Abstract
Changes in genome architecture often have a significant effect on ecological specialization and speciation. This effect may be further enhanced by involvement of sex chromosomes playing a disproportionate role in reproductive isolation. We have physically mapped the Z chromosome of the major pome fruit pest, the codling moth, Cydia pomonella (Tortricidae), and show that it arose by fusion between an ancestral Z chromosome and an autosome corresponding to chromosome 15 in the Bombyx mori reference genome. We further show that the fusion originated in a common ancestor of the main tortricid subfamilies, Olethreutinae and Tortricinae, comprising almost 700 pest species worldwide. The Z–autosome fusion brought two major genes conferring insecticide resistance and clusters of genes involved in detoxification of plant secondary metabolites under sex-linked inheritance. We suggest that this fusion significantly increased the adaptive potential of tortricid moths and thus contributed to their radiation and subsequent speciation.
Keywords: adaptive evolution, leaf-rollers, performance genes, sex chromosome–autosome fusion, sex-linkage
Karyotype differences observed between closely related species have stimulated long-standing debates over the role of chromosome rearrangements in speciation. Recently, new empirical evidence has inspired the development of theoretical models that offer an explanation of how changes in genome architecture may facilitate speciation in the face of gene flow. It has been suggested that selection can favor chromosome rearrangements that decrease the incidence of recombination between alleles contributing to local adaptations, which in turn can enhance fixation of karyotype differences within local populations (1). Of all such chromosomal rearrangements, the scope of these models is limited to inversion polymorphisms that directly suppress recombination. However, another significant mode of karyotype change that often leads to speciation is intraspecific differences in chromosome numbers, altered by chromosome fusions and fissions (2). These rearrangements have the potential to limit gene flow although their effect is presumably smaller (1). Indeed, chromosome fusions have been shown to influence recombination by decreasing the number of chiasmata via their interference and, more importantly, by coupling previously unlinked loci (3). Similar to chromosomal rearrangements, genetic linkage between traits contributing to reproductive and ecological isolation has been found to impede breakdown of linkage disequilibria following recombination (4–7).
Both linkage disequilibrium and chromosome rearrangements are important forces in the rise of sex chromosomes and their subsequent differentiation. Natural selection appears to favor the linkage of sexually antagonistic alleles to sex-determining loci and inversion-mediated suppression of recombination in sex-specific W or Y chromosomes (8). The lack of recombination ultimately causes degeneration of sex-specific chromosomes via accumulation of repetitive sequences and gene loss. In contrast, recombining X and Z chromosomes are known to undergo fast adaptive evolution and play a special role in speciation due to their involvement in postzygotic reproductive isolation (8–10). Furthermore, recent reports on the turnover of sex chromosomes have contributed to the idea that sex chromosome–autosome fusions might actually promote speciation (11).
Moths and butterflies (Lepidoptera) have a WZ/ZZ sex chromosome system with female heterogamety. Although sex chromosomes have been identified in only a handful of species, derived variants W1W2Z/ZZ and WZ1Z2/Z1Z1Z2Z2 were observed in nine genera, suggesting a relatively high incidence of neo-sex chromosomes in this species-rich group (12). Neo-sex chromosome evolution via multiple sex chromosome–autosome fusions was described in moths with highly derived karyotypes, Orgyia antiqua and Orgyia thyellina (Lymantriidae), and in geographical subspecies of Samia cynthia (Saturniidae) (13). Recently, it has been suggested that the sex chromosome rearrangements in S. cynthia populations may contribute to the formation of reproductive barriers and facilitate divergence toward speciation (14).
A previous study predicted a translocation of an autosome onto the Z chromosome in the family Tortricidae (15). To test this hypothesis, we performed comparative physical mapping of the Z chromosome in the major pome fruit pest, the codling moth, Cydia pomonella (Tortricidae: Olethreutinae), and found that a neo-Z chromosome formed following fusion between an ancestral Z chromosome and an autosome corresponding to chromosome 15 in the Bombyx mori reference genome. Furthermore, we show that the fusion originated in a common ancestor of the main subfamilies Olethreutinae and Tortricinae, which comprise 97% of extant species of tortricids. We discuss the relevance of our findings for adaptive evolution and radiation of tortricid moths.
Results
BAC-FISH Mapping of the Codling Moth Z Chromosome.
Partial sequences of 17 C. pomonella genes linked to the chromosomes Z and 15 in the reference genome of B. mori (Table S1) were cloned and deposited in GenBank (see Table S2 for accession numbers). These genes included three major genes linked to insecticide resistance (ABCC2, Ace-1, and Rdl), four enzyme-coding genes (Idh-2, Ldh, Pgd, and Tpi), and 10 protein-coding genes without enzymatic function (ABCF2, apterous, kettin, mago, nanchung, Notch, RpL10, RpP0, RpS5, and Shaker). In the case of Ace-1 gene, a comparison of the obtained sequence showed 100% identity with the corresponding part of Ace-1 isolated earlier from C. pomonella susceptible strain [accession no. DQ267977 (16)]. Additionally, a partial sequence of the C. pomonella circadian gene period (per) was acquired from GenBank [accession no. JX996071 (17)]. Hybridization probes generated from the cloned gene fragments were used for screening of the C. pomonella bacterial artificial chromosome (BAC) library. Positive BAC clones were identified and confirmed by PCR for all genes except Ace-1 (Table S3). For full gene names and their abbreviated symbols, see Table S1.
