Abstract
Conjugation is one of the most common ways bacteria acquire antibiotic resistance, contributing to the emergence of multidrug-resistant “superbugs.” Bacteria of the genus Enterococcus faecalis are highly antibiotic-resistant nosocomial pathogens that use the mechanism of conjugation to spread antibiotic resistance between resistance-bearing donor cells and resistance-deficient recipient cells. Here, we report a unique quorum sensing-based communication system that uses two antagonistic signaling molecules to regulate conjugative transfer of tetracycline-resistance plasmid pCF10 in E. faecalis. A “mate-sensing” peptide sex pheromone produced by recipient cells is detected by donor cells to induce conjugative genetic transfer. Using mathematical modeling and experimentation, we show that a second antagonistic “self-sensing” signaling peptide, previously known to suppress self-induction of donor cells, also serves as a classic quorum-sensing signal for donors that functions to reduce antibiotic-resistance transfer at high donor density. This unique form of quorum sensing may provide a means of limiting the spread of the plasmid and present opportunities to control antibiotic-resistance transfer through manipulation of intercellular signaling, with implications in the clinical setting.
During the past two decades, Enterococcus faecalis, a normal commensal in the intestinal tract (1), has emerged as a major nosocomial pathogen largely because of acquisition of genetic determinants for antibiotic resistance and virulence via horizontal gene transfer, especially through the efficient transfer of the conjugative pheromone-responsive plasmids, exemplified by pCF10 (2, 3). Donor cells carrying pCF10 are induced to high expression of conjugative transfer and virulence genes by a heptapeptide (LVTLVFV)-mating pheromone cCF10 (C) (4). Donor cells import C into the cytoplasm, where its binding to the pCF10-encoded master regulator Prg X [the protein product of pheromone responsive gene X (prgX)] abolishes repression of transcription of the prgQ operon encoding the conjugation genes (Fig. 1) (5, 6). The direct effect of C on pheromone responsive gene Q (prgQ) transcription is enhanced greatly by several co- as well as posttranscriptional mechanisms, whose cumulative effects cause the system to function as a bistable genetic switch (7). It also is noteworthy that pheromone induction increases the virulence of donor strains (8).
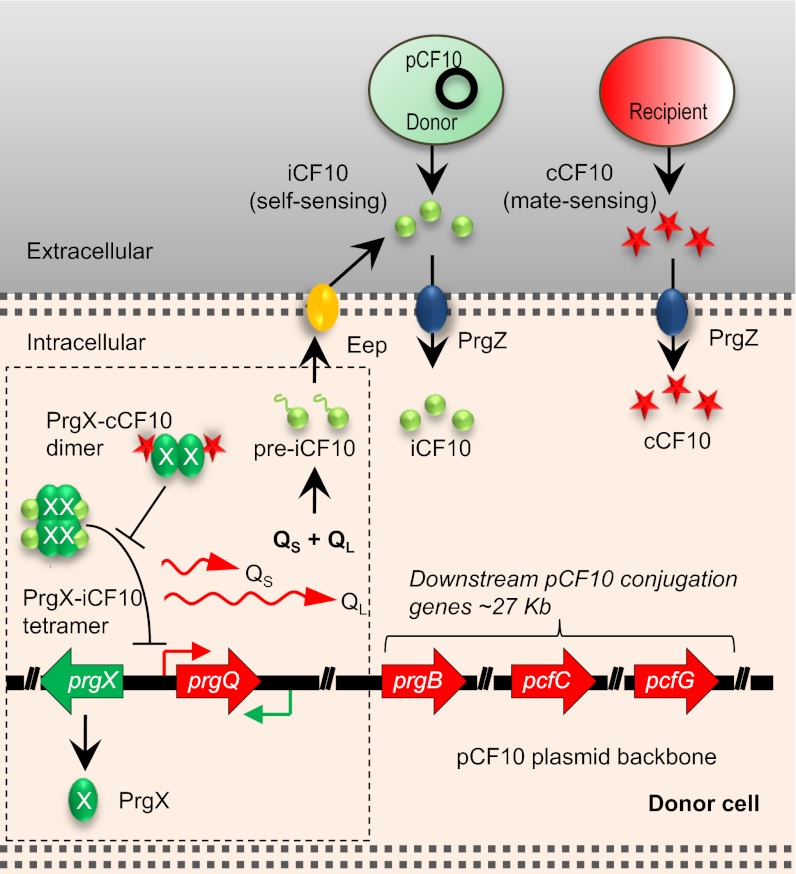
Fig. 1.
