Abstract
Over the two past decades, a significant number of studies have observed animal growth traits to examine animal genetic mechanisms due to their ease of measurement and high heritability. Chicken which has a significant impact on fundamental biology is a major source of protein worldwide, making it an ideal model for examining animal growth trait development. The genetic mechanisms of chicken growth traits have been studied using quantitative trait loci mapping through genome-scan and candidate gene approaches, genome-wide association studies (GWAS), comparative genomic strategies, microRNA (miRNA) regulation of growth development analysis, and epigenomic analysis. This review focuses on chicken GWAS and miRNA regulation of growth traits. Several recently published GWAS reports showed that most genome-wide significant single nucleotide polymorphisms are located on chromosomes 1 and 4 in chickens. Chicken growth, particularly skeletal muscle growth and development, is greatly regulated by miRNA. Using dwarf and normal chickens, let-7b was found to be involved in determining chicken dwarf phenotypes by regulating growth hormone receptor gene expression.
Keywords: Chicken, Genome-wide association study (GWAS), Growth traits, microRNA (miRNA) regulation, Quantitative trait loci (QTL), Single nucleotide polymorphisms (SNPs).
1. INTRODUCTION
The growth and development of livestock is a complicated life process and is subject to a significant amount of genetic control. Several studies have examined “hotspot” areas, including candidate gene mutation, effects of quantitative trait loci (QTL), genome-wide association studies (GWAS), microRNA (miRNA) regulation, and epigenetic inheritance. Gene expression patterns can be used to explain phenotype variations and to illustrate epigenetic mechanisms such as consensus methylation-rich promoters that correspond to down-regulation of gene expression and transgenerational stability of epigenetic variants that simultaneously appeared in mammals, birds, and plants [1-3]. Focusing on growth traits and miRNA regulation, this review discusses three main areas related to chicken growth.
First, growth trait-correlated mutations in the genes of the somatotropic axis (Table 1) will be discussed before focusing on GWAS and miRNA regulation of growth traits. A number of single nucleotide polymorphisms (SNPs) located within genes encoding for growth hormone (GH) and its receptor (GHR), ghrelin (GHRL) and its receptor (GHSR), insulin-like growth factor 1 (IGF-1) and its receptor (IGF-1R), insulin (INS), insulin-like growth factor binding protein2 (IGFBP2), adipose triglyceride lipase (ATGL), and pituitary-specific transcription factor 1 (PIT1) have been associated with chicken growth traits [4-17]. Moreover, SNPs of the high mobility group AT-hook 2 gene (HMGA2), which is located on chicken (Gallus gallus) chromosome (GGA) 1, should be noted for their significant associations with body weight. Common insertion-deletion (indel) variations in growth-related genes have also been shown to act on growth diversity. The 1,773 bp deletion located at exon 10 and the 3′ untranslated region (UTR) of the GHR gene resulted in a dysfunctional precursor, leading to sex-linked dwarf chickens (SLD). One 57-bp indel in intron 2 of the PIT1 gene was reported to be significantly associated with hatch weight and shank length at 84 d of age [8]. Another 6-bp indel of the GHSR gene was found to be related to crude fatty content of the leg muscle, although no evidence for its association with growth traits was identified [7]. An 8-bp indel in exon1 of the GHRL gene may have a negative effect on chicken growth [13]. Some SNPs are likely fixed and must transmit in a high frequency format through haplotypes such as strong linkage disequilibrium. Using haplotypes and linkage disequilibrium, important SNPs and QTL were identified. For instance, no indication of an association between either SNP A17299834G or SNP C17293932T and the IGF-IR gene was observed for growth traits; however, haplotypes could be constructed from these two SNPs, which were shown to be significantly associated with body weights, daily weight gains, and leg length [9]. Additionally, SNP C6540334T and SNP C6542011T of the GHR [10] gene and SNP A+428G of the INS gene [14] exhibit a similar pattern.
Table 1.
Significant Associations of the SNPs Within Candidate Genes with their Growth Traits (P<0.01)
| Gene | Chr | SNPa | Growth Traitsb | Ref. |
|---|---|---|---|---|
| GHRL | 7 | C71T | BW: 7 wk, 16 wk; | 11 |
| SG: 7 wk, 16 wk; SL: 16 wk | ||||
| G1215A | BW: 16 wk | 13 | ||
| 8bp indel (exon) | BW: 2 wk, 3 wk, 5 wk, 6 wk, 7 wk, 13 wk; BL | |||
| INS | 5 | T+3737C (intron) | SIL | 14 |
| A+3971G (intron) | BW: 4 wk; HW; BA; SIL | |||
| C+1549T (intron) | BW: 4 wk, 12 wk; BD | |||
| GH | 1 | G+119A (intron) | SIL | 15 |
| G+1705A (intron) | BW: 3 wk, 4 wk, 10 wk, 12 wk; ADG: 0-4 wk; | |||
| G+3037T (intron) | SL: 10 wk, 12 wk | |||
| IGF1R | 10 | A17299834G (5’UTR) | LL: 8 wk | 9 |
| GHR | Z | G6631778A (3’UTR) | HW; BW: 5 wk, 6 wk, 7 wk | 10 |
| PIT1 | 1 | 57 bp indel (intron) | HW; SL: 12 wk | 8 |
| rs13687127 (intron) | SD: 11 wk | |||
| rs13687128 (exon) | BW: 4 wk; ADG: 0-4 wk | |||
| ATGL | 5 | c.782G>A (exon) | BW: 6 wk, 7 wk, 8 wk | 6 |
| IGFBP2 | 7 | G729T (intron) | BW: 7 wk | 16 |
| C1032T (intron) | HW; BW: 2 wk | |||
| A663T (exon) | BW:1 wk, 2 wk, 3 wk, 5 wk, 6 wk, 7 wk; HW | |||
| G738A (exon) | BW:1 wk, 2 wk, 3 wk, 4 wk, 5 wk, 6 wk, 7 wk, 8 wk, 13 wk; HW | |||
| RB1 | 1 | g.39692 G>A | BW: 6 wk, 11 wk | 24 |
| g.77260 A>G | BW: 5 wk, 6 wk, 7 wk, 8 wk, 9 wk, 10 wk, 11 wk, 12 wk | |||
| HMGA2 | 1 | rs15231472 (intron) | BW:6 wk | 5 |
| rs13849381 (intron) | BW: 1 wk, 2 wk, 3 wk, 5 wk, 6 wk; HW | |||
Names of SNPs are cited from original papers.
BW: body weight; ADG: average daily gain; SL: shank length; BL: body length; LL: leg length; SD: shank diameter; HW: hatch weight; SIL: length of small intestine; BA: breast angle; BD: breast depth; SG: shank girth.
In addition to candidate gene approaches, identifying QTL underlying traits is a useful strategy. Continuous improvement in molecular genetics technology and DNA marker availability, identifying QTL controlling growth traits for marker-assisted selection applications has rapidly increased [18, 19]. More QTL have been identified on GGA1 and GGA4 than on other chromosomes. Among growth traits correlated with known QTL, body weight accounts for 30% (207), growth rate for 6% (40), and abdominal fat weight for 5% (35), while other traits constitute the remaining traits (http://www.animal genome.org/cgi-bin/QTL db/GG/index). For GGA4, most reported QTL are growth-related. Although several QTL have been successfully identified, most are orientated in dispersive regions of distinct chromosomes. They typically span a large range. For example, QTL affects abdominal fat weight on GGA1, which ranges from 73.6 to 215.4 cM [20] and 454 to 276 cM [21]. This makes applying these results to various commercial lines infeasible because imprecise mapping. Increasing marker density or population size under tightly restricted conditions is used for trouble-shooting. In a study by Liu et al., 10 QTL were identified at the 1% chromosome-wide level for body weight from 4 to 12 weeks [22]. Shortly after, the authors refined the previously reported QTL interval by increasing nine novel microsatellites between LEI0079 and ROS0025 and enlarging family sizes from 4 to 12 and F2 individual numbers from 369 to 1011, leading to more precise QTL identification for these two traits across this region [23]. For fine mapping of this QTL, 14 additional SNPs close to ADL328 were examined and the most likely QTL for body weight was orientated between SNP G173196313A and SNP T173539900C over a 400-kb interval [24]. Finally, two SNPs (g.39692 G>A and g.77260 A>G) of the retinoblastoma 1 (RB1) gene were confirmed to be related to growth. Using high-resolution QTL is effective for identifying quantitative traits near genes.