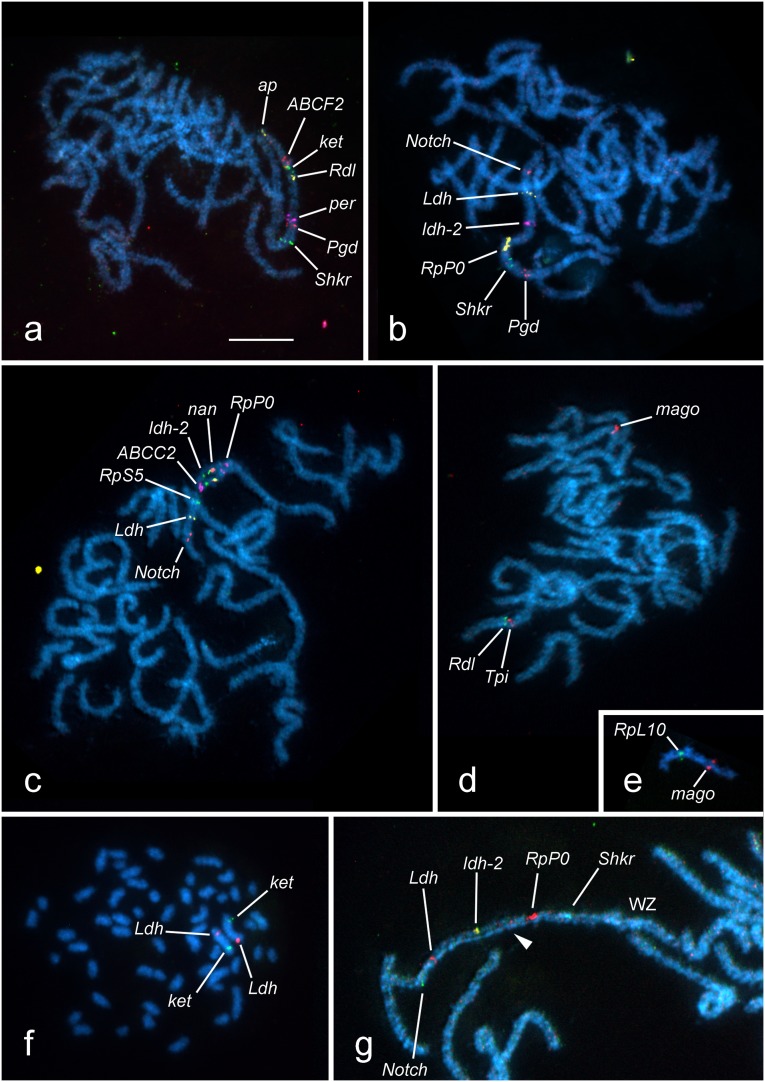
Fluorescent in situ hybridization (FISH) of BAC-derived probes on pachytene nuclei of the codling moth confirmed conserved synteny of all nine tested orthologs of the B. mori Z-linked genes. Eight of these orthologs mapped to about one half of a long pachytene bivalent (Fig. 1 A and D). Also, the gene order of all but one marker (Ldh, see below) was conserved. However, a terminal position of the apterous gene and its distance from its closest neighbor ABCF2 (Figs. 1A and 2) suggested a possible inversion in the subterminal chromosome region. The only exception to a strong colinearity was a BAC clone containing the Ldh gene, which hybridized to the other half of the same bivalent instead of its expected position between Pgd and Shaker (Fig. 1B). Six out of eight orthologs of the chromosome 15 genes of B. mori mapped to the same codling moth bivalent as the Z-linked markers. In this case, the genes retained the same gene order as their B. mori orthologs in chromosome 15, with Ldh inserted between RpS5 and Notch (Fig. 1 B and C). The results of gene mapping indicate that a large chromosome rearrangement, probably a fusion event involving chromosome regions corresponding to the B. mori linkage groups (LG) Z and 15, differentiated karyotypes of the two species from a common ancestor. Two remaining orthologs of B. mori LG15 genes, namely RpL10 and mago, mapped to another chromosome pair (Fig. 1 D and E), revealing a translocation corresponding to a 0.5- to 2.8-Mb segment of the B. mori chromosome 15. However, the distance between hybridization signals of the RpL10 and mago genes on the codling moth autosome seems to greatly exceed the expected size of the translocated segment. A plausible explanation could be that the two originally closely linked genes were separated from each other by a subsequent inversion. All mapping data are integrated in Fig. 2.
Fig. 1.
BAC-FISH mapping of genes on chromosome preparations of the codling moth, C. pomonella. Chromosomes were counterstained with DAPI (light blue). Hybridization signals of BAC probes (yellow, green, red, and violet) indicate the physical positions of loci marked by abbreviated names. (A–D) Pachytene spermatocyte complements. (A) Three runs of BAC-FISH localized seven orthologs of B. mori Z-linked genes (ap, ABCF2, ket, Rdl, per, Pgd, and Shkr) to a single bivalent, the anticipated sex chromosome pair ZZ. (B) Two runs of BAC-FISH with orthologs of B. mori genes of the chromosomes Z (Pgd, Shkr, and Ldh) and 15 (RpP0, Idh-2, and Notch) revealed their positions on the same chromosome bivalent of C. pomonella. (C) Three runs of BAC-FISH localized six orthologs of B. mori chromosome 15 genes (RpP0, nan, Idh-2, ABCC2, RpS5, and Notch) and the Z-linked Ldh gene to the same bivalent. Note the position of Ldh between RpS5 and Notch genes. (D) BAC-FISH localized two orthologs of B. mori Z-linked genes, Rdl and Tpi, to the anticipated Z chromosome bivalent, whereas mago (an ortholog of B. mori chromosome 15 gene) mapped to an autosome bivalent. (E) An autosome bivalent bearing hybridization signals of two orthologs of B. mori chromosome 15 genes, RpL10 and mago. (F) Male mitotic metaphase consisting of 2n = 56 chromosomes showing two BAC probes containing ket and Ldh genes, respectively, hybridized to two largest elements earlier identified as Z chromosomes. (G) A part of pachytene oocyte with the sex chromosome bivalent (WZ) easily discernible by DAPI-positive staining of a heterochromatic thread of the W chromosome (arrowhead) and characteristic twisting of paired chromosomes. Hybridization signals of BAC probes confined the Shkr, RpP0, Idh-2, Ldh, and Notch loci to the Z chromosome. (Scale bar: 10 μm.)
