Introduction
Although the area of miRNA research in the nervous system is still in its infancy, it has come quite a long way in the last ten years. Much has been learned from diverse sources, experimental systems, and approaches, making it easy to become dazzled and intimidated. We are confronted with new breakthroughs almost on a weekly basis! Most of the future theoretical advancements will be created in parallel with, or even secondary to, technical innovations. Already, miRNA researchers experience a challenging array of technical tools. Just for miRNA expression analyses, one could use microarrays, PCR, Northern blot, deep sequencing, and dozens of more specialized analytic platforms (Nelson et al., 2006; Roy et al., 2011; Streichert et al., 2011), and it is not always easy to select a reliable choice, much less a “gold standard”.
By means of introduction to this exciting special issue of Experimental Neurology, dedicated to the study of miRNAs in neurons and neurodegeneration, we provide for our readers an admittedly subjective list of ten “issues” that have been learned through experience in this demanding field. The described topics are mainly technical in nature, but in many ways applied methodological considerations interact with theoretical aspects of miRNA biology, for instance the tendency of miRNAs to work through different mechanisms in different biological (and thus experimental) contexts. We discuss these “issues” that we have experienced in our own work and recognized in the field in general. It is hoped that these discussions may in some way be useful, or may complement our colleagues’ research efforts, as we embark in the next 10 years of miRNA research in the brain.
Issue #1. The number of highly expressed miRNAs in the adult brain is relatively low, and modestly expressed “annotated” miRNAs are sometimes not validated when evaluated by Northern blot analyses
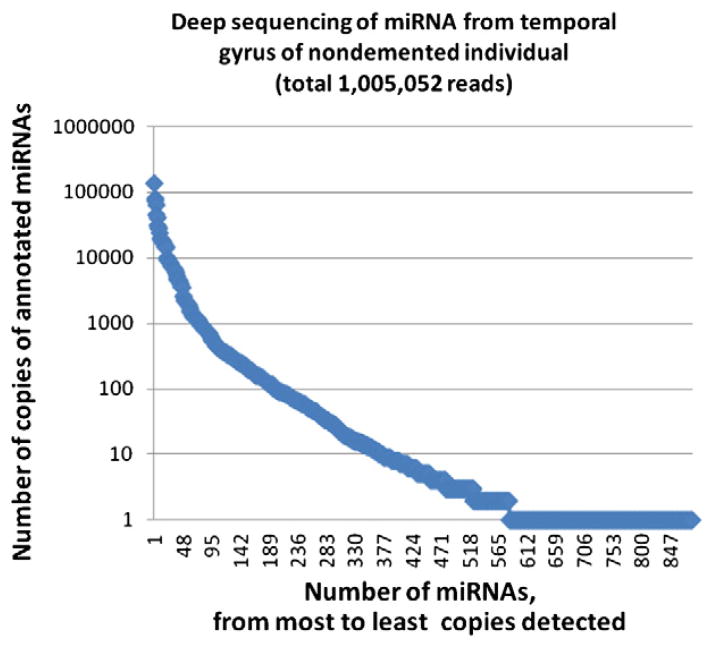
Although it seems as though new miRNAs are constantly being discovered and annotated, the characterization of the human miRNome has probably reached a point of diminishing returns. This perspective derives from expression profiling involving a number of different platforms (see for example Nelson et al., 2008b) and recent deep sequencing efforts providing base level resolution of small RNA populations. We have found while studying the human brain miRNA repertoire that the probable number of miRNAs that are expressed in moderate-to-high copy number is under 200, and the truly high-copy miRNAs (depending on how these are defined) are only in the few dozens (Table 1 and Fig. 1). Fig. 1 shows the partial results of deep sequencing of small RNA from nondemented human brain temporal gyri. It is noteworthy that only a handful of miRNAs represent the great majority of total counted small RNAs in this sample. More detailed description of our analyses of deep sequencing in human brain will be described elsewhere. This is an important idea to keep in mind because the biological activities of the highly expressed miRNAs may be quantitatively more impactful than the lowly expressed miRNAs (see Issue #6 below). Yet, we still do not know the exact relationships between miRNA “expression” (i.e., the copy number of a given miRNA in a cell) and miRNA “activity”.