2. GWAS ON CHICKEN GROWTH TRAITS
The genome-wide association study, which shows a remarkable success rate, can be used to detect sequence variations influencing growth traits. GWAS on chicken growth traits showed that most significant SNPs were centered near GGA1 and GGA4 [25]. Growth traits including aggregate body weight at 0–90 d of age measured weekly, biweekly average daily gains derived from weekly body weight, and breast muscle weight, leg muscle weight, and wing weight at 90 d of age were identified in a 1.5-Mb region (173.5–175 Mb) of chicken GGA1, which was a narrow range compared to other SNPs evaluated. Of effective SNPs, five SNPs in the 1.5-Mb karyopherin alpha 3 (KPNA3)-forkhead box O1 (FOXO1A) regions showed the highest significant effects for all growth traits, while two miRNA genes putatively targeting mRNA of IGF-1, FOXO1A, and KPNA3 genes were contained. Another GWAS study showed that most significant SNPs associated with late growth during weeks 7–12 were present in an 8.6-Mb region (71.6–80.2 Mb) of chicken GGA4, whereas only one SNP on GGA18 was significantly associated with early growth (body weight at 2 w of age). This may be due to that more genetic variance during early growth is caused by epistatic interactions rather than by single point effect [26, 27]. According to the results of artificial selection revealed in another study, a region (60–80 Mb on GGA4) may have been under intense recent selection in divergent chicken lines, where 50 generations of selection showed 9-fold differences in body weights. Similarly on GGA1, several regions respond highly to selection [28]. This implies that these two confined regions are important for understanding the molecular basis of growth traits. Although a lack of conformity between the two GWAS studies was caused by breed diversity (different materials), previously reported QTL offer powerful evidence supporting that these two regions and chicken growth are related (http://www. animalgenome.org/cgi-bin/QTLdb/GG/index). Because the described DNA variants identified through GWAS may influence growth traits, subsequent experiments should be performed [29, 30]. Of the 15 validated SNPs in the region from 173.5 to 175Mb, 12 SNPs with P<0.0033 were significant between fast-growing and slow-growing groups, indicating that these SNPs play a role in a causal mechanism or in coupling linkage phase with causal mutations [25].
The most promising genes harboring growth-related SNPs, including the FOXO1A gene, in which two highly significant SNPs at 8.9 Kb upstream and 1.9 Kb downstream were identified, may contribute to myogenic growth and differentiation. FOXO1A expression is notably up-regulated in energy-deprived skeletal muscle. Transgenic mice overexpressing Foxo1a in skeletal muscle weighed less than wild-type mice and showed reduced skeletal muscle mass [31]. The integrator complex subunit 6 (INTS6) gene, containing the most significant SNP effecting aggregate body weight at 90 d of age, may act as a tumor suppressor gene by inhibiting growth of prostate cancer by altering the cell cycle profile and Wnt signaling, resulting in down-regulated expression in human multiple prostate cells [32]. The LIM domain-binding factor 2 (LDB2)gene, in which SNPs important for late growth are located, can bind to several transcription factors and drive vascular maturation [33].
SNPs in GGA1 and GGA4, which are reliably predicted using powerful statistical methods, can be used to understand chicken growth. Phenotypic differences should not only be ascribed to genetic differences or epigenetic changes as a result of sequence mutations, but also to miRNAs which affect this intricate phenomenon.
3. MICRORNA REGULATION OF CHICKEN GROWTH
3.1. MicroRNA Regulation of Biological Processes
MicroRNAs are key elements in several cell processes, including proliferation, differentiation, and apoptosis. They are a class of small non-coding RNAs approximately 22 nucleotides in length and are generally expressed in different tissues, regulating expression of specific genes at the post-transcriptional level by targeting mRNAs for degradation or inhibiting translation [34]. During miRNA formation, miRNA genes are initially transcribed as long primary transcripts (pre-miRNAs) by RNaseII or III. Mature miRNAs are generated after processing by Dicer, an RNase III endonuclease, and subsequently incorporate into the RNA-induced silencing complex (RISC) [35]. Mature miRNA always binds to the 3' UTR of target mRNAs, which mainly induces translational inhibition and exonucleolytic mRNA decay; targets can also be cleaved endonucleolytically as a result of highly complementary base-pairing [36]. Each miRNA has a specific structure and formation mechanism. For example, subtle sequence variations exist within the gga-miR-1 and gga-miR-133 genes, but none exist for gga-miR-206 (http://www.mirbase.org/). Gga-miR-1a and gga-miR-1c differ in their 3′ ends and middle sequences by one nucleotide, while gga-miR-133a and gga-miR-133c differ in their 3′ end sequences by only one nucleotide. These sequence polymorphisms result in differential cleavage of the same miRNA precursor to produce a larger number of miRNA target sites. Gga-miR-1a-1 and miR-1a-2 share identical mature sequences but are located on different chromosomes. Many mRNAs can be targets of one miRNA, and different combinations of miRNAs may coordinate to regulate specific target genes [37]. For example, miR-1 and miR-206 have four mutual targets: histone deacetylase 4 (HDAC4) [38, 39], connexin43 (Cx43) [40], paired box 7 (Pax7 ) [41], and cMet [42]. Additionally, miR-206 can control myocyte differentiation by inhibiting fibroblast suppressors such as follistatin-1 (Fstl1), utrophin (Utrn) [43], and a subunit of DNA polymerase alpha (Pola1) [44], indicating its extensive role the complicated gene network.
Some miRNAs are expressed ubiquitously in different tissues, while some are expressed in a tissue-specific manner; miR-1, miR-133, and miR-206 belong to the latter category. These muscle-specific miRNAs have been demonstrated to regulate skeletal development in different manners [38, 45]and were dramatically up-regulated during myoblast differentiation [39, 46]. Up-regulation of miR-1 is primarily due to accumulation of myocyte enhancer factor-2 (MEF2) resulting from decreased expression of HDAC4; interestingly, this confined expression of HDAC4 is caused by high miR-1 expression. Thus, when MEF2 expression increases accompanied by up-regulation of miR-1, HDAC4 activity is repressed, resulting in further expression of MEF2 and control of myocyte differentiation.
3.2. Identification and Characterization of miRNAs Affecting Chicken Growth
Previous studies have been focused on identifying and characterizing a large number of miRNAs associated with embryo development [47, 48], skeletal muscle development [49], lipogenesis and cell proliferation [50], lung and trachea with avian influenza virus infection [51], and embryo fibroblasts infected with Marek's disease [52]. In the absence of differentially expressed miRNAs, gene expression would be dominated by monotony at the post-transcriptional and translational levels, and the resulting message would be unreadable and meaningless. Differential miRNAs found at different time points in the developing embryo and in discriminating skeletal muscle are of significant concern with regard to chicken growth. Generally, skeletal muscle goes through two stages of myoblast proliferation and differentiation during the embryonic phase and postnatal muscle fiber hypertrophy. During the embryonic phase, the appearance of somites represents muscle development. Progenitor cells derived from somites generate myoblasts, which differentiate into mature muscle fibers via proliferation, migration, and fusion [53]. Muscle growth after birth is accomplished primarily through resizing of muscle fibers resulting from satellite cell fusion to existing fibers, rather than changing their numbers, as the final number of muscle fibers is determined at the end of embryogenesis [54].