Fig. 2.
A gene-based scheme of the Z chromosome of the codling moth, C. pomonella, integrating all BAC-FISH mapping data (Fig. 1) and its comparison with the B. mori chromosomes Z and 15. Locations of B. mori genes were retrieved from KAIKObase (Table S1). The mean relative positions of loci in the codling moth were calculated from data obtained by measuring physical distances between hybridization signals and the chromosome end in at least 10 ZZ bivalents; the distances were then related to the total length of the Z chromosome. Note the conserved synteny and conserved gene order between Z-linked genes of B. mori and the corresponding segment of the codling moth Z chromosome, except for Ldh, which moved to the part corresponding to B. mori chromosome 15. Carboxylesterases (CCE) and ABC transporters (ABC) with putative role in detoxification of synthetic and natural xenobiotics are annotated on the left of the B. mori chromosomes. Major genes conferring insecticide resistance are in red (for details, see Discussion).
BAC-FISH with selected probes on male mitotic chromosomes of the codling moth identified the rearranged chromosome as the largest element in the karyotype (Fig. 1F) reported earlier as the sex chromosome Z (18). Furthermore, in female preparations of pachytene oocytes, the BAC-derived probes hybridized to the WZ bivalent, which was easily discernible according to the DAPI-positive heterochromatic thread of the W chromosome. In this case, hybridization signals were confined only to the Z chromosome thread (Fig. 1G), which is in accordance with overall degeneration of the codling moth W chromosome (19). Taken together, we conclude that the codling moth Z chromosome is composed of two sets of genes, one originating from the ancestral Z chromosome and the other from an autosome referred to as chromosome 15 in the model species, B. mori.
Sex-Linkage Analysis of Selected Genes by qPCR.
Because no BAC clone containing Ace-1 was identified in the codling moth BAC library, quantitative real-time PCR (qPCR) using genomic DNA as template was used to determine a gene dose, i.e., copy number, of Ace-1 in the codling moth males and females. The results clearly showed a twofold difference in the Ace-1 gene dose between males and females, thus establishing its linkage to the Z sex chromosome (Fig. 3, SI Text, Fig. S1, Fig. S2, and Table S4).
Fig. 3.
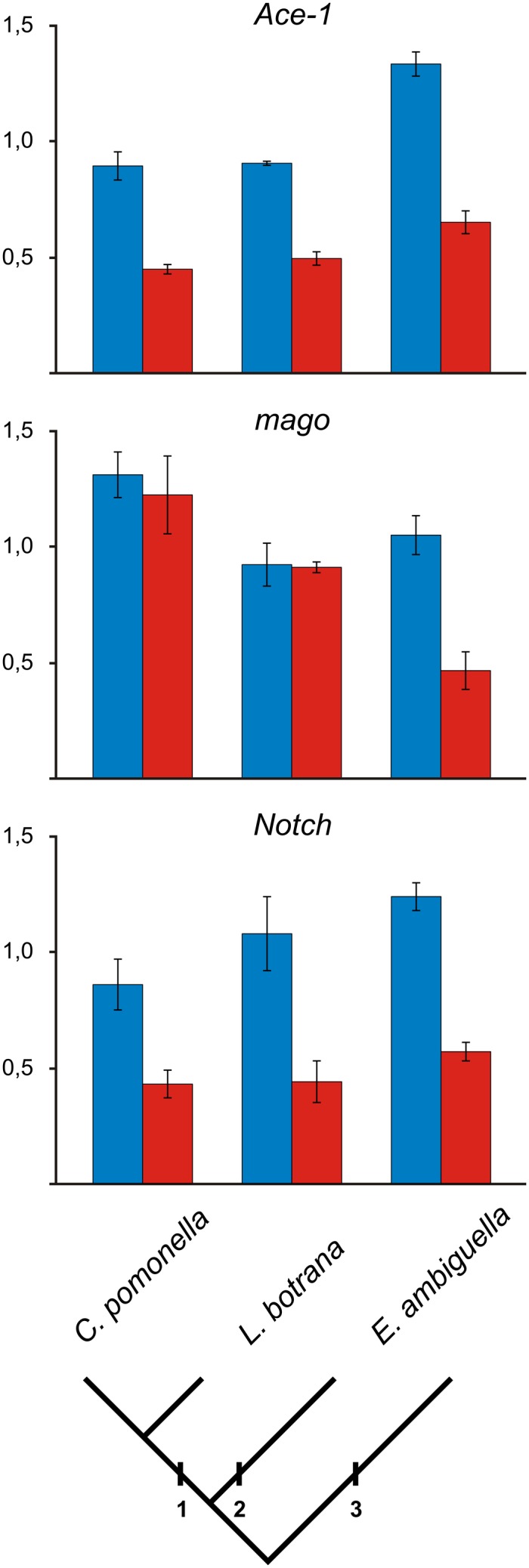
Quantitative PCR comparison of male (blue columns) and female (red columns) doses of Ace-1, mago, and Notch genes normalized to the autosomal reference gene EF-1α in C. pomonella (Olethreutinae, Grapholitini), L. botrana (Olethreutinae, Olethreutini), and E. ambiguella (Tortricinae, Tortricini). Male and female genomic DNAs were used as templates. Error bars represent SDs calculated from three independent samples (Table S4). Twofold differences in both Ace-1 and Notch gene doses between males and females suggest a Z-linkage of the genes in all three tortricids examined. However, mago gene doses did not differ significantly between males and females in both members of the subfamily Olethreutinae, C. pomonella, and L. botrana, thus indicating an autosomal location of the mago gene, in contrast with E. ambiguella where a two times higher dose of this gene in males compared with females suggests its Z-linkage. Phylogenetic relationships are based on ref. 20. 1, Olethreutinae; 2, Tortricinae; 3, Chlidanotinae.