Table 1.
Comparison of microarray vs. deep sequencing data. miRNA expression analyses from two different expression profiling platforms. For both platforms, RNA was isolated, using Trizol LS, from superior and mid-temporal gyrus (Brodmann Areas 21/22) of nondemented elderly human brain. See text for more details.
| Top 20 miRNAs | Microarray | Deep sequencing |
|---|---|---|
| 1 | miR-125b | miR-26a |
| 2 | miR-124 | let-7a |
| 3 | miR-29a | miR-125b |
| 4 | miR-26a | miR-22 |
| 5 | let-7b | let-7f |
| 6 | miR-107 | miR-27b |
| 7 | let-7c | miR-127-3p |
| 8 | miR-103 | miR-9 |
| 9 | miR-128 | miR-125a-5p |
| 10 | let-7a | miR-143 |
| 11 | miR-221 | miR-99b |
| 12 | miR-23b | miR-100 |
| 13 | miR-125a | miR-30a |
| 14 | miR-145 | miR-128 |
| 15 | miR-24-1,2 | let-7g |
| 16 | miR-99a | let-7c |
| 17 | let-7d | miR-191 |
| 18 | miR-143 | miR-138 |
| 19 | miR-143a | miR-30e |
| 20 | miR-129-3p | miR-126-3p |
Fig. 1.
miRNA deep sequencing of the human brain. RNA isolated from the superior and middle temporal gyrus was analyzed using deep sequencing (see text for more details), resulting in slightly over 1,000,000 reads of annotated miRNAs. Note the Y axis uses a logarithmic scale to describe the number of a particular miRNA species; each blue diamond is a particular annotated miRNA. Fewer than 900 total annotated miRNAs were detected, of which fewer than 200 were detected with more than 100 reads in this sample. More than one-third (358,073) of the total miRNAs were represented in the top four high copy number miRNAs. These data underscore the point that relatively few miRNAs are highly expressed in brain.
What we do know is that a number of annotated miRNAs, when evaluated in the brain, do not show Northern blot patterns compatible with classic pre-miRNAs and mature miRNAs (Nelson et al., 2010; Wang et al., 2011). This is even the case for some miRNAs that show high “expression” on a miRNA microarray (Wang et al., 2011). Thus, while some small RNAs populations remain to be fully characterized (e.g., snoRNAs, piRNAs, rasiRNAs, crasiRNAs, sdRNAs, etc.), the number of moderate-to-high expressed brain miRNAs – as currently defined – has likely reached a plateau.
Issue #2. Computational methodology needs to be viewed with critical scrutiny, and we need more experimental validation to refine the computational algorithms for miRNA:target predictions
Computational methods have provided vitally helpful tools for miRNA research. However, the results of predictive logarithms should not always be conflated with experimental outcomes. The current miRNA prediction programs such as TargetScan (Lewis et al., 2005), MicroCosm (Griffiths-Jones et al., 2008), Pictar (Krek et al., 2005), rna22 (Miranda et al., 2006), and others are impressive, but it should be noted that each has different assumptions (e.g., percentage of target conservation, structural conformation, free energy of duplex formation, and whether limited to 3′UTR targeting), and, for any given target prediction, they tend to arrive at differing predictions. In some circumstances the assumptions overlap, and thus the predictive results overlap, but these do not necessarily strengthen the validity of their conclusions. While important and necessary, it is difficult for any algorithm to capture all of the biological complexities of miRNA function, particularly in an organ as complex as the human brain. It has become clear that different miRNAs may work according to idiosyncratic ‘binding rules’. Specifically, miR-107 tends to systematically recognize open reading frame of some targets, in contradistinction to miR-124 and other miRNAs (Nelson et al., 2010, 2011; Wang et al., 2010a, 2010b, 2010c), and this level of complexity will need to be incorporated into future predictive algorithms. Furthermore, many miRNAs appear to be trafficked into the nucleus where the targeting functions may be quite different from the cytoplasmic miRNAs (Hwang et al., 2007; Marcon et al., 2008; Tan et al., 2009; Weinmann et al., 2009).