Control of conjugation by two antagonistic peptide signals in E. faecalis. Signaling molecules cCF10 (C) and iCF10 (I) are released by recipient and donor cells, respectively, and imported into the donor cell, where these signals compete for binding to PrgX; PrgX negatively regulates the promoter for the prgQ operon, encoding conjugation functions. Whereas PrgX/I complexes repress transcription initiation, PrgX/C complexes do not. In uninduced donor cells, basal transcription of prgQ generates a short-transcript QS. Induced cells express higher levels of QS, as well as extended transcripts such as QL and other longer RNAs. Both QS and QL encode for the 22-aa preiCF10 polypeptide, which is processed into I and secreted into the growth medium. The opposing and partially overlapping prgX operon encodes PrgX protein and a small RNA Anti-Q that promotes termination of prgQ transcription. The unique organization of these two operons provides several layers of co- as well as posttranscriptional regulation that allow the system to function as a sensitive biological switch. In mixed cultures at low recipient cell density, the switch is off, whereas at high recipient cell density, sufficient C is produced to induce prgQ transcription, generating QL RNA and longer transcripts encoding downstream genes, including prgB (donor/recipient aggregation), pcfC (coupling of transferred DNA to the transfer machinery), and pcfG (relaxase, which nicks the plasmid DNA to initiate the transfer process).
The mating response of donor cells to C is encoded by pCF10, whereas C is produced from a conserved chromosomal gene present in most if not all E. faecalis strains (4). To suppress self-induction by endogenously produced C in donors, a heptapeptide (AITLIFI) inhibitor molecule iCF10 (I) is encoded by the first gene in the polycistronic prgQ operon of pCF10 (9, 10). Both C and I are synthesized initially as prepeptides that, after cleavage of the leader sequence, are secreted into the extracellular environment as active peptides (9) (Fig. 1 and Fig. S1A). Once taken up by donor cells, I competes with C for binding to PrgX; the two peptides bind to the same subdomain of PrgX but have opposing effects on PrgX structure and function (11). It has been suggested that in the absence of recipient cells, the basal level expression of I keeps the expression of conjugation genes OFF in donor cells (11, 12). When recipient cells are in close proximity, the increased C level overcomes the inhibition of I, thus allowing donor cells to mount a mating response.
Although many studies of the pheromone induction process have been done, relatively little is known about the requirements for the return of the system to the “off” state following an induction cycle. In this study, we describe the results of experiments analyzing the turning off of conjugation, especially the role of I in the process. These results led to the discovery of the much broader role of I in the social behavior of donor cells, including its function as a unique quorum-sensing signal that inhibits conjugation. Our results suggest that these two antagonistic signals function in constraining the dissemination of the plasmid in mixed populations of donors and recipients. Manipulation of the newly described quorum-sensing circuit has potential as a therapeutic strategy to reduce resistance transfer and virulence in vivo.
Results
Pheromone Induction Generates a Burst of Transcription Through the prgQ Operon.
Conjugative transfer of pCF10 is initiated by the induction of the prgQ operon that encodes I and the conjugative machinery required for plasmid transfer (Fig. 1). In uninduced cells, basal transcription from the PQ promoter generates a 380-nt transcript, QS, which includes a single ORF (prgQ) encoding a 22-aa polypeptide processed into I and secreted into the growth medium (10). An increased intracellular level of C, resulting from addition of exogenous C or C-producing recipient cells, shifts the PrgX structure and oligomerization state from a repressing to a nonrepressing conformation, resulting in induction (11, 12). Induced donor cells contain increased levels of QS, as well as longer transcripts (Fig. 1), including the 530-nt QL and other mRNAs extending >10 kb into the operon (13, 14). These longer transcripts encode conjugation proteins (Fig. 1).