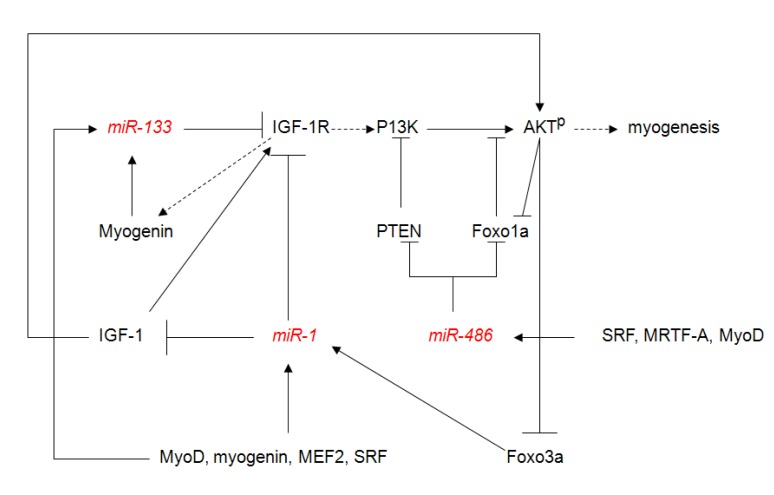
As described above, miR-1 and miR-133 are involved in skeletal muscle growth and development. A bicistronic gene cluster encoding miR-1 and miR-133a, and another encoding miR-133b and miR-206 are transcribed from non-coding regions on mouse chromosomes 2 and 1, respectively. Transcription of these miRNAs is mainly achieved by myogenic transcription factors, such as myogenic differentiation 1 (MyoD), serum response factor (SRF), and MEF2 by acting through cis-regulatory elements [55-57]. For instance, transcription regulation of miR-1/133/206 can be implemented by mammalian target of rapamycin (mTOR) signaling in a Myo-dependent manner. With the aid of MyoD, which lies downstream of mTOR, transcription of miR-1 is triggered and in turn degrades HDAC4, a follistatin suppressor, affecting myocyte fusion. Similarly, inhibiting mTOR by rapamycin elicits a sharp decrease in miR-133 and miR-206 [58]. Moreover, miR-1 and miR-133 can regulate myogenesis via phosphatidylinositol 3-kinase/v-AKT murine thymoma viral oncogene (Akt) (PI3K/Akt) signaling Fig. (1), which at least partially participates in muscle growth and hypertrophy [59]. Through IGF-1 stimulation in mouse neonatal cardiomyocytes (CMCs) and during C2C12 skeletal muscle cell differentiation, binding of IGF-1 and IGF-1R results in activation of the PI3K/Akt pathways via PI3K-dependent phosphorylation and activation of Akt [60], stimulating hypertrophic growth and glucose uptake. In turn, active Akt phosphorylates and depresses transcription factor forkhead box O3 (Foxo3a), a negative regulator of protein synthesis and muscle growth which regulates miR-1 promoter activity; the declining miR-1 reciprocally regulates IGF-1 and IGF-1R expression since they are targets of miR-1. This inverse correlation between IGF-1 protein levels and miR-1 is also found in myocardial biopsies of acromegalic patients [61]. Moreover, inverse regulation of miR-133 and IGF-1R was observed during C2C12 skeletal muscle cell differentiation [62]. Expression of miR-133 is up-regulated in response to increased expression of myogenin, a myogenic transcription factor which transactivates miR-133 in the presence of exogenous IGF-1. Overexpression of miR-133 has important implications on the IGF-1R/PI3K/Akt signaling pathway due to its ability to block IGF-1R expression and sequentially decrease regulation of Akt phosphorylation, miR-1 overexpression gives rise to pronounced down-regulation of phospho-Akt supported by a generalized decrease in the IGF-1 signal transduction pathway. Another muscle-enriched miRNA, miR-486, reportedly indirectly and positively inhibits activation of PI3K/Akt signaling in rat CMCs through translation repression of two crucial negative regulators, phosphatase and tensin homolog (PTEN) and Foxo1a [63]. If activated by MyoD and myocardin-related transcription factor-A (MRTF-A), accumulation of miR-486 will rise and activate PI3K, target PTEN, or activate the phosphorylation of Akt by targeting Foxo1a to activate pathway. Active Akt in reverse inhibits Foxo1a activity. Thus, miR-1 and miR-206 play important roles in promoting differentiation. In contrast, miR-133 overexpression inhibits myoblast differentiation, but promotes myoblast proliferation by targeting SRF, a regulator of miR-133 transcription [39].
Fig. (1).
MicroRNAs regulating myogenesis via PI3K/Akt signaling. This schematic diagram is a reference for [60-63]. The dash arrows indicates the favorable pathway has not been well clarified. IGF-1: insulin-like growth factor 1; IGF-1R: insulin-like growth factor 1 receptor; AKTP: phospho-AKT; Foxo1a: forkhead box O1; Foxo3a: forkhead box O3; MEF2: myocyte enhancer factor-2; MRTF-A: myocardin-related transcription factor-A; SRF: serum response factor; PTEN: phosphatase and tensin homolog; PI3K: phosphoinositide-3-kinase; MyoD: myogenic differentiation 1.
3.3. MicroRNAs and Embryonic Development
Dynamic sets of miRNAs have been demonstrated to express, which influence embryogenesis and organogenesis in chicks at 11 days of incubation [48]. By constructing and sequencing a small RNA library, a many known miRNAs (approximately 38% of the embryonic chick small RNA library), along with small regions of homologous miRNAs with other species and novel miRNAs, were identified. Identified miRNAs such as miR-143, miR-214, miR-363, miR-10a, and miR-22 show a significant degree of conservation compared to mouse, gorilla, bovine, and human. At least 27 existing miRNA clusters were discovered in the chicken genome, and nearly all are conserved within vertebrate species, suggesting that miRNAs both have an evolutionally ancient origin and share similar roles among species [64]. Novel miRNAs represented 1% of identified miRNAs, while miR-125b accounted for 11% of the total library [48]. The most abundant miR-125b is significant. Most recently, miR-125b was implicated in myoblast differentiation in vitro and muscle regeneration in vivo. Expression of IGF-2 can be directly regulated by enhanced mTOR signaling at the transcriptional level [65]. A recent study complemented miR-125b to correlate IGF-2 and mTOR [66], i.e., expression of miR-125b is negatively controlled by mTOR signaling in an mTOR kinase-independent manner. When mTOR signaling is enhanced, miR-125b is reduced, and the inhibitory effect of miR-125b on IGF-2 is weakened, leading to high IGF-2 expression. IGF-2 is an autocrine factor that initiates myoblast differentiation in vitro [67]; its high expression is imperative for injury-induced muscle regeneration [68].