Issue #3. 3′UTR luciferase assays are not unassailable methods for target validation
It is easy to be impressed by the robustness and functionality of the experimental approaches that can be obtained using 3′UTR luciferase assays. These assays are clearly among the most widespread and (in some senses at least) reliable experimental contexts for miRNA:target validation. The merits of this system relate both to the well-optimized reagents, and the fact that the cancer cell lines that are usually used (e.g., HEK293, HeLa) are also robust biological models that generally have a functional Argonate:miRNA apparatus, although the full relevance of the protein and RNA components to actual neural tissue is debatable (see Issue #8 below). Other points of caution are the physiological relevance of these assays and choice of miRNA candidates.
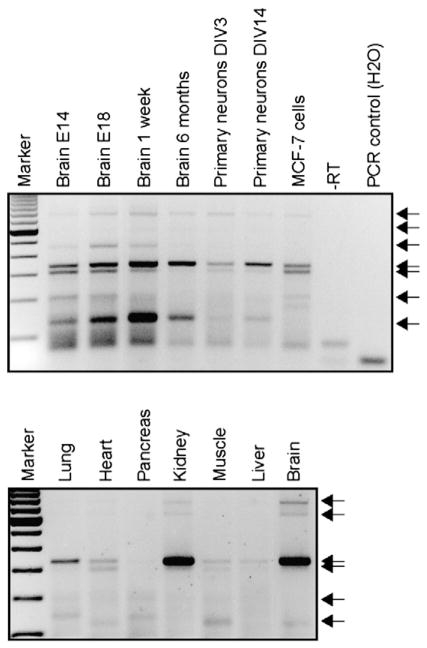
The complexity of mRNA processing in human brain is even more extreme due to the fact that a significant (~30%) of miRNA:mRNA effects are not on the 3′UTR according to high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) data (Chi et al., 2009). In addition, human 3′UTRs are long with complex cis- and trans-regulators (Dreyfuss et al., 2002; Mazumder et al., 2003), and the transfection reagents used in some 3′UTR studies may have secondary effects on the miRNA processing. In our experience, up to 50% of candidate miRNAs identified using bioinformatics can modulate luciferase activity under co-overexpression paradigms (Delay et al., 2011; Hebert et al., 2008, 2009). However, a positive signal may not necessarily reflect a natural phenomenon, as candidate miRNAs are not necessarily co-expressed with their targets and many other parameters affect 3′UTRs. For example, 3′UTR length may vary according to developmental stage, tissue, and/or cell type, and thus the “canonical” 3′UTR generally used in luciferase experiments may be non-representative. A specific example is shown in Fig. 2 using the Amyloid precursor protein (APP) 3′UTR. In sum, although 3′UTR reporter assays are powerful technical tools, they should be complemented with neuronal cell-based studies at minimum, ideally on endogenously expressed proteins.
Fig. 2.
3′UTR length heterogeneity as observed using 3′RACE-PCR. RNA was isolated from wildtype mouse organs, mouse primary cortical neurons, and human MCF-7 cells were used to perform 3′ Rapid Amplification of cDNA 3′ Ends (RACE) PCR using APP-specific oligonucleotides. These assays reveal changes in APP 3′UTR length and abundance depending on developmental stage, age, cell type and organ. Arrows indicate various 3′UTR products. Different 3′UTRs for a given transcript is probably one of many ways that miRNA targeting is regulated in the brain.