Furthermore, two other tortricid species, the European grapevine moth Lobesia botrana (Olethreutinae) and the vine moth Eupoecilia ambiguella (Tortricinae), were studied to trace the evolutionary origin of the rearrangement between the sex chromosome Z and an autosome corresponding to B. mori chromosome 15. Partial sequences of L. botrana and E. ambiguella orthologs of the Ace-1, EF-1α, mago, and Notch genes were cloned and sequenced (see Table S2 for their accession nos.). Sex-linkage of Ace-1, mago, and Notch was then tested using qPCR with the EF-1α as a reference in all three tortricid species examined. The Ace-1 and Notch gene doses differ significantly between males and females, suggesting their linkage to the Z chromosome (Fig. 3, Table S4). Therefore, Z chromosome–autosome fusion appears to be common to all species of subfamilies Olethreutinae and Tortricinae. Consistent with the results of BAC-FISH, the C. pomonella mago gene doses did not differ between males and females. Similar results were obtained by comparison of the mago to EF-1α gene dose ratios in L. botrana, suggesting that the mago gene is located on an autosome in both members of the subfamily Olethreutinae. However, different doses of the mago gene in males and females of E. ambiguella, a representative of the subfamily Tortricinae, indicate that this gene is located on the Z chromosome (Fig. 3, Table S4). Thus, the translocation of a chromosomal region containing the mago and RpL10 genes to an autosome, identified in the codling moth by BAC-FISH (Figs. 1 D and E and 2), has no causal link with the Z chromosome–autosome fusion. The translocation event originated independently and much later, after the divergence of the subfamilies Olethreutinae and Tortricinae.
Discussion
We performed physical mapping of the Z sex chromosome in a major pest of pome fruit, the codling moth, Cydia pomonella (Tortricidae: Olethreutinae) (Figs. 1 A–E and 2). Although genome organization of the nontineoid Ditrysia (21) was shown to be highly conserved (22–24), our results revealed that a neo-Z chromosome formed following fusion between chromosomes corresponding to the linkage groups Z and 15 of the Bombyx mori reference genome, henceforth referred to as F(Z;15), thus supporting an earlier anecdotal prediction (15). Sex-linkage of the Acetylcholinesterase 1 (Ace-1) and Notch orthologs of the B. mori chromosome 15 genes in two other tortricid pests (Fig. 3), L. botrana (Olethreutinae) and E. ambiguella (Tortricinae), strongly suggests that the F(Z;15) fusion occurred in a common ancestor of these lineages, which comprise about 97% of the tortricid species (25). The fate of the maternally inherited homolog of chromosome 15 cannot be conclusively resolved with current data sets. However, a previous molecular analysis of the codling moth W chromosome sequence library (19) along with the results of BAC-FISH (Fig. 1G) support the existence of extensive molecular degeneration of the codling moth W chromosome, ultimately leading to the loss of W-linked alleles.
Recently, resistance of the codling moth to a highly specific and virulent pathogen, Cydia pomonella granulovirus (CpGV) (Baculoviridae), has been reported. The CpGV resistance is mediated by a major gene with concentration-dependent dominance linked to the Z chromosome (26). Although other CpGV isolates were shown to overcome CpGV resistance (27, 28) caused by an early blockage of virus replication (29), its genetic basis remains elusive possibly due to false assumption of conserved gene content of the Z chromosome between B. mori and C. pomonella.
We found that three other targets for either chemical or biological insecticides, namely Resistance to dieldrin (Rdl), Ace-1, and ABC transporter C2 (ABCC2), are linked to chromosome Z in the codling moth (Figs. 2 and 3), and presumably in all other species of the tortricid subfamilies Olethreutinae and Tortricinae, which comprise almost 700 economically important pests worldwide (30). Whereas Rdl orthologs conferring resistance to cyclodiene insecticides are also Z-linked in other Lepidoptera (31, 32), the Ace-1 and ABCC2 associated with insensitivity to organophosphates and carbamates, and resistance to Bacillus thuringiensis toxin Cry1Ab, respectively, are assignable to the autosomal linkage group corresponding to B. mori chromosome 15 in distantly related species (15, 33–35). By contrast, in most tortricids, the sex-linkage of these two genes is thus a direct consequence of F(Z;15). Theory predicts that recessive mutations conferring resistance spread faster in a pest population if they are Z-linked due to their hemizygosity in the females (36).
Although ABCC2 mutations are reported to be recessive (33–35), the resistance conferred by insensitive Ace is in most cases semidominant. However, dominance levels of insensitive Ace alleles were shown to vary from recessivity to dominance and correlate with the activity of insensitive Ace forms in mosquito Culex pipiens. When activity of the resistant allele is low, heterozygotes, which possess only half the amount of insensitive Ace present in resistant homozygotes, display a lower tolerance to insecticide (37). This explanation seems to exclude the occurrence of recessive Ace-1 conferred resistance in tortricids because there would be no difference in Ace-1 activity between heterozygous males and hemizygous females due to absence of global dosage compensation in Lepidoptera (38). However, Kanga et al. (39) reported that Ace-1 insenstitivity, the major mechanism of carbamate resistance in a tortricid pest, Grapholita molesta, is both sex-linked and recessive. The recessivity of G. molesta Ace-1 insensitivity was probably facilitated by a female-specific modifier compensating for lower dosage of Ace-1, which evolved independently before resistance as suggested by Ace-1 activity ratios between sexes in both susceptible and resistant strains. Thus, the Z-linkage of both ABCC2 and Ace-1 is of importance to pest management programs attempting to delay the onset of insecticide resistance in tortricid pests.