In situ hybridization of chicken embryos during early stages of incubation revealed that miR-1, which facilitates differentiation of mesodermal progenitors to the muscle lineage [69], was detected in the somatic myotome beginning at stage 14 [70], which corresponds to the onset of skeletal muscle cell differentiation. In contrast, miR-133a was detected in the myocardium and myotome at stage 15, coincident with rapid progress in limb development, and miR-206 was detected in somites at stage 20, which is characterized by limb-bud enlargement [71]. Furthermore, restricted expression of miR-206 steadily increased over 1.5 to 5 d of incubation and was exclusive in developing somites, particularly in the developing myotome. An opposite trend in expression was observed following fibroblast growth factor (FGF) bead implantation due to FGF-mediated signaling, which negatively regulated the onset of miR-206 expression [72]. FGF signals can appoint some sets of progenitor cells in the ventral somite to ribs and tendons by acting through the mitogen-activated protein kinase (MAPK) signal transduction cascade [73]. Temporal expression of specific miRNA is necessary for organ development; therefore, growth and development of the embryo can effectively occur. However, during embryogenesis and organogenesis, formation of the somite is a key step.
Somites are short-lived mesodermal structures for which ventral somite cells engender the sclerotome containing progenitor cells for cartilage and bone, and dorsal somite cells form the dermomyotome containing progenitor cells for skeletal muscle. This acts as a visible symbol for the segmented nature of the vertebrate body plan [74]. Individual vertebrae are derived from the somite, which is generated following sequential segmentation of a region of the presomatic mesoderm. Because few novel miRNAs were identified in somites using traditional sequencing, Solexa sequencing was used to globally explore novel miRNAs involving chicken somites [75]. Conventional sequencing of cDNA is a cumbersome process and cannot easily detect miRNAs present in relatively low abundance; deep sequencing or massively parallel signature sequencing (MPSS) can be used to overcome these limitations [76]. Rathjen et al. obtained 651,273 reads from dissected somite tissue from 3-, 4-, and 5-day old embryos and identified 42 new miRNA candidates [75]. Eighteen were confirmed using Northern blot analysis. Notably, the novel miR-10a was confirmed with a high number of reads (28,660), and miR-10b was the most abundant (113,106) among known miRNAs, implying that these two miRNAs are related during somite development. Effects caused by miRNAs on somite development and embryonic morphology have been reported previously, such as those caused by miR-196 [77]. miR-196 acts upstream of the homeobox (Hox) gene to define the boundary of Hox gene expression along the anterior-posterior embryonic axis. A central function of the spatio-temporal expression of Hox gene in embryonic development is to pattern of the vertebrate axial skeleton. Unilaterally localized injection of antagomiR196 into the presomatic mesoderm in ova leads to vertebral transformations at the cervical-thoracic boundary, and thus high-frequency ectopia of the last cervical vertebra (c14) occurs as decreased miR-196 expression gives expands the limit of homeoboxB8 (Hoxb8) transcripts in more anterior somites. Intriguingly, miR-10 has been shown decreases expression of several Hox genes in zebrafish [78]. Whether this data agrees with the mechanism of regulation of Hox targets with miR-196 requires further investigation since little evidence for its role in embryogenesis has been found.
Additionally, Glazov et al. found that the total number of let-7 family reads increased significantly during embryonic stages on days 5, 7, and 9, particularly let-7b, with maximum reads on day 9 [47]. After identification in C. elegans, let-7 miRNAs have been shown to play vital roles in mediating cell proliferation and differentiation. As a lung tumor suppressor in humans, overexpression of let-7 results in inverse expression of RAS, a potential oncogene, and inhibits lung tumor cell growth [79]. It was reported to exert a negative effect on cell number and positive effect on the fraction of cells in the G2/M cell cycle phase following peripheral introduction of let-7 in primary fibroblasts by targeting and down-regulating the cell division cycle 34 (Cdc34) gene, indicating its crucial influence on cell cycle control [80]. Introduction of let-7 family members in mouse embryonic stem cells (ESCs) can suppress continuous self-renewal resulting from a lack of DiGeorge syndrome critical region gene 8 (Dgcr8), which enables silencing of this program. Moreover, inhibition of let-7b boosts de-differentiation of somatic cells into induced pluripotent stem (iPS) cells [81]. Overexpression of let-7b elevates mouse neural stem cell differentiation but reduces proliferation by targeting the stem cell regulator nuclear receptor subfamily 2, group E, member 1 (Nr2e1) and the cell cycle regulator cyclin D1[82]. This information underscores its specific role in stabilizing the self-renewing program and differentiating stem cells. Two miRNAs, miR-1623 and miR-181b, are differentially expressed in 14-day-old embryos of diverse growth-rate dwarf and normal chickens [83].
In summary, miRNAs, such as miR-101,play an indispensable role in establishing certain organs by localizing or fast shifting expression in appropriate timing with embryonic development. miR-101 may monitor gonadal development to determine the gender of a chicken [84]. Expression increases between 5.5 d and 9.5 d of the embryonic phase in both sexes, with higher expression in males at E5.5 and E7.5, but a significant increase in females is observed at E9.5. It is likely that the high abundance of miR-101 after gonadal differentiation in females is crucial for determining the nature of ovarian cells due to its inhibitory effect on SRY (sex determining region Y)-box 9 (SOX9), a key component of testes differentiation. Additionally, upon suppression of some signaling inhibitory factors, transforming growth factor beta/ Anti-Müllerian hormone (TGF-β/AMH) signaling is reinforced to regulate follicle activation. In males, miR-101 may be responsible for testes formation by fine tuning SOX9. As a sensitive indicator, spatio-temporal expression of miRNAs reflects the accurate developmental conditions of some organs.
3.4. MicroRNAs and Growth of Skeletal Muscle
Wang et al. identified 32 known miRNAs from skeletal muscle of Arbor Acres commercial chickens, of which 12 form five clusters: miR-133a-1-miR-1a-2, miR-23b-miR-24, miR-99a-let-7c, miR-92-miR-19b-miR-18a-miR-17, and miR-30e-miR-30c-1, suggesting that most miRNAs co-express in skeletal muscle [85]. Because of the complexity of muscle development based upon spatiotemporal networks regulating mRNA transcription and translation, miRNA expression during eight different developmental stages (from the birth to 7 w of age) was determined by using quantitative RT-PCR. Both miR-133a and miR-1a were more important during later stages of development than during early stages of development since expression of both increased from 14 to 49 d and decreased from 0 to 14 d.
Divergent muscle growth rate is the most common characteristic in commercial chicken lines, particularly in broilers and layers, normal, and SLD chickens. Broilers are raised for meat while layers are used for egg production, which is a consequence of the higher growth rate and larger muscle mass as well as bigger body size of broilers compared to layers. Both exterior selection pressure and intrinsic genetic patterns collectively elicit lager batteries of differences between these two breeds. For SLD chickens, aberrant muscle development attributed to a defective GHR gene show typical features of dwarf, serious weight loss, and decreased number of muscle fibers and fiber diameter.
