points of a branching diffusion process. J R Stat Soc Ser B. 1970;32:155–174. [Google Scholar]
- Ross S. Introduction to Probability Models. San Diego: Academic Press; 2007. [Google Scholar]
- Tavaré S. Line-of-descent and genealogical processes, and their applications in population genetics models. Theor Popul Biol. 1984;26:119–164. doi: 10.1016/0040-5809(84)90027-3. [DOI] [PubMed] [Google Scholar]
- Wakeley J. Coalescent Theory. Greenwood Village: Roberts & Company; 2008. [Google Scholar]
- Pamilo P, Nei M. Relationships between gene trees and species trees. Mol Biol Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. The probability of topological concordance of gene trees and species trees. Theor Pop Biol. 2002;61:225–247. doi: 10.1006/tpbi.2001.1568. [DOI] [PubMed] [Google Scholar]
- Semple C, Steel M. Phylogenetics, vol. 24 of Oxford Lecture Series in Mathematics and its Applications. Oxford: Oxford University Press; 2003. [Google Scholar]
- Harel D, Tarjan RE. Fast algorithms for finding nearest common ancestors. SIAM J Comput. 1984;13:338–355. doi: 10.1137/0213024. [DOI] [Google Scholar]
- Schiever B, Vishkin U. On finding lowest common ancestors: simplification and parallelization. SIAM J Comput. 1988;17:1253–1262. doi: 10.1137/0217079. [DOI] [Google Scholar]
- Than C, Ruths D, Nakhleh L. Phylonet: A software package for analyzing and reconstructing reticulate evolutionary relationships. BMC Bioinformatics. 2008;9:322. doi: 10.1186/1471-2105-9-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evolut Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman ES, Degnan JH, Rhodes JA. Identifying the rooted species tree from the distribution of unrooted gene trees under the coalescent. J Math Biol. 2011;62:833–862. doi: 10.1007/s00285-010-0355-7. [DOI] [PubMed] [Google Scholar]