Because donor cells become competent for conjugation only when QL and longer prgQ transcripts are expressed (13–15), we examined the dynamics of turning ON and OFF of conjugation by using quantitative RT-PCR (qRT-PCR, Table S1) to measure levels of these transcripts following exposure to various levels of C. QL and transcripts of three downstream conjugation genes, prgB, pcfC, and pcfG (Fig. 1), showed similar rapid increases to maximum levels within 15–30 min of exposure of pCF10-containing donor cells to various amounts of C (Fig. 2 A–D). The expression of all four transcripts increased with increasing concentrations of C, but expression of all the genes began to decrease after 30 min. It is remarkable that all four genes showed a similar rapid return to basal expression levels following induction. The rapid turning off is not the result of a low level of inducer, as even at a saturation level of 50 ng/mL C (about 100× the concentration normally seen in mating experiments), the transcription was turned off after a short burst (4). The dynamic response after induction was also affirmed by RNA sequencing (Figs. S2 and S3) and Isobaric tag for relative and absolute quantitation (iTRAQ) (Figs. S4 and S5).
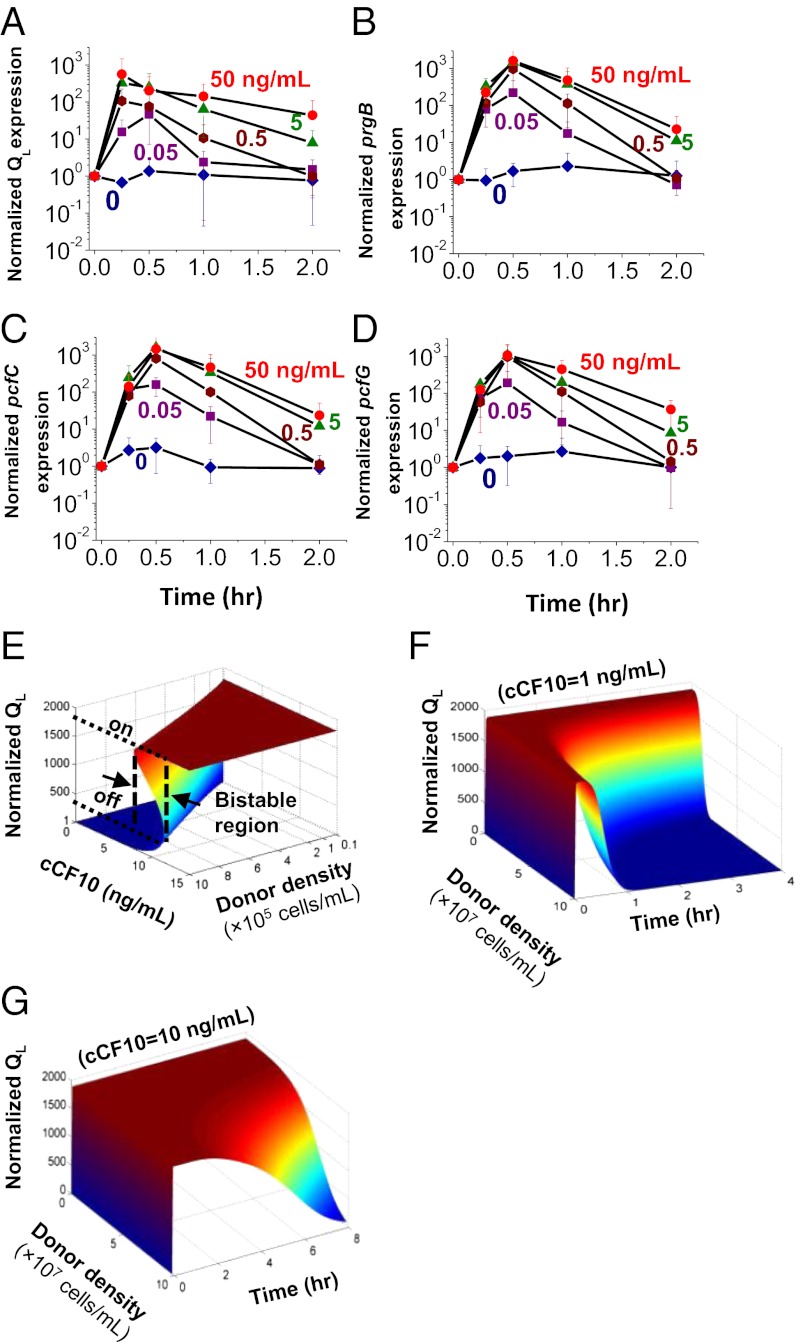
Fig. 2.