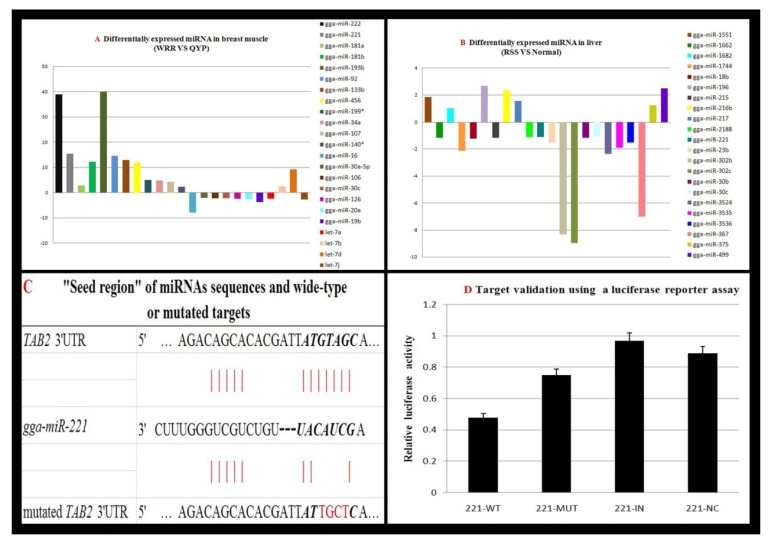
A recent preliminary study examined the miRNA transcriptome to identify differentially expressed miRNAs involved in skeletal muscle growth between broilers and layers [49]. A total of 33 novel chicken miRNAs were obtained, and 102 miRNAs showed significant differences between broilers and layers. To confirm the accuracy of these results, 17 of these miRNAs were randomly validated using microarray analysis as well as real-time RT-PCR. Expression patterns of 15 miRNAs identified using RT-PCR agreed with those identified using deep sequencing, miR-101, miR-10a, miR-10b, miR-1677, let-7f, and miR-31 were higher in layers, while miR-200b, let-7c, miR-16c, miR15b, miR-15c, miR460, miR-429, miR-2188, and the novel miR-N2 were higher in broilers. Of note, miR-206 was the most abundant miRNA in both broilers and layers (131,609 reads and 222,998 reads, respectively), but significant differences in expression were observed. Expression of miR-133 was lower than that of miR-206 and showed significant differences, miR-1a expression was high in both libraries and showed significant differences in expression as well. Based on this information, the corresponding mRNA transcriptome was investigated and 57 candidate targets for 16 miRNAs were identified. Additionally, the final integrated network was constructed to depict miRNA-protein interactions and protein-protein interactions. Although three miRNAs, miR-1, miR-101, and miR-499, were predicted to target the activin A receptor type IIB (ACVR2B) gene, validation was only performed for miR-1 and ACVR2B. We examined miRNA expression in fast-growing White Recessive Rock chickens in contrast to slow-growing Qingyuan Partridge chickens at 7 w of age. We identified 23 differentially expressed miRNAs in the breast muscle of slow-growing chickens compared to in fast-growing chicken Fig. (2A). Wnt and insulin signaling pathways were two main pathways according to pathway analysis (data not published).
Fig. (2).
Differentially expressed miRNAs in chickens with different growth performance and target validation. A: Expression pattern of 23 miRNAs in breast muscles of White Recessive Rock (WRR) and Qingyuan Partridge chickens (QYP) based on microarray experiments. The bars represent the fold change in WRR/QYP. B: Relative abundance of 22 miRNAs in the livers of normal and RSS chickens based on Solexa sequencing data. The bars represent the fold change of RSS/normal. C: Schema of gga-miR-221 binding sites in chicken wide-type and mutated TAB2 (TGF-beta activated kinase 1/MAP3K7 binding protein 2) 3'-UTR sequence. Seed binding sites and mutated bases are highlighted in red. Target prediction was performed using TargetScan 5.0 software (http://targetscan.org/). D: To verify whether gga-miR-221 targets TAB2, a luciferase reporter assay was conducted. A gga-miR-221 overexpression plasmid was constructed using pcDNA3.1 (Promega, Fitchburg, WI, USA). Wild-type (WT) or mutated TAB2 3'-UTR sequence (MUT) was cloned into the pmirGLO luciferase reporter vector at downstream of Renilla luciferase, the internal Firefly luciferase gene was used to normalize for transfection efficiency. DF-1 cells were used for transfection, and 600 ng of the gga-miR-221 overexpression plasmid was co-transfected with 200 ng of WT or MUT reporter plasmid (221-WT/-MUT). A parallel inhibition or negative control experiment was conducted using30 ng IN or NC vector plasmid cotransfected with 200 ng of WT reporter plasmid (221-IN/-NC). Dual-luciferase assays were conducted in triplicate in 24-well plates. Cells were lysed and assayed for luciferase activity 36h after transfection. Error bars represent the standard error of three independent experiments per group.
The sex-linked dwarfism chicken is characterized by its relatively small body size, lower feed intake, lower basal metabolism, reduced IGF-1 levels, and high concentrations of GH in the plasma compared to in normal chickens. Previous studies suggested that mutations in the GHR gene may result in phenotypic variation of SLD [86-88]. These mutations induced either different transcripts in terms of transcription level or chaotic GHR proteins in terms of translation level. Comparison of mRNA expression for GH, GHR, and IGF-1 genes at 56 d of age in the livers of sex-linked dwarf chickens with normal chickens showed that GH expression between these chickens showed equally high transcription levels. However, expression of GHR in dwarf chickens was significantly higher than that in normal chickens, and very little IGF-1 gene expression was observed in dwarf chickens, further illustrating that the GH-independent dwarfism is due to dysfunctional GHR protein in accordance with a mutant GHR gene [89]. Notably, GHR-deficient mice showed reductions in myofiber number, coupling with defective muscle skeletal development due to diminished myoblast fusion [90]. Based upon our previous study, miRNA regulation, which implicates GHR as well, may be important in dwarfism formation [83]. A significant difference in expression of let-7b in skeletal muscle was observed between SLD chickens due to the identical mutant type of GHR with Connecticut SLD [88] and normal chickens at 7 w of age. Comparison of differential expression profiles of miRNA and mRNA in SLD and normal chickens showed that, in addition to let-7b, mRNA expression of GHR genes increased SLD compared to in normal chicken. Because aberrant GHR elicits the dwarfism phenotype, it is important in the functional mechanism that how GHR acts on skeletal muscle in SLD in response to let-7b. The relationship between let-7b and GHR has been confirmed through a dual reporter assay and overexpression experiment in vitro. As expected, the complementary sequence of let-7b to normal GHR matched the deleted region of mutant GHR, indicating that let-7b suppresses expression of GHR in normal chickens but not in SLD chickens. The primary pathway in which GHR participates is for the janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling pathway. Interestingly, mRNA expression of suppressor of cytokine signaling 3 (SOCS3) was markedly up-regulated in SLD in contrast to other genes involved in this pathway. Thus, tight control of let-7b may block expression of SOCS3 in a non-mutant GHR-dependent manner.
SOCS3 is a suppressor of cytokine signaling which blocks the JAK/STAT pathway relying on binding to janus kinase and to specific cytokine receptors, which affects cell proliferation, differentiation, apoptosis, and immunoregulation [91]. SOCS3 also blocks insulin signaling by targeting insulin receptor substrate 1 (IRS1) and insulin receptor substrate 2 (IRS2), two critical signaling molecules for insulin action, and forces their ubiquitination and degradation, contributing to insulin resistance [92]. Leptin typically influences the feeding and neuroendocrine function depending on binding to and consequently activating its receptor (LEPR). For SLD, the indirect, acute up-regulation of SOCS3 impairs the effect of leptin by inhibiting LEPR production. Thus, leptin suppression increases feeding and glucocorticoid production, but decreases energy expenditure of reproductive, growth, and metabolic rate. SLD chickens eat less than normal chickens, perhaps for because of their smaller somatotypes or due to unknown regulators compensating for the effect induced by SOCS3. However, the data provide insight into the regulatory mechanism.