It has recently been suggested that gene content might be relevant for maintenance of neo-sex chromosomes (40). The Ace-1 and ABCC2 genes belong to insect carboxylesterase and ATP-binding cassette (ABC) transporter gene families, whose members are involved in metabolism and regulated absorption of both insecticides and plant secondary metabolites, respectively (41–44). Along with glutathione S-transferases and cytochrome P450 monooxygenases (P450s), they represent the so-called performance genes affecting growth and survival of insect larvae on host plants (45). Recent analyses revealed an uneven distribution of performance gene clusters in the B. mori genome. In particular, chromosome 15 was shown to bear two clusters of Lepidoptera-specific esterases and a major cluster of ABC transporters (Fig. 2) (46, 47). Functions of these genes are largely unknown. However, sex-related response to organophosphates (48) correlating with sex-specific levels of general esterase activities (49) reported in G. molesta is consistent with sex-linkage and the absence of dosage compensation of involved genes. These findings suggest that the sex-linked esterases of tortricids play a role in detoxification of xenobiotics. Moreover, expansion of ABC transporters, including two genes located in B. mori chromosome 15, observed in the genome of the diamondback moth, Plutella xylostella (Yponomeutoidea), suggests their potential role in detoxification of plant secondary metabolites (50). Therefore, it is reasonable to assume that F(Z;15) physically linked a battery of performance genes to the tortricid Z chromosome.
Physical linkage between performance and either preference or host-independent isolation genes, shown to be disproportionately associated with the lepidopteran Z chromosome (51–54), is expected to generate genetic covariance between traits and thus facilitate ecological speciation under divergent selection (4, 55). Furthermore, performance genes are importantly associated with shifts in host plant utilization. Duplications and subsequent functional divergence of P450s have been reported to play a crucial role in dietary specialization of swallowtail butterflies of the genus Papilio (56). In general, duplications of performance genes are thought to be an adaptive response to environmental stress (57), a scenario well-supported by the role of gene amplification in metabolic resistance to insecticides (42, 58). Following this line of reasoning, we hereby hypothesize that duplicates of tortricid sex-linked performance genes, compensating for the loss of the W-linked alleles, were in all probability fixed as beneficial and acquired novel functions increasing the detoxification capacity of tortricid larvae. Therefore, F(Z;15) constitutes an evolutionary key innovation, potentially conferring physiological advantage in plant–herbivore interactions (59) and resulting in adaptive radiation of the species-rich tortricid subfamilies Tortricinae and Olethreutinae. Our findings thus not only contribute to management of tortricid pests but also allow a unique perspective concerning the role of neo-sex chromosomes in the adaptive radiation and ultimately speciation of phytophagous insects, a huge group of the class Insecta.
Materials and Methods
Insects.
A laboratory strain (Krym-61) of the codling moth, C. pomonella (Olethreutinae) (for its origin and diet, see ref. 18) was used. Laboratory cultures of the European grapevine moth L. botrana (Olethreutinae) and the vine moth E. ambiguella (Tortricinae), both originating from field collections in wine-growing regions in Germany, were obtained from Annette Reineke (Research Center Geisenheim, Geisenheim, Germany) along with a rearing protocol and the composition of an artificial diet. The diet was prepared according to the recipe of Christoph Hoffmann (Julius Kühn Institute, Siebeldingen, Germany). All three tortricid species were reared in a constant-temperature room under nondiapausing conditions (25 ± 1 °C; 16:8 light:dark).
Isolation of Genes for Comparative Mapping.
Genes of interest were selected from a public genome database of the silkworm, B. mori, KAIKOBase (http://sgp.dna.affrc.go.jp/KAIKO) (Table S1). Degenerate primers were designed for regions of coding sequences conserved between the B. mori genes and other insect species and used for RT-PCR amplification of partial orthologous sequences in the tortricids examined (Table S2). The primer concentrations in RT-PCR were increased to 5 µM to compensate for their degeneration. First-strand cDNA synthesized from larval total RNA by oligo-dT primed SuperScript III Reverse Transcriptase (Invitrogen) was used as a template. Amplified fragments were cloned into pGEM-T Easy Vector (Promega) and confirmed by Sanger sequencing.
Identification of BAC Clones Containing Selected Genes.
We used a copy of the codling moth BAC library constructed by GENEfinder Genomic Resource Laboratory (Texas A&M University, College Station, TX). Partial sequences of codling moth orthologs of selected B. mori genes were used as hybridization probes for screening of 18,432 C. pomonella BAC clones of average insert size 140 kbp, spotted as duplicates on high-density colony filters (obtained from GENEfinder Genomic Resources). Probes were labeled with alkali labile DIG-11-dUTP (Roche Diagnostics) using PCR and purified by gel filtration. Screening procedure followed a standard Southern hybridization protocol as described in ref. 19. Hybridization was carried out overnight at 42 °C. Positive BAC clones were confirmed by PCR with specific primers (Table S3). BAC-DNA was extracted using Qiagen Plasmid Midi Kit (Qiagen) according to the manufacturer’s instructions.
BAC-FISH Mapping.
Meiotic chromosomes were prepared from gonads of male and female larvae by the spreading technique as described in ref. 60. For FISH, BAC-DNA was labeled using a Nick Translation Kit (Abbott Molecular). Fifty microliters of labeling reaction mixture containing 1 μg of BAC-DNA and 25 μM dATP, dCTP, and dGTP each, 9 μM dTTP, and 16 μM fluorochrome-conjugated dUTP was incubated for 4 h at 15 °C. Two-color BAC-FISH with Cy3-dUTP (GE Healthcare) and ChromaTide Fluorescein-12-dUTP (Invitrogen)-labeled probes was performed following ref. 61, with some modifications. The same procedure was used for multicolor BAC-FISH, except that the probes that were labeled with Green-dUTP, Orange-dUTP, Red-dUTP (Abbott Molecular) and Cy5-dUTP (GE Healthcare). For BAC-FISH mapping, we used a reprobing protocol as described in ref. 62. Briefly, chromosome preparations were postfixed for 5 min in freshly prepared 4% formaldehyde in 2× SSC, washed twice in 2× SSC for 3 min, and incubated for 30 min in 5× Denhardt’s solution in 2× SSC shortly before their denaturation in the first FISH round. The preparations were reprobed repeatedly with different probe mixtures. After each FISH round, the chromosomes were denatured during a stripping step, and the next probe mixture was applied directly to the dehydrated and air-dried slides.