Issue #4. Tissue culture miRNA experiments need adequate experimental controls
Many miRNA experiments are performed downstream of bioinformatics, so a researcher is often evaluating evidence for or against a particular hypothesis that was predicted to occur. Experiments in this context should be performed with the aim of disproving a hypothesis, not with the aim of proving it. Thus, for miRNA tissue culture experiments a high bar should be maintained with regard to the number and subtypes of experimental controls. As a practical example, it is probably optimal to include multiple control miRNAs in transfection experiments, multiple reporter constructs in luciferase assays, multiple potential targets in some circumstances where the specificity of miRNA:target interactions is being hypothesized, and an evaluation of more than a single cell line to prove a given phenomenon can be expected to occur in a broad context. And of course it is an ethical obligation to report the results of all the controls that were evaluated.
Issue #5. For miRNA expression analyses, there is simply no gold standard; every method has technical biases that should be incorporated into study design
The assessment of miRNA expression is deceptively challenging. Each high-throughput method for quantifying the amount of multiple miRNAs in a particular sample has its own biases. For example, deep sequencing harbors biases related to the necessity of “tagging” (with RNA ligase) and amplifying the small RNA products (Hafner et al., 2011). Real-time quantitative polymerase chain reaction (RTqPCR) entails biases related to the method itself (e.g., cDNA production and biases related to the universal probes) and do not take into consideration the 5′ and 3′ end biological variation in miRNAs. Other hybridization-based methods (e.g., microarrays) are limited by the fact that one only sees output related to the probes on the array, and there is always some degree of cross-reaction, with both false-negatives and false-positives. Northern blots, which may be as close as possible to a “gold standard”, are semi-quantitative in nature and limited by the low throughput and the amount of material required. Other miRNA expression platforms are more individualized and have not received the validation from sufficient number of independent laboratories to date. In our experience, very “credible” (in terms of theoretical underpinning and near-perfect replicability for biological replicates) expression data can be produced using a particular sequencing platform, but the results still differ substantially between platforms. Note for example in Table 1 that miR-124 is not among the top 20 miRNAs detected using small RNA deep sequencing, in contrast to the results using tissue from the same source but profiled using a microarray exactly as described previously (Wang et al., 2008). Thus, no individual method can be used exclusively, and the result of one technique needs to be validated when possible using at least one independent method. There is also a substantial issue related to the normalization methods, which require important assumptions, as has been previously discussed (Meyer et al., 2010; Nelson et al., 2008b; Sarver, 2010).
Issue #6. In terms of miRNA expression changes, the current emphasis of “fold-change” is problematic; it is likely more biologically relevant to have a small % change in a highly expressed miRNA than a large % change in a negligibly expressed miRNA
Focusing on “fold-change” without commensurate attention to the baseline expression patterns could lead to faulty conclusions. There is extreme variation in RNA copy numbers among transcripts; this is even truer for miRNAs than for messenger RNAs (mRNAs). For example, the highest copy number for abundant neuronal mRNAs is generally below 2500 copies per cell and over 90% of cells have fewer than 100 mRNA copies per cell (Carter et al., 2005; Femino et al., 1998; Latham et al., 1994). By contrast, a high-copy miRNA may be present in over 25,000 copies per cell (Chen et al., 2005; Liang et al., 2007; Lim et al., 2003) (and see Lu and Tsourkas, 2009). This may reflect the fact that a miRNA’s activity is more sensitive to “mass effect” (passively occupying a mRNA target with imperfect target specificity) in contrast to an mRNA in which a single copy can be recruited to produce hundreds or thousands of polypeptides per day. A 10% perturbation in an abundant neuronal miRNA, such as miR-125b, would probably translate to a change in ~2000 copies of the miRNA per cell, whereas for a miRNA with 10 copies/cell, an increase to 30 copies per cell may not necessarily have a profound effect on that cell’s function.