Switch behavior of pCF10-carrying donor cells in response to pheromone induction. (A–D) qRT-PCR analysis of the dynamics of expression of conjugation determinants encoded by the prgQ operon in response to pheromone induction. mRNA was purified from cultures of pCF10-containing donors exposed to different concentrations of C for various periods. Transcript levels are shown for QL (A), prgB (B), pcfC (C), and pcfG (D), normalized relative to a transcript from a constitutive chromosomal gene, gyrB, and to time t = 0. Data shown in A–D are averages of three independent experiments (error bars are SDs from mean values). (E–G) Mathematical modeling of the pCF10-encoded pheromone response. (E) The steady-state response of QL (normalized to the off state) to induction demonstrates a characteristic bistable switch behavior. The bistable region shifts to a higher level of cCF10 with increasing donor density, accompanied by an increasing threshold level of cCF10 to turn on the conjugative genes. (F and G) Simulated dynamic response of QL transcript level (normalized to the off state at time t = 0) to C induction at various donor cell densities and at two different C concentrations: 1 ng/mL (F), and 10 ng/mL (G). At low donor density, QL is sustained at a high level over a longer period. At a high donor density, it decreases rapidly after a short period of high-level expression. At a high level of induction (10 ng/mL) corresponding to high a level of recipient cell density, QL stays high for a longer period.
Mathematical Modeling of the Pheromone Response Dynamics.
The fact that I first is secreted by donor cells then reimported to exert its conjugation suppression effect prompted us to hypothesize that I is a quorum-sensing signal (16, 17). To explore this notion further, we used a mathematical model describing the gene-regulatory circuit as well as population dynamics of donor, recipient, and transconjugant (recipient cells that acquire pCF10 by conjugation) cells (Fig. 1 and Fig. S1 A–C). We note that our previous modeling work reporting bistable switch characteristics of the prgQ operon in response to C (7) did not incorporate a role for I as a quorum sensor. In the current model, the concentration of I is allowed to vary, as in the case of changing donor cell density. The state of conjugation induction then is evaluated by the transcript level of QL (Fig. 2 E and F). The mathematical model incorporated the underlying genetic regulation of pCF10, which entails nonlinear interactions of sense:antisense transcripts within prgQ and prgX operons and interactions of PrgX with C and I (7, 11, 18) and is described in detail in Supporting Information. The parameter values were obtained from experimental results and from the literature (Table S2) (7).
The steady-state simulation showed a bistable switch-like response of QL RNA to induction with pheromone, over a wide range of donor densities. The bistable region, or the region in which two steady states coexist, changes with donor cell density (Fig. 2E). For donors originally at an off state and a cell density of 106 cells per milliliter, QL transcription remains at basal level (off) with increasing C, until C reaches 12 ng/mL; at that point, the steady-state transcript level of QL increases to that of an “on” state. On shifting down of donor cells from the on state, QL remains at an on level until C decreases to 8 ng/mL. As donor density increases, so does the concentration of I, causing the bistable region to shift toward a higher concentration of C; thus, a higher C concentration is required for donors to switch to an on state (Fig. 2E).