Notably, disease in chicken populations can also give rise to a dramatic drop in skeletal muscle mass and result in runting and stunting syndrome (RSS). This is one of the most common syndromes causing serious growth obstruction, and chickens with this syndrome consume large amounts of feed, show poor growth performance, and can be easily affected by a disease. Thus far, its aetiology remains unclear. To identify the extensive role of miRNAs in balancing this complicated pathological mechanism, Solexa sequencing was performed to determine differential expression profiles of 22 miRNAs in the livers of normal and RSS chickens at 7 w of age Fig. (2B). Subsequently, we found that TGF-beta activated kinase 1/MAP3K7 binding protein 2 (TAB2) was target of gga-miR-221 using a luciferase reporter gene assay Fig. (2C and 2D). Interestingly, miR-221 affected skeletal muscle cell growth and disease. It was down-regulated during myogenic differentiation, with assistance from the RAS-mitogen-activated protein kinase (MAPK) pathway. Ectopic expression of miR-221 in differentiated myoblasts extended the cell cycle and delayed myogenin expression, which had a suppressive role on cell cycle inhibitor p27 [93]. In chronic myeloid leukemia (CML) patients, miR-221was significantly up-regulated during the blast crisis phase and was predicted to target v-crk sarcoma virus CT10 oncogene homolog (avian)-like (CRKL) in the MAPK signaling and phosphoinositide-3-kinase, regulatory subunit 1 (PIK3R1) in epidermal growth factor receptor (EGFR) signaling associated with CML [94]. The roles of these two miRNAs and their targets underlying RSS must be further examined. In general, miRNAs are thought to be involved in regulating multiple genes regardless of consistently expressing or differentially expressing in different skeletal muscle by targeting and inhibiting their mRNAs. Thus, it is necessary to examine the function and characteristics of each miRNA.
4. CONCLUSION AND PROSPECTS
Various studies have been conducted to identify connections between genotype and phenotype. Chicken growth is complex and orchestrated through precise control of multiple genes. Association analysis using candidate gene and genome scan using linkage analysis are two primary means of detecting QTL related to growth, but this method exhibits some limitations. Because candidate genes are limited when using candidate gene approaches and QTL mapping is imprecise due to insufficient marker density when using linkage analysis, major genes leading to phenotype differences can not be determined. With the development of biotechnology and bioinformatic approaches, GWAS can be used to identify genes using tens of thousands of SNP markers at the whole-genome level. GGA1 and GGA4 are two critical chromosomes affecting chicken growth, particularly body weight. As integral members of gene networks, miRNAs explain the complexity of life. Many growth-related miRNAs have been discovered, including miR-1, miR-133, miR-206, miR-101, and let-7b, the biochemical roles of which have been demonstrated through experimental validation. Indepth knowledge of miRNAs will increase the understanding of the animals’ growth molecular mechanisms.
ACKNOWLEDGEMENTS
This work was supported by the China Agriculture Research System (CARS-42-G05), China High-Tech Programs (2011AA100301), and Natural Scientific Foundation of China (31172200).
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Natt D, Rubin CJ, Wright D, Johnsson M, Belteky J, Andersson L, Jensen P. Heritable genome-wide variation of gene expression and promoter methylation between wild and domesticated chickens. BMC Genomics. 2012;13(59 ) doi: 10.1186/1471-2164-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Franklin TB, Mansuy IM. Epigenetic inheritance in mammals: evidence for the impact of adverse environmental effects. Neurobiol. Dis. 2010;39:61–65. doi: 10.1016/j.nbd.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 3.Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- 4.Sato S, Ohtake T, Uemoto Y, Okumura Y, Kobayashi E. Polymorphism of insulin-like growth factor 1 gene is associated with breast muscle yields in chickens. Anim Sci J . 2012;83:1–6. doi: 10.1111/j.1740-0929.2011.00917.x. [DOI] [PubMed] [Google Scholar]
- 5.Song C, Gu X, Feng C, Wang Y, Gao Y, Hu X, Li N. Evaluation of SNPs in the chicken HMGA2 gene as markers for body weight gain. Anim. Genet. 2011;42:333–336. doi: 10.1111/j.1365-2052.2010.02141.x. [DOI] [PubMed] [Google Scholar]
- 6.Nie QH, Fang MX, Xie L, Shen X, Liu J, Luo ZP, Shi JJ, Zhang XQ. Associations of ATGL gene polymorphisms with chicken growth and fat traits. J Appl Genet. 2010;51:185–191. doi: 10.1007/BF03195726. [DOI] [PubMed] [Google Scholar]
- 7.Fang M, Nie Q, Luo C, Zhang D, Zhang X. Associations of GHSR gene polymorphisms with chicken growth and carcass traits. Mol. Biol. Rep. 2010;37:423–428. doi: 10.1007/s11033-009-9556-9. [DOI] [PubMed] [Google Scholar]
- 8.Nie Q, Fang M, Xie L, Zhou M, Liang Z, Luo Z, Wang G, Bi W, Liang C, Zhang W, Zhang X. The PIT1 gene polymorphisms were associated with chicken growth traits. BMC Genet. 2008;9:20. doi: 10.1186/1471-2156-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei M, Peng X, Zhou M, Luo C, Nie Q, Zhang X. Polymorphisms of the IGF1R gene and their genetic effects on chicken early growth and carcass traits. BMC Genet. 2008;9:70. doi: 10.1186/1471-2156-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouyang JH, Xie L, Nie Q, Luo C, Liang Y, Zeng H, Zhang X. Single nucleotide polymorphism (SNP) at the GHR gene and its associations with chicken growth and fat deposition traits. Br Poult Sci. 2008;49:87–95. doi: 10.1080/00071660801938817. [DOI] [PubMed] [Google Scholar]
- 11.Li CC, Li K, Li J, Mo DL, Xu RF, Chen GH, Qiangba YZ, Ji SL, Tang XH, Fan B, Zhu MJ, Xiong TA, Guan X, Liu B. Polymorphism of ghrelin gene in twelve Chinese indigenous chicken breeds and its relationship with chicken growth traits. Asian-Aust. J. Anim. 2006:153–159. [Google Scholar]
- 12.Lei M, Luo C, Peng X, Fang M, Nie Q, Zhang D, Yang G, Zhang X. Polymorphism of growth-correlated genes associated with fatness and muscle fiber traits in chickens. Poult Sci. 2007;86:835–842. doi: 10.1093/ps/86.5.835. [DOI] [PubMed] [Google Scholar]
- 13.Fang M, Nie Q, Luo C, Zhang D, Zhang X. An 8bp indel in exon 1 of Ghrelin gene associated with chicken growth. Domest Anim Endocrinol. 2007;32:216–225. doi: 10.1016/j.domaniend.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Qiu FF, Nie QH, Luo CL, Zhang DX, Lin SM, Zhang XQ. Association of single nucleotide polymorphisms of the insulin gene with chicken early growth and fat deposition. Poult Sci. 2006;85:980–985. doi: 10.1093/ps/85.6.980. [DOI] [PubMed] [Google Scholar]
- 15.Nie Q, Sun B, Zhang D, Luo C, Ishag NA, Lei M, Yang G, Zhang X. High diversity of the chicken growth hormone gene and effects on growth and carcass traits. J. Hered. 2005;96:698–703. doi: 10.1093/jhered/esi114. [DOI] [PubMed] [Google Scholar]
- 16.Lei MM, Nie QH, Peng X, Zhang DX, Zhang XQ. Single nucleotide polymorphisms of the chicken insulin-like factor binding protein 2 gene associated with chicken growth and carcass traits. Poult Sci. 2005;84:1191–1198. doi: 10.1093/ps/84.8.1191. [DOI] [PubMed] [Google Scholar]
- 17.Amills M, Jimenez N, Villalba D, Tor M, Molina E, Cubilo D, Marcos C, Francesch A, Sanchez A, Estany J. Identification of three single nucleotide polymorphisms in the chicken insulin-like growth factor 1 and 2 genes and their associations with growth and feeding traits. Poult Sci. 2003;82:1485–1493. doi: 10.1093/ps/82.10.1485. [DOI] [PubMed] [Google Scholar]