Issue #7. It can be advantageous to complement tissue-level miRNA expression changes using single cell-level methods
Many miRNAs are expressed in a cell-type specific manner, and this is particularly true in the central nervous system where neurons, glial cells, and vascular cells express differing retinues of miRNAs. This fact makes it important to use in situ hybridization (ISH) or some other single-cell method to better understand which cells express which miRNAs. In some diseases, and in other contexts that may be used for comparison’s sake at the tissue level (e.g., developmental stages), there can be a replacement and/or a loss/gain of a particular cell population that can make tissue-level profiling problematic. For example, at the tissue level, miR-124 is “down-regulated” in oligodendroglial brain tumors (Nelson et al., 2006). However, using ISH it was found that the decreased miR-124 that was observed in brain tumor tissue was an artifact; higher-grade neoplasms had fewer neurons intermingled with the tumor cells and miR-124 was therefore just a bystander. By contrast, ISH showed that miR-9, which was up-regulated at the tissue level, was indeed expressed in the tumor cells (Nelson et al., 2006). This is just one example to illustrate the necessity of corroborating tissue-level miRNA profiling with ISH (Nelson and Wilfred, 2009).
Issue #8. More in vivo models are required; the mechanistic reliance on cancer cell lines has some downsides because living tissues, particularly the brain, may well have different mechanisms at play
After productive miRNA research was performed in plant species, C. elegans, and D. melanogaster, much of the work on mammalian miRNA function and targeting, and many of the particular highly-expressed human miRNAs, were first performed in human cancer cell lines. As has been commented previously (Nelson et al., 2010) there are compelling reason to use these robust, easy to transfect, well-characterized, clonal cells for easily replicable hypothesis testing. However, there are also good reasons to reflect on the essential characteristics of tumor cell lines: they are neoplastic, they tend to have extensive chromosomal abnormalities, and they are selected for particular characteristics (robustness, transfectability, plate adherence, etc.) that are inimical to normal biological processes. The very survival and passage number of cancer cells in culture is evidence of stark differences relative to most normal cells, and in particular neurons. In sum, tumor cell lines are non-physiologic. Of direct relevance to the present review, cancer cells tend to have suppression of miRNA expression and/or modification of miRNA processing proteins (Adams et al., 2009; Chung et al., 2009; Ciafre et al., 2005; Guo et al., 2009; Wijnhoven et al., 2007). It truly seems likely that in vivo mammalian brain cells will have evolved mechanisms that differ in some important ways relative to these cultured cells. Some in vivo systems have been developed already (Delay and Hebert, 2011). However, we anticipate future technically improved and validated in vivo experimental systems for mechanistic manipulations of miRNAs and the miRNA-processing machinery.
Issue #9. Post-mortem brain studies should always include detailed biochemical, neuropathological, and clinical details
Most human neurodegenerative diseases (e.g., Alzheimer’s disease, synucleinopathies, hippocampal sclerosis, and fronto-temporal lobe dementia) are essentially human specific (Rapoport, 1990; Rapoport and Nelson, 2011), so miRNA research devoted to studying these diseases must at some point assess actual human brain tissues. This is a necessary stage for validating any hypothesis, providing clinical relevance to findings, and also is important for developing new hypotheses to test in other systems. Confidence in the value of high-quality RNA requires complementary data about clinical and pathological parameters. This is particularly true for disease tissues from Alzheimer’s disease, in which disease progression itself contributes to RNA deterioration (Hebert and De Strooper, 2009; Nelson and Keller, 2007; Nelson et al., 2008a). Further, it has been shown that miRNAs can rapidly degrade postmortem (Sethi and Lukiw, 2009). In our experience, a fairly low percentage (depending on brain bank origin) of processed post-mortem tissues processed is of suitable quality for RNA-related studies. Thus, biochemical (e.g., RNA integrity number, 260/280 absorbance ratio, pH measurement), neuropathological (e.g. amyloid load, tangles) and clinical profiles (e.g., gender, memory score, MRI scans) are appropriate data to include in novel publications.