Upon induction, the rapid increase in prgQ transcription increases production of I, shifting the relative concentrations of C and I, and ultimately returning PrgX to the repressive conformation. At higher donor densities, the increase in I levels is faster. Donor cell density therefore also affects the duration of the on state: at a low densities, donor cells remained on for a long time (Fig. 2 F and G). At high densities, conjugation was turned off rapidly after induction (Fig. 2 F and G). As expected, the donor response also was influenced by recipient cell concentration, as illustrated by simulation at different C concentrations (Fig. 2 F and G). For any given donor cell density, a faster turning-off response was predicted at low recipient densities. Taken together, the model simulation results suggest that I is pivotal in regulating the turning off of conjugation and that I might function as a quorum-sensing signal of donor population density.
iCF10 Functions as a Quorum Sensor of Donor Population Density.
Next, we carried out experiments to test the predictions of the model by examining the effects of donor density on the induction of conjugation. Fig. 3 depicts the results of mating experiments between donors and recipients in which the recipient population density was at the same high level in all cases but the donor populations were varied over a 100-fold range. The donors were uninduced initially, and the emergence of transconjugants was measured over time. In all three conditions, the appearance of transconjugants lagged for about an hour as C accumulated in the growth medium. At 3 h, the ratio of transconjugant to donor increased to 0.12 in matings with an equal number of recipients and donors. Remarkably, increasing the donor concentration by 10-fold suppressed the transfer of pCF10—no significant induction of transfer occurred—whereas in decreasing donor concentration to a recipient-to-donor ratio of 10, the number of transconjugants per donor increased. Thus, a high donor concentration had a suppressive effect on the conjugative transfer of pCF10 (Fig. 3).
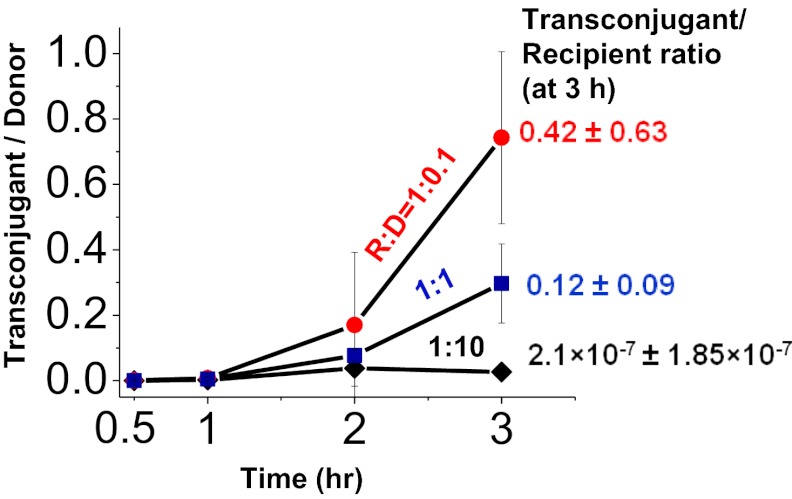
Fig. 3.
Conjugation efficiency decreases at high donor densities. Recipient cells (∼3.5 × 107 cells per milliliter) were mixed with different amounts of donor cells such that the recipient-to-donor ratios (R:D) ranged from ∼1:10 to 1:0.1. Transconjugants, recipients, and donors were enumerated on selective agar medium after 0.5, 1, 2, and 3 h of coculture. The number of transconjugants formed per donor cell decreased with increasing donor density. Data shown are the average of at least three independent experiments (error bars are SDs from mean values).
The reduced efficiency of conjugation at a high donor-to-recipient cell ratio likely is the result of a faster turning off of induction. We analyzed the time dynamics of induction at high and low donor densities. Low-density donor cultures showed a stronger and more sustained response to C over a concentration range spanning 100-fold (Fig. 4A). In high-density cultures, the induction subsided rapidly, especially when the inducer C level was low (Fig. 4B). This is similar to the results shown in Fig. 2, which also were obtained at relatively high population densities.