- 18.Ankra-Badu GA, Shriner D, Le Bihan-Duval E, Mignon-Grasteau S, Pitel F, Beaumont C, Duclos MJ, Simon J, Porter TE, Vignal A, Cogburn LA, Allison DB, Yi N, Aggrey SE. Mapping main, epistatic and sex-specific QTL for body composition in a chicken population divergently selected for low or high growth rate. BMC Genomics. 2010;11:107. doi: 10.1186/1471-2164-11-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao Y, Shen X, Xia M, Luo C, Nie Q, Zhang D, Zhang X. SNP mapping of QTL affecting growth and fatness on chicken GGA1. Genet. Sel. Evol. 2007;39:569–582. doi: 10.1186/1297-9686-39-5-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jennen DG, Vereijken AL, Bovenhuis H, Crooijmans RM, van der Poel JJ, Groenen MA. Confirmation of quantitative trait loci affecting fatness in chickens. Genet. Sel. Evol. 2005;37:215–228. doi: 10.1186/1297-9686-37-3-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uemoto Y, Sato S, Odawara S, Nokata H, Oyamada Y, Taguchi Y, Yanai S, Sasaki O, Takahashi H, Nirasawa K, Kobayashi E. Genetic mapping of quantitative trait loci affecting growth and carcass traits in F2 intercross chickens. Poult Sci. 2009;88:477–482. doi: 10.3382/ps.2008-00296. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Li H, Wang S, Hu X, Gao Y, Wang Q, Li N, Wang Y, Zhang H. Mapping quantitative trait loci affecting body weight and abdominal fat weight on chicken chromosome one. Poult Sci. 2007;86:1084–1089. doi: 10.1093/ps/86.6.1084. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Zhang H, Li H, Li N, Zhang Y, Zhang Q, Wang S, Wang Q, Wang H. Fine-mapping quantitative trait loci for body weight and abdominal fat traits: effects of marker density and sample size. Poult Sci. 2008;87:1314–1319. doi: 10.3382/ps.2007-00512. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Liu SH, Zhang Q, Zhang YD, Wang SZ, Wang QG, Wang YX, Tang ZQ, Li H. Fine-mapping of quantitative trait loci for body weight and bone traits and positional cloning of the RB1 gene in chicken. J. Anim. Breed. Genet. 2011;128:366–375. doi: 10.1111/j.1439-0388.2011.00927.x. [DOI] [PubMed] [Google Scholar]
- 25.Xie L, Luo C, Zhang C, Zhang R, Tang J, Nie Q, Ma L, Hu X, Li N, Da Y, Zhang X. Genome-wide association study identified a narrow chromosome 1 region associated with chicken growth traits. PLoS One. 2012;7:e30910. doi: 10.1371/journal.pone.0030910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlborg O, Kerje S, Schutz K, Jacobsson L, Jensen P, Andersson L. A global search reveals epistatic interaction between QTL for early growth in the chicken. Genome Res. 2003;13:413–421. doi: 10.1101/gr.528003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu X, Feng C, Ma L, Song C, Wang Y, Da Y, Li H, Chen K, Ye S, Ge C, Hu X, Li N. Genome-wide association study of body weight in chicken F2 resource population. PLoS One. 2011;6:e21872. doi: 10.1371/journal.pone.0021872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson AM, Pettersson ME, Siegel PB, Carlborg O. Genome-wide effects of long-term divergent selection. PLoS Genet. 2010;6:e1001188. doi: 10.1371/journal.pgen.1001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 30.Zeggini E, McCarthy MI. Identifying susceptibility variants for type 2 diabetes. Methods Mol Biol. 2007;376:235–250. doi: 10.1007/978-1-59745-389-9_16. [DOI] [PubMed] [Google Scholar]
- 31.Kamei Y, Miura S, Suzuki M, Kai Y, Mizukami J, Taniguchi T, Mochida K, Hata T, Matsuda J, Aburatani H, Nishino I, Ezaki O. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes and impaired glycemic control. J. Biol. Chem. 2004;279:41114–41123. doi: 10.1074/jbc.M400674200. [DOI] [PubMed] [Google Scholar]
- 32.Filleur S, Hirsch J, Wille A, Schon M, Sell C, Shearer MH, Nelius T, Wieland I. INTS6/DICE1 inhibits growth of human androgen-independent prostate cancer cells by altering the cell cycle profile and Wnt signaling. Cancer Cell Int. 2009;9:28. doi: 10.1186/1475-2867-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Javerzat S, Franco M, Herbert J, Platonova N, Peille AL, Pantesco V, De Vos J, Assou S, Bicknell R, Bikfalvi A, Hagedorn M. Correlating global gene regulation to angiogenesis in the developing chick extra-embryonic vascular system. PLoS One. 2009;4:e7856. doi: 10.1371/journal.pone.0007856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.le Sage C, Agami R. Immense promises for tiny molecules uncovering miRNA functions. Cell Cycle. 2006;5:1415–1421. doi: 10.4161/cc.5.13.2890. [DOI] [PubMed] [Google Scholar]
- 35.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 36.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 37.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, Da PI, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 38.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr. Opin. Cell Biol. 2009;21:461–469. doi: 10.1016/j.ceb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson C, Catoe H, Werner R. MIR-206 regulates con-nexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34:5863–5871. doi: 10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao X, Wang DZ. microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 2010;190:867–879. doi: 10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan D, Dong XE, Chen X, Wang L, Lu C, Wang J, Qu J, Tu L. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J. Biol. Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, Tapscott SJ. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J. Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen JF, Callis TE, Wang DZ. microRNAs and muscle disorders. J. Cell Sci. 2009;122:13–20. doi: 10.1242/jcs.041723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J. Biol. Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 47.Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hicks JA, Tembhurne P, Liu HC. MicroRNA expression in chicken embryos. Poult Sci. 2008;87:2335–2343. doi: 10.3382/ps.2008-00114. [DOI] [PubMed] [Google Scholar]
- 49.Li T, Wu R, Zhang Y, Zhu D. A systematic analysis of the skeletal muscle miRNA transcriptome of chicken varieties with divergent skeletal muscle growth identifies novel miRNAs and differentially expressed miRNAs. BMC Genomics. 2011;12:186. doi: 10.1186/1471-2164-12-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hicks JA, Trakooljul N, Liu HC. Discovery of chicken microRNAs associated with lipogenesis and cell proliferation. Physiol. Genomics. 2010;41:185–193. doi: 10.1152/physiolgenomics.00156.2009. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, Brahmakshatriya V, Zhu H, Lupiani B, Reddy SM, Yoon BJ, Gunaratne PH, Kim JH, Chen R, Wang J, Zhou H. Identification of differentially expressed miRNAs in chicken lung and trachea with avian influenza virus infection by a deep sequencing approach. BMC Genomics. 2009;10:512. doi: 10.1186/1471-2164-10-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burnside J, Ouyang M, Anderson A, Bernberg E, Lu C, Meyers BC, Green PJ, Markis M, Isaacs G, Huang E, Morgan RW. Deep sequencing of chicken microRNAs. BMC Genomics. 2008;9:185. doi: 10.1186/1471-2164-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle from somite to limb. J. Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moss FP, Leblond CP. Satellite cells as the source of nuclei in muscles of growing rats. Anat Rec. 1971;170:421–435. doi: 10.1002/ar.1091700405. [DOI] [PubMed] [Google Scholar]