Issue #10. MiRNAs may have more roles to play: the big(ger) picture
Current technologies such as high-throughput RNA sequencing and mapping of Ago-miRNA binding sites in vivo have taught us that miRNA gene regulatory networks are immensely complex, and the “one-miRNA-one-target” concept is extremely over-simplistic. Some miRNAs may work in the nucleus possibly affecting transcription and RNA processing (Hwang et al., 2007; Maniataki and Mourelatos, 2005; Tan et al., 2009), miRNAs may in some circumstances upregulate translation (Lytle et al., 2007; Steitz and Vasudevan, 2009), antisense RNAs can interact dynamically with miRNAs (Faghihi and Wahlestedt, 2009; Modarresi et al., 2011), and many miRNA:mRNA interactions are probably relatively subtle “tuning” rather than a dichotomous effect (Bartel, 2009). Further, downstream effects of miRNAs can include drastic global changes in mRNA splicing that are relevant to neuronal differentiation and AD pathogenesis (Makeyev et al., 2007; Smith et al., 2011a, 2011b).
As an indication of the complexity of miRNA:mRNA interactions, we refer our readers to databases such as starBase (starbase.sysu.edu.cn/), which allow searching for validated miRNA targets in vivo, including in the brain. Existing microarray and cell-based studies provide a strong “theoretical” foundation to move forward and place these data into biological context. Also, recent advances in bioinformatics programs such as IPA (ingenuity.com) allow for global analyses of mRNA:miRNA networks using microarray data. Future research should therefore consider these new technologies – from which most data are publicly available – to develop new ideas while integrating the now multi-factorial aspects of miRNA research, for instance by complementing in “wet lab” data with bioinformatics.
Conclusion
The human brain is incredibly complex with ~1018 synapses that derive from a template of only ~105 protein-coding genes (Nelson and Keller, 2007). That is 1000-fold more synapses per gene than mice overall, and approximately 1.5 times more synapses per neuron than mice (DeFelipe et al., 2002). It has been hypothesized that non-coding RNA-based gene expression regulation – very much including miRNAs – is likely an important component of evolutionary change in mammalian nervous systems (Hausser et al., 2009; Heimberg et al., 2008; Nelson and Keller, 2007). As bench researchers studying miRNAs in the brain, we should constantly push our critical thinking, and not necessarily defer to newly minted dogmas (“what we now know”) because what we don’t know is so much vaster. Having said this, we anticipate that the next 10 years of miRNA brain research will engender exciting new technologies, ideas, and breakthroughs. The development of new tools will be necessary to catch a better glimpse at the complex networks involved in brain function and, importantly, dysfunction. We hope that the issues presented herein, among many others, will help pave the way to understanding the molecular mechanisms underlying neurodegenerative diseases, a global concern in our aging world.
Contributor Information
Sébastien S. Hébert, Centre de recherche du CHUQ (CHUL), Neurosciences, 2705 boul. Laurier, RC-9800, Québec, Qc, Canada G1V 4G2
Peter T. Nelson, Sanders-Brown Center on Aging, Rm 311, Sanders-Brown Center, 800 S. Limestone, University of Kentucky, Lexington, KY 40536-0230, USA
References
- Adams BD, Claffey KP, White BA. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150:14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MG, Sharov AA, VanBuren V, Dudekula DB, Carmack CE, Nelson C, Ko MS. Transcript copy number estimation using a mouse whole-genome oligonucleotide microarray. Genome Biol. 2005;6:R61. doi: 10.1186/gb-2005-6-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung TK, Cheung TH, Huen NY, Wong KW, Lo KW, Yim SF, Siu NS, Wong YM, Tsang PT, Pang MW, Yu MY, To KF, Mok SC, Wang VW, Li C, Cheung AY, Doran G, Birrer MJ, Smith DI, Wong YF. Dysregulated microRNAs and their predicted targets associated with endometrioid endometrial adenocarcinoma in Hong Kong women. Int J Cancer. 2009;124:1358–1365. doi: 10.1002/ijc.24071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of micro- RNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Alonso-Nanclares L, Arellano JI. Microstructure of the neocortex: comparative aspects. J Neurocytol. 2002;31:299–316. doi: 10.1023/a:1024130211265. [DOI] [PubMed] [Google Scholar]