Chromosome preparations were observed either in a Zeiss Axioplan 2 microscope (Carl Zeiss) or DM6000B fluorescence microscope (Leica Microsystems) equipped with appropriate fluorescence filter sets. Black-and-white images were captured with a cooled F-View CCD camera equipped with AnalySIS software, version 3.2 (Soft Imaging System), and a DFC350FX CCD camera with Leica LAS Image Analysis software (Leica Microsystems), respectively. The images were pseudocolored and superimposed with Adobe Photoshop CS3. Image analysis was carried out using freeware ImageJ (National Institutes of Health).
Quantitative Analysis of Gene Dose.
qPCR using genomic DNA as a template was used to test sex-linkage of selected genes in the tortricid species studied. Gene doses of the target genes were compared with a single-copy autosomal (AA) reference gene, elongation factor 1α (EF-1α), in the male (AA, ZZ) and female (AA, WZ) genomes. If the target gene is autosomal, its copy number ratio to the autosomal reference gene is expected to be 1:1 in both sexes. In the case of Z-linkage, a target to autosomal reference gene dose ratio is expected to be 1:1 in males (ZZ) but 1:2 in females (WZ) (SI Text). W-linked genes should be missing completely in males.
Quantitative analysis was carried out in iQ 96-Well PCR Plates covered by Microseal “B” Adhesive Seals using the C1000 Thermal cycler CFX96 Real-Time System (Bio-Rad). Each qPCR reaction contained 1× SYBR Premix Ex Taq II (Perfect Real Time) (Takara), 0.4 µM forward and reverse primer (Table S5), and 100–150 ng of either male or female genomic DNA (gDNA) isolated from adult moths by a DNeasy Blood Tissue Kit (Qiagen). The target and reference genes were analyzed simultaneously in triplicates of three independent samples of both male and female gDNA. Default amplification efficiencies (E) of 1 were used to calculate target-to-reference gene dose ratio (R) using the formula R = (1+Etarget)CtTarget/(1+ERef)CtReference. However, if R deviated considerably from the expected value of 1:1 in males, the PCR efficiencies were determined from the slope of the standard curve generated by plotting the threshold cycle (Ct) values against the log-concentrations of serial dilutions of male genomic DNA. The obtained data were processed using CFX Manager Software (Bio-Rad), and their significance was statistically assessed by unpaired two-tailed t test for unequal variances. The t test was used to test null hypothesis of no difference or a twofold difference in the means between males and females.
Supplementary Material
Acknowledgments
We thank A. Reineke for cultures of L. botrana and E. ambiguella, D. G. Heckel for encouraging discussion at the early stage of this work, H. D. Loxdale for critical reading, and M. Korchová for technical assistance. This research was part of Entomology Institute Project Z50070508 and part of a Coordinated Research Project of the International Atomic Energy Agency, Vienna (Research Agreement 15838). Experiments were funded by Grant 523/09/2106 of the Czech Science Foundation and Grant IAA600960925 of the Grant Agency of the Academy of Sciences of the Czech Republic. Additional support was provided from the Building up Modern Biotechnologies for Agriculture project (Registration 229518, European Union Program FP7-REGPOT-2008-1) and Grant 137/2010/P of the Grant Agency of the University of South Bohemia. K.S. was supported from Japan Society for the Promotion of Science Grant 23380030 and L.G.N. from US Department of Agriculture-Agricultural Research Service base funds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JQ771334, JQ771335, JQ771337, JQ771338, JQ771339, JQ771341, JQ771343, JQ771344, JQ771346, JQ771353, JQ771354, JQ771355, JQ771357, JQ771358, JQ771360–JQ771363, JQ771368, JQ771369, JX258662, JX258665–JX258668, and JX307647).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220372110/-/DCSupplemental.
References
- 1.Faria R, Navarro A. Chromosomal speciation revisited: Rearranging theory with pieces of evidence. Trends Ecol Evol. 2010;25(11):660–669. doi: 10.1016/j.tree.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Lukhtanov VA, Dincă V, Talavera G, Vila R. Unprecedented within-species chromosome number cline in the Wood White butterfly Leptidea sinapis and its significance for karyotype evolution and speciation. BMC Evol Biol. 2011;11:109. doi: 10.1186/1471-2148-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butlin RK. Recombination and speciation. Mol Ecol. 2005;14(9):2621–2635. doi: 10.1111/j.1365-294X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 4.Hawthorne DJ, Via S. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature. 2001;412(6850):904–907. doi: 10.1038/35091062. [DOI] [PubMed] [Google Scholar]
- 5.Servedio MR. The role of linkage disequilibrium in the evolution of premating isolation. Heredity (Edinb) 2009;102(1):51–56. doi: 10.1038/hdy.2008.98. [DOI] [PubMed] [Google Scholar]
- 6.Wiley C, Shaw KL. Multiple genetic linkages between female preference and male signal in rapidly speciating Hawaiian crickets. Evolution. 2010;64(8):2238–2245. doi: 10.1111/j.1558-5646.2010.01007.x. [DOI] [PubMed] [Google Scholar]
- 7.Merrill RM, Van Schooten B, Scott JA, Jiggins CD. Pervasive genetic associations between traits causing reproductive isolation in Heliconius butterflies. Proc Biol Sci. 2011;278(1705):511–518. doi: 10.1098/rspb.2010.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellegren H. Sex-chromosome evolution: Recent progress and the influence of male and female heterogamety. Nat Rev Genet. 2011;12(3):157–166. doi: 10.1038/nrg2948. [DOI] [PubMed] [Google Scholar]