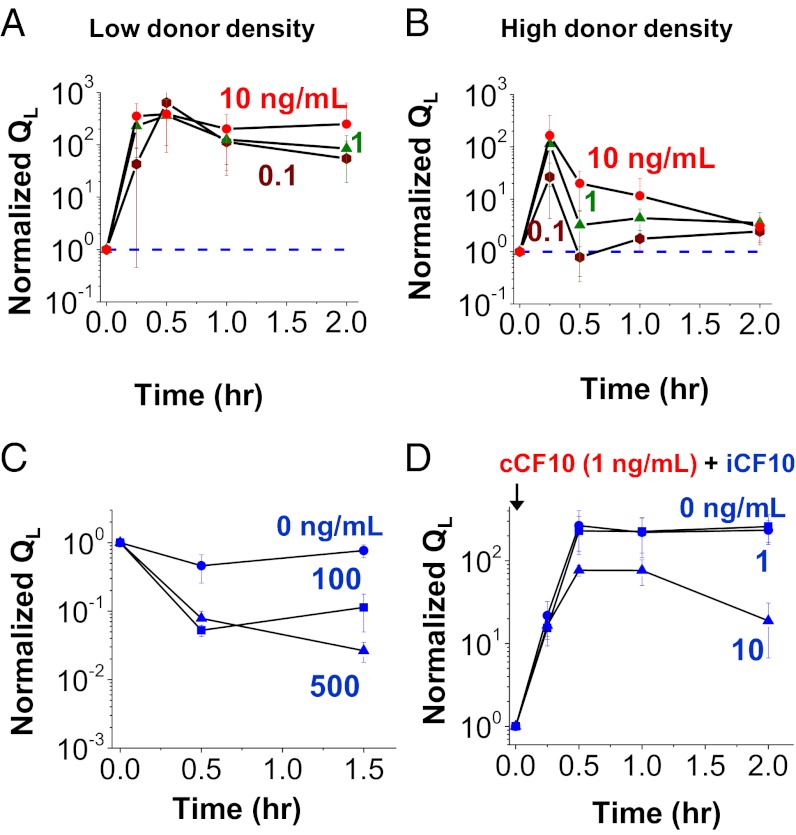
Fig. 4.
I is responsible for the decreased pheromone response of high-density donor cultures and the return of donor conjugation to the off state. (A and B) The dynamics of turning on and turning off of conjugation in wild-type donor cells are altered by donor population density. Low-density (A) and high-density (B) cultures of wild-type donors were induced with various concentrations of C, and QL expression levels were measured by qRT-PCR. (C) Expression of QL in a donor with pCF10IdT. I was added at different concentrations. (D) Expression of QL in JRC101 carrying pCF10IdT. JRC101 does not produce C. C was added to induce conjugation gene expression. I was added at different concentrations along with C. At high I (10 ng/mL), the QL expression at the end of 2 h was significantly lower than at 1 h (P = 0.049), whereas at low I (1 ng/mL), QL expression did not decrease significantly compared with 1 h (P = 0.279).
We hypothesized that the rapid shut-off of the response at high density was caused by increased levels of I in the high-density cultures. We demonstrated this by evaluating the effect of I concentration directly. We constructed plasmid pCF10IdT, which is identical to pCF10, except it carries a deletion of the codon for the third amino acid residue of I peptide; this abolished I activity. When introduced into a wild-type host, the resulting OG1RF-pCF10IdT donor exhibited a de-repressed phenotype with a high QL expression because of the basal level of C production from the chromosome. Cultures of the mutant strain aggregated as the result of constitutive expression of PrgB and grew poorly unless a basal level of I was added exogenously (19). Upon induction with exogenous C, QL remained at the induced level in the absence of I. With a sufficiently high level of I, QL decreased rapidly (Fig. 4C). We then introduced pCF10IdT into JRC101, a host strain that does not produce C. The resulting donor strain did not express QL in the absence of exogenously added C, and the cells grew normally. Induction with 1 ng/mL of C caused QL to increase more than 400-fold. QL remained at the induced level in the absence or with a low level of I (1 ng/mL), but decreased rapidly when I concentrations of 10 ng/mL were added exogenously (Fig. 4D). A similar effect of exogenous I in suppressing QL after induction also was observed in a strain containing a mutation disrupting the Eep protease, which abolishes processing the preI to produce mature I (Fig. S6).