- Delay C, Hebert SS. MicroRNAs and Alzheimer’s disease mouse models: current insights and future research avenues. Int J Alzheimers Dis. 2011;2011:894938. doi: 10.4061/2011/894938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delay C, Calon F, Mathews P, Hebert SS. Alzheimer-specific variants in the 3′ UTR of Amyloid precursor protein affect microRNA function. Mol Neurodegener. 2011;6:70. doi: 10.1186/1750-1326-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Science. Vol. 280. New York, N.Y: 1998. Visualization of single RNA transcripts in situ; pp. 585–590. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- Hafner M, Renwick N, Brown M, Mihailovic A, Holoch D, Lin C, Pena JT, Nusbaum JD, Morozov P, Ludwig J, Ojo T, Luo S, Schroth G, Tuschl T. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011;17:1697–1712. doi: 10.1261/rna.2799511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausser J, Landthaler M, Jaskiewicz L, Gaidatzis D, Zavolan M. Relative contribution of sequence and structure features to the mRNA binding of Argonaute/EIF2C-miRNA complexes and the degradation of miRNA targets. Genome Res. 2009;19:2009–2020. doi: 10.1101/gr.091181.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Bergmans B, Papadopoulou AS, Delacourte A, De Strooper B. MicroRNA regulation of Alzheimer’s Amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. Micro-RNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008;105:2946–2950. doi: 10.1073/pnas.0712259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. Science. Vol. 315. New York, N.Y: 2007. A hexanucleotide element directs microRNA nuclear import; pp. 97–100. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Latham VM, Jr, Kislauskis EH, Singer RH, Ross AF. Beta-actin mRNA localization is regulated by signal transduction mechanisms. J Cell Biol. 1994;126:1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Tsourkas A. Imaging individual microRNAs in single mammalian cells in situ. Nucleic Acids Res. 2009;37:e100. doi: 10.1093/nar/gkp482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA. 2005;11:849–852. doi: 10.1261/rna.2210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon E, Babak T, Chua G, Hughes T, Moens PB. miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosome Res. 2008;16:243–260. doi: 10.1007/s10577-007-1190-6. [DOI] [PubMed] [Google Scholar]
- Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- Meyer SU, Pfaffl MW, Ulbrich SE. Normalization strategies for microRNA profiling experiments: a ‘normal’ way to a hidden layer of complexity? Biotechnol Lett. 2010;32:1777–1788. doi: 10.1007/s10529-010-0380-z. [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Modarresi F, Faghihi MA, Patel NS, Sahagan BG, Wahlestedt C, Lopez-Toledano MA. Knockdown of BACE1-AS nonprotein-coding transcript modulates beta-amyloid-related hippocampal neurogenesis. Int J Alzheimers Dis. 2011;2011:929042. doi: 10.4061/2011/929042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Keller JN. RNA in brain disease: no longer just "the messenger in the middle". J Neuropathol Exp Neurol. 2007;66:461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- Nelson PT, Wilfred BR. In situ hybridization is a necessary experimental complement to microRNA (miRNA) expression profiling in the human brain. Neurosci Lett. 2009;466:69–72. doi: 10.1016/j.neulet.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA. 2006;12:187–191. doi: 10.1261/rna.2258506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008a;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Wilfred BR, Tang G. Technical variables in high-throughput miRNA expression profiling: much work remains to be done. Biochim Biophys Acta. 2008b;1779:758–765. doi: 10.1016/j.bbagrm.