- 9.Presgraves DC. Patterns of postzygotic isolation in Lepidoptera. Evolution. 2002;56(6):1168–1183. doi: 10.1111/j.0014-3820.2002.tb01430.x. [DOI] [PubMed] [Google Scholar]
- 10.Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24(7):336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitano J, Peichel CL. Turnover of sex chromosomes and speciation in fishes. Environ Biol Fishes. 2012;94(3):549–558. doi: 10.1007/s10641-011-9853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traut W, Sahara K, Marec F. Sex chromosomes and sex determination in Lepidoptera. Sex Dev. 2007;1(6):332–346. doi: 10.1159/000111765. [DOI] [PubMed] [Google Scholar]
- 13.Yoshido A, Marec F, Sahara K. Resolution of sex chromosome constitution by genomic in situ hybridization and fluorescence in situ hybridization with (TTAGG)(n) telomeric probe in some species of Lepidoptera. Chromosoma. 2005;114(3):193–202. doi: 10.1007/s00412-005-0013-9. [DOI] [PubMed] [Google Scholar]
- 14.Yoshido A, Sahara K, Marec F, Matsuda Y. Step-by-step evolution of neo-sex chromosomes in geographical populations of wild silkmoths, Samia cynthia ssp. Heredity (Edinb) 2011;106(4):614–624. doi: 10.1038/hdy.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heckel DG, Bryson PK, Brown TM. Linkage analysis of insecticide-resistant acetylcholinesterase in Heliothis virescens. J Hered. 1998;89:71–78. [Google Scholar]
- 16.Cassanelli S, Reyes M, Rault M, Carlo Manicardi G, Sauphanor B. Acetylcholinesterase mutation in an insecticide-resistant population of the codling moth Cydia pomonella (L.) Insect Biochem Mol Biol. 2006;36(8):642–653. doi: 10.1016/j.ibmb.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 17.Fuková I, et al. Rapid assessment of the sex of codling moth Cydia pomonella (Linnaeus) (Lepidoptera: Tortricidae) eggs and larvae. J Appl Entomol. 2009;133(4):249–261. [Google Scholar]
- 18.Fuková I, Nguyen P, Marec F. Codling moth cytogenetics: Karyotype, chromosomal location of rDNA, and molecular differentiation of sex chromosomes. Genome. 2005;48(6):1083–1092. doi: 10.1139/g05-063. [DOI] [PubMed] [Google Scholar]
- 19.Fuková I, et al. Probing the W chromosome of the codling moth, Cydia pomonella, with sequences from microdissected sex chromatin. Chromosoma. 2007;116(2):135–145. doi: 10.1007/s00412-006-0086-0. [DOI] [PubMed] [Google Scholar]
- 20.Regier JC, et al. A molecular phylogeny for the leaf-roller moths (Lepidoptera: Tortricidae) and its implications for classification and life history evolution. PLoS ONE. 2012;7(4):e35574. doi: 10.1371/journal.pone.0035574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mutanen M, Wahlberg N, Kaila L. Comprehensive gene and taxon coverage elucidates radiation patterns in moths and butterflies. Proc Biol Sci. 2010;277(1695):2839–2848. doi: 10.1098/rspb.2010.0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baxter SW, et al. Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS ONE. 2011;6(4):e19315. doi: 10.1371/journal.pone.0019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heliconius Genome Consortium Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487(7405):94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van't Hof AE, et al. Linkage map of the peppered moth, Biston betularia (Lepidoptera, Geometridae): A model of industrial melanism. Heredity (Edinb) 2012;110(3):283–295. doi: 10.1038/hdy.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilligan TM, Baixeras J, Brown JW, Tuck KR. T@RTS: Online World Catalogue of the Tortricidae (Ver. 2.0) 2012. Available at www.tortricid.net/catalogue.asp. Accessed February 6, 2013.
- 26.Asser-Kaiser S, et al. Rapid emergence of baculovirus resistance in codling moth due to dominant, sex-linked inheritance. Science. 2007;317(5846):1916–1918. doi: 10.1126/science.1146542. [DOI] [PubMed] [Google Scholar]
- 27.Eberle KE, Asser-Kaiser S, Sayed SM, Nguyen HT, Jehle JA. Overcoming the resistance of codling moth against conventional Cydia pomonella granulovirus (CpGV-M) by a new isolate CpGV-I12. J Invertebr Pathol. 2008;98(3):293–298. doi: 10.1016/j.jip.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Berling M, et al. Cydia pomonella granulovirus genotypes overcome virus resistance in the codling moth and improve virus efficiency by selection against resistant hosts. Appl Environ Microbiol. 2009;75(4):925–930. doi: 10.1128/AEM.01998-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asser-Kaiser S, Radtke P, El-Salamouny S, Winstanley D, Jehle JA. Baculovirus resistance in codling moth (Cydia pomonella L.) caused by early block of virus replication. Virology. 2011;410(2):360–367. doi: 10.1016/j.virol.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Zhang BC. Index of Economically Important Lepidoptera. Wallingford, UK: CAB International; 1994. [Google Scholar]
- 31.Daly JC, Fisk JH. Sex-linked inheritance of endosulphan resistance in Helicoverpa armigera. Heredity (Edinb) 1998;81(1):55–62. [Google Scholar]
- 32.Yuan GR, Gao WY, Yang YH, Wu YD. Molecular cloning, genomic structure, and genetic mapping of two Rdl-orthologous genes of GABA receptors in the diamondback moth, Plutella xylostella. Arch Insect Biochem Physiol. 2010;74(2):81–90. doi: 10.1002/arch.20361. [DOI] [PubMed] [Google Scholar]