These results strongly support the hypothesis that I is responsible for the suppressive effect of donor density on conjugation, acting both to interfere with induction and to increase the rate at which the system is shut off following induction. I thus functions as a classic quorum-sensing signal that allows donor cells to monitor their own density and calibrate their response to pheromone induction according to the relative abundance of recipients and donors.
Discussion
We demonstrate here that both the induction of conjugation and its subsequent shutting off in the E. faecalis sex pheromone response are affected by donor cell densities in an I-dependent fashion. We propose a model in which pCF10 conjugation is controlled by a dual cell–cell signaling system encompassing a traditional self-sensing signaling molecule (I) and a mate-sensing signaling molecule (C).
Quorum sensing in bacteria controls a wide variety of cellular processes (16, 17), including sporulation (20), competence (21), biofilm development (22), and virulence (23, 24). Quorum sensing is known to enhance conjugation in other bacteria, such as Agrobacterium tumefaciens, where an acyl-homoserine lactone signaling circuit activates conjugation at high cell density (25). In E. faecalis, we demonstrated the contrary, discovering a critical role for I as both an essential downstream effector in turning off the mating response to C and in blocking the ability of donors to initiate a mating response at high population density. Thus, in E. faecalis, quorum sensing has the ability to negatively affect conjugation frequencies.
At first glance, the existence of a plasmid-encoded product that hinders the proliferation of the plasmid seems counterintuitive if the plasmid is viewed as a “selfish” mobile genetic element evolved to disseminate via lateral transfer. Although the resistance and virulence genes carried by plasmids such as pCF10 may confer strong fitness advantages under selective conditions (e.g., during an opportunistic infection of an antibiotic-treated hospital patient), significant fitness costs likely are associated with pCF10 carriage under less selective conditions (e.g., during commensal intestinal growth in a healthy individual) in terms of plasmid carriage, expression of antibiotic resistance, and expression of conjugation genes (26). Replication and maintenance of the plasmid comprises some metabolic burden, and induction of a conjugation response requires the transcription and translation of more than 25 genes, as well as the assembly of a complex, ATP-dependent type IV secretion system to transfer the plasmid to recipients (27). Uncontrolled induction of conjugative genes not only is wasteful but also causes donor cells to aggregate because of the expression of PrgB protein. Whereas aggregation with recipient cells enhances mating, self-aggregation by donor cells at low recipient density only reduces growth rate and extracts extra metabolic costs. The calibrated response to both donor and recipient density in both turning on and shutting off of conjugative gene expression might provide a fitness advantage.
The self-sensing I signal also may play a role in restricting the spread of the plasmid in natural populations containing mixtures of donors and recipients. This would ensure that the population is endowed with antibiotic resistance while maintaining a subpopulation free from the metabolic burden of harboring a particular plasmid. Although this hypothesis requires further validation, the idea of a balanced, mixed population is supported by mathematical modeling (Fig. S7) and experimental evidence (Fig. S8) for the lack of complete conversion of recipients to donors in prolonged matings.
The dual signaling mechanism reported here may be rather common in nature, as it confers fitness advantages to the bacterial species in the face of multiple and sometimes opposing selective pressures. A new understanding of the interplay between the two signals may reveal means of interrupting its control circuit and offer new possibilities for suppressing virulence and resistance transfer in the clinical setting.
Materials and Methods
Bacterial Strains.
The bacterial strains used in this study all were derived from E. faecalis strain OG1RF, whose complete genome sequence has been reported (28). In mating studies, OG1RFSSp was used as the recipient cell and strain OG1RF carrying plasmid pCF10 (denoted as OG1RF-pCF10) was used as the donor cell. Strain OG1RF∆Eep, containing an in-frame deletion in the eep gene, and strain JRC101, in which C synthesis is rendered inoperative, were described previously (29). The construction of plasmid pCF10IdT, which produces an inactive I because of a T deletion, and the strains OG1RF-pCF10IdT and JRC101- pCF10IdT are described in detail in SI Materials and Methods.
Culture Conditions