- 55.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 58.Sun Y, Ge Y, Drnevich J, Zhao Y, Band M, Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J. Cell Biol. 2010;189:1157–1169. doi: 10.1083/jcb.200912093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- 60.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 61.Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, Russo MA, Cimino V, De Marinis L, Frustaci A, Catalucci D, Condorelli G. Reciprocal regulation of microRNA-1 and insulin-like growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation . 2009;120:2377–2385. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang MB, Xu H, Xie SJ, Zhou H, Qu LH. Insulin-like growth factor-1 receptor is regulated by microRNA-133 during skeletal myogenesis. PLoS One. 2011;6:e29173. doi: 10.1371/journal.pone.0029173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Small EM, O'Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, Richardson JA, Olson EN. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci U S A. 2010;107:4218–4223. doi: 10.1073/pnas.1000300107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shao P, Zhou H, Xiao ZD, He JH, Huang MB, Chen YQ, Qu LH. Identification of novel chicken microRNAs and analysis of their genomic organization. Gene . 2008;418:34–40. doi: 10.1016/j.gene.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 65.Erbay E, Park IH, Nuzzi PD, Schoenherr CJ, Chen J. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J. Cell Biol. 2003;163:931–936. doi: 10.1083/jcb.200307158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ge Y, Sun Y, Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol. 2011;192:69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Florini JR, Magri KA, Ewton DZ, James PL, Grindstaff K, Rotwein PS. "Spontaneous" differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J. Biol. Chem. 1991;266:15917–15923. [PubMed] [Google Scholar]
- 68.Paoni NF, Peale F, Wang F, Errett-Baroncini C, Steinmetz H, Toy K, Bai W, Williams PM, Bunting S, Gerritsen ME, Powell-Braxton L. Time course of skeletal muscle repair and gene expression following acute hind limb ischemia in mice. Physiol. Genomics. 2002;11:263–272. doi: 10.1152/physiolgenomics.00110.2002. [DOI] [PubMed] [Google Scholar]
- 69.Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell . 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 71.Darnell DK, Kaur S, Stanislaw S, Konieczka JH, Yatskievych TA, Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- 72.Sweetman D, Rathjen T, Jefferson M, Wheeler G, Smith TG, Wheeler GN, Munsterberg A, Dalmay T. FGF-4 signaling is involved in mir-206 expression in developing somites of chicken embryos. Dev Dyn. 2006;235:2185–2191. doi: 10.1002/dvdy.20881. [DOI] [PubMed] [Google Scholar]
- 73.Smith TG, Sweetman D, Patterson M, Keyse SM, Munsterberg A. Feedback interactions between MKP3 and ERK MAP kinase control scleraxis expression and the specification of rib progenitors in the developing chick somite. Development. 2005;132:1305–1314. doi: 10.1242/dev.01699. [DOI] [PubMed] [Google Scholar]
- 74.Aulehla A, Herrmann BG. Segmentation in vertebrates clock and gradient finally joined. Genes Dev. 2004;18:2060–2067. doi: 10.1101/gad.1217404. [DOI] [PubMed] [Google Scholar]
- 75.Rathjen T, Pais H, Sweetman D, Moulton V, Munsterberg A, Dalmay T. High throughput sequencing of microRNAs in chicken somites. Febs Lett. 2009;583:1422–1426. doi: 10.1016/j.febslet.2009.03.048. [DOI] [PubMed] [Google Scholar]
- 76.Moxon S, Jing R, Szittya G, Schwach F, Rusholme PR, Moulton V, Dalmay T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18:1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGlinn E, Yekta S, Mansfield JH, Soutschek J, Bartel DP, Tabin CJ. In ovo application of antagomiRs indicates a role for miR-196 in patterning the chick axial skeleton through Hox gene regulation. Proc Natl Acad Sci U S A. 2009;106:18610–18615. doi: 10.1073/pnas.0910374106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS One. 2008;3:e1396. doi: 10.1371/journal.pone.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 80.Legesse-Miller A, Elemento O, Pfau SJ, Forman JJ, Tavazoie S, Coller HA. let-7 Overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J. Biol. Chem. 2009;284:6605–6609. doi: 10.1074/jbc.C900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci U S A. 2010;107:1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin S, Li H, Mu H, Luo W, Li Y, Jia X, Wang S, Jia X, Nie Q, Li Y, Zhang X. Let-7b regulates the expression of the growth hormone receptor gene in deletion-type dwarf chickens. BMC Genomics. 2012;13:306. doi: 10.1186/1471-2164-13-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cutting AD, Bannister SC, Doran TJ, Sinclair AH, Tizard MV, Smith CA. The potential role of microRNAs in regulating gonadal sex differentiation in the chicken embryo. Chromosome Res. 2012;20:201–213. doi: 10.1007/s10577-011-9263-y. [DOI] [PubMed] [Google Scholar]
- 85.Wang XG, Yu JF, Zhang Y, Gong DQ, Gu ZL. Identification and characterization of microRNA from chicken adipose tissue and skeletal muscle. Poult Sci. 2012;91:139–149. doi: 10.3382/ps.2011-01656. [DOI] [PubMed] [Google Scholar]
- 86.Hull KL, Marsh JA, Harvey S. A missense mutation in the GHR gene of Cornell sex-linked dwarf chickens does not abolish serum GH binding. J. Endocrinol. 1999;161:495–501. doi: 10.1677/joe.0.1610495. [DOI] [PubMed] [Google Scholar]
- 87.Tanaka M, Hayashida Y, Wakita M, Hoshino S, Nakashima K. Expression of aberrantly spliced growth hormone receptor mRNA in the sex-linked dwarf chicken, Gifu 20. Growth Regul . 1995;5:218–223. [PubMed] [Google Scholar]
- 88.Agarwal SK, Cogburn LA, Burnside J. Dysfunctional growth hormone receptor in a strain of sex-linked dwarf chicken evidence for a mutation in the intracellular domain. J. Endocrinol. 1994;142:427–434. doi: 10.1677/joe.0.1420427. [DOI] [PubMed] [Google Scholar]
- 89.Wu GQ, Zheng JX, Yang N. [Expression profiling of GH, GHR, and IGF-1 genes in sex-linked dwarf chickens] Yi Chuan. 2007;29:989–994. doi: 10.1360/yc-007-0989. [DOI] [PubMed] [Google Scholar]
- 90.Mavalli MD, DiGirolamo DJ, Fan Y, Riddle RC, Campbell KS, van Groen T, Frank SJ, Sperling MA, Esser KA, Bamman MM, Clemens TL. Distinct growth hormone receptor signaling modes regulate skeletal muscle development and insulin sensitivity in mice. J. Clin. Invest. 2010;120:4007–4020. doi: 10.1172/JCI42447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tamiya T, Kashiwagi I, Takahashi R, Yasukawa H, Yoshimura A. Suppressors of cytokine signaling (SOCS) proteins and JAK/STAT pathways: regulation of T-cell inflammation by SOCS1 and SOCS3. Arterioscler Thromb Vasc Biol. 2011;31:980–985. doi: 10.1161/ATVBAHA.110.207464. [DOI] [PubMed] [Google Scholar]
- 92.Rui L, Yuan M, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J. Biol. Chem. 2002;277:42394–42398. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- 93.Cardinali B, Castellani L, Fasanaro P, Basso A, Alema S, Martelli F, Falcone G. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PLoS One. 2009;4:e7607. doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Machova PK, Lopotova T, Klamova H, Burda P, Trneny M, Stopka T, Moravcova J. Expression patterns of microRNAs associated with CML phases and their disease related targets. Mol Cancer. 2011;10:41. doi: 10.1186/1476-4598-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]