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Kiriakidou M, Mourelatos Z, Tan GS, Jennings MH, Xie K, Wang WX. High-throughput experimental studies to identify miRNA targets directly, with special focus on the mammalian brain. Brain Res. 2010;1338:122–130. doi: 10.1016/j.brainres.2010.03.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Mao G, Wilfred BR, Xie K, Jennings MH, Gao Z, Wang X. Specific sequence determinants of miR-15/107 microRNA gene group targets. Nucleic Acids Res. 2011 Oct;39(18):8163–8172. doi: 10.1093/nar/gkr532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport SI. Integrated phylogeny of the primate brain, with special reference to humans and their diseases. Brain Res Brain Res Rev. 1990;15:267–294. doi: 10.1016/0165-0173(90)90004-8. [DOI] [PubMed] [Google Scholar]
- Rapoport SI, Nelson PT. Biomarkers and evolution in Alzheimer disease. Prog Neurobiol. 2011 Dec;95(4):510–513. doi: 10.1016/j.pneurobio.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Soh JH, Gao Z. A microfluidic-assisted microarray for ultrasensitive detection of miRNA under an optical microscope. Lab Chip. 2011;11:1886–1894. doi: 10.1039/c0lc00638f. [DOI] [PubMed] [Google Scholar]
- Sarver AL. Toward understanding the informatics and statistical aspects of micro-RNA profiling. J Cardiovasc Transl Res. 2010;3:204–211. doi: 10.1007/s12265-010-9180-z. [DOI] [PubMed] [Google Scholar]
- Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- Smith P, Al Hashimi A, Girard J, Delay C, Hebert SS. In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J Neurochem. 2011a;116:240–247. doi: 10.1111/j.1471-4159.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- Smith PY, Delay C, Girard J, Papon MA, Planel E, Sergeant N, Buee L, Hebert SS. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum Mol Genet. 2011b;20:4016–4024. doi: 10.1093/hmg/ddr330. [DOI] [PubMed] [Google Scholar]
- Steitz JA, Vasudevan S. miRNPs: versatile regulators of gene expression in vertebrate cells. Biochem Soc Trans. 2009;37:931–935. doi: 10.1042/BST0370931. [DOI] [PubMed] [Google Scholar]
- Streichert T, Otto B, Lehmann U. microRNA expression profiling in archival tissue specimens: methods and data processing. Mol Biotechnol. 2011 Jun 21; doi: 10.1007/s12033-011-9427-1. [DOI] [PubMed] [Google Scholar]
- Tan GS, Garchow BG, Liu X, Yeung J, Morris JPt, Cuellar TL, McManus MT, Kiriakidou M. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Rajeev BW, Stromberg AJ, Ren N, Tang G, Huang Q, Rigoutsos I, Nelson PT. The expression of microRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of betasite amyloid precursor protein-cleaving enzyme 1. J Neurosci. 2008;28:1213–1223. doi: 10.1523/JNEUROSCI.5065-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Wilfred BR, Hu Y, Stromberg AJ, Nelson PT. Anti-Argonaute RIP-Chip shows that miRNA transfections alter global patterns of mRNA recruitment to microribonucleoprotein complexes. RNA. 2010a;16:394–404. doi: 10.1261/rna.1905910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Wilfred BR, Madathil SK, Tang G, Hu Y, Dimayuga J, Stromberg AJ, Huang Q, Saatman KE, Nelson PT. miR-107 regulates granulin/progranulin with implications for traumatic brain injury and neurodegenerative disease. Am J Pathol. 2010b;177:334–345. doi: 10.2353/ajpath.2010.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Wilfred BR, Xie K, Jennings MH, Hu Y, Stromberg AJ, Nelson PT. Individual microRNAs (miRNAs) display distinct mRNA targeting "rules". RNA Biol. 2010c;7 doi: 10.4161/rna.7.3.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WX, Huang Q, Hu Y, Stromberg AJ, Nelson PT. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: white matter versus gray matter. Acta Neuropathol. 2011;121:193–205. doi: 10.1007/s00401-010-0756-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- Wijnhoven BP, Michael MZ, Watson DI. MicroRNAs and cancer. Br J Surg. 2007;94:23–30. doi: 10.1002/bjs.5673. [DOI] [PubMed] [Google Scholar]