- 33.Gahan LJ, Pauchet Y, Vogel H, Heckel DG. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010;6(12):e1001248. doi: 10.1371/journal.pgen.1001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baxter SW, et al. Parallel evolution of Bacillus thuringiensis toxin resistance in Lepidoptera. Genetics. 2011;189(2):675–679. doi: 10.1534/genetics.111.130971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atsumi S, et al. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc Natl Acad Sci USA. 2012;109(25):E1591–E1598. doi: 10.1073/pnas.1120698109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orr HA. The population genetics of beneficial mutations. Philos Trans R Soc Lond B Biol Sci. 2010;365(1544):1195–1201. doi: 10.1098/rstb.2009.0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bourguet D, et al. Variation of dominance of newly arisen adaptive genes. Genetics. 1997;147(3):1225–1234. doi: 10.1093/genetics/147.3.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison PW, Mank JE, Wedell N. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol Evol. 2012;4(11):1118–1126. doi: 10.1093/gbe/evs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanga LHB, Pree DJ, Plapp FW, van Lier JL. Sex-linked altered acetylcholinesterase resistance to carbamate insecticides in adults of the oriental fruit moth, Grapholita molesta (Lepidoptera:Tortricidae) Pestic Biochem Physiol. 2001;71(1):29–39. [Google Scholar]
- 40.Pala I, et al. Evidence of a neo-sex chromosome in birds. Heredity (Edinb) 2012;108(3):264–272. doi: 10.1038/hdy.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorensen JS, Dearing MD. Efflux transporters as a novel herbivore countermechanism to plant chemical defenses. J Chem Ecol. 2006;32(6):1181–1196. doi: 10.1007/s10886-006-9079-y. [DOI] [PubMed] [Google Scholar]
- 42.Li XC, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 43.Buss DS, Callaghan A. Interaction of pesticides with p-glycoprotein and other ABC proteins: A survey of the possible importance to insecticide, herbicide and fungicide resistance. Pestic Biochem Physiol. 2008;90(3):141–153. [Google Scholar]
- 44.Zangerl AR, Liao LH, Jogesh T, Berenbaum MR. Aliphatic esters as targets of esterase activity in the parsnip webworm (Depressaria pastinacella) J Chem Ecol. 2012;38(2):188–194. doi: 10.1007/s10886-012-0073-2. [DOI] [PubMed] [Google Scholar]
- 45.Berenbaum MR, Feeny PP. In: Specialization, Speciation and Radiation: The Evolutionary Biology of Herbivorous Insects. Tilmon KJ, editor. Berkeley, CA: Univ California Press; 2008. pp. 3–19. [Google Scholar]
- 46.Yu QY, Lu C, Li WL, Xiang ZH, Zhang Z. Annotation and expression of carboxylesterases in the silkworm, Bombyx mori. BMC Genomics. 2009;10:553. doi: 10.1186/1471-2164-10-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie X, et al. Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in the silkworm, Bombyx mori. Mol Biol Rep. 2012;39(7):7281–7291. doi: 10.1007/s11033-012-1558-3. [DOI] [PubMed] [Google Scholar]
- 48.Shearer PW, Usmani KA. Sex-related response to organophosphorus and carbamate insecticides in adult Oriental fruit moth, Grapholita molesta. Pest Manag Sci. 2001;57(9):822–826. doi: 10.1002/ps.367. [DOI] [PubMed] [Google Scholar]
- 49.de Lame FM, Hong JJ, Shearer PW, Brattsten LB. Sex-related differences in the tolerance of Oriental fruit moth (Grapholita molesta) to organophosphate insecticides. Pest Manag Sci. 2001;57(9):827–832. doi: 10.1002/ps.368. [DOI] [PubMed] [Google Scholar]
- 50.You M, et al. A heterozygous moth genome provides insights into herbivory and detoxification. Nat Genet. 2013;45(2):220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- 51.Thompson JN. Evolutionary genetics of oviposition preference in swallowtail butterflies. Evolution. 1988;42(6):1223–1234. doi: 10.1111/j.1558-5646.1988.tb04182.x. [DOI] [PubMed] [Google Scholar]
- 52.Sperling FAH. Sex-linked genes and species-differences in Lepidoptera. Can Entomol. 1994;126(3):807–818. [Google Scholar]
- 53.Prowell DP. In: Endless Forms: Species and Speciation. Howard DJ, Berlocher SH, editors. Oxford: Oxford Univ Press; 1998. pp. 309–319. [Google Scholar]
- 54.Nygren GH, Nylin S, Stefanescu C. Genetics of host plant use and life history in the comma butterfly across Europe: Varying modes of inheritance as a potential reproductive barrier. J Evol Biol. 2006;19(6):1882–1893. doi: 10.1111/j.1420-9101.2006.01174.x. [DOI] [PubMed] [Google Scholar]
- 55.Matsubayashi KW, Ohshima I, Nosil P. Ecological speciation in phytophagous insects. Entomol Exp Appl. 2010;134(1):1–27. [Google Scholar]
- 56.Li WM, Schuler MA, Berenbaum MR. Diversification of furanocoumarin-metabolizing cytochrome P450 monooxygenases in two papilionids: Specificity and substrate encounter rate. Proc Natl Acad Sci USA. 2003;100(Suppl 2):14593–14598. doi: 10.1073/pnas.1934643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3:0008.1. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bass C, Field LM. Gene amplification and insecticide resistance. Pest Manag Sci. 2011;67(8):886–890. doi: 10.1002/ps.2189. [DOI] [PubMed] [Google Scholar]
- 59.Ehrlich PR, Raven PH. Butterflies and plants: A study in coevolution. Evolution. 1964;18(4):586–608. [Google Scholar]
- 60.Mediouni J, Fuková I, Frydrychová R, Dhouibi MH, Marec F. Karyotype, sex chromatin and sex chromosome differentiation in the carob moth, Ectomyelois ceratoniae (Lepidoptera: Pyralidae) Caryologia. 2004;57(2):184–194. [Google Scholar]
- 61.Yoshido A, Bando H, Yasukochi Y, Sahara K. The Bombyx mori karyotype and the assignment of linkage groups. Genetics. 2005;170(2):675–685. doi: 10.1534/genetics.104.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shibata F, Sahara K, Naito Y, Yasukochi Y. Reprobing multicolor FISH preparations in lepidopteran chromosome. Zoolog Sci. 2009;26(3):187–190. doi: 10.2108/zsj.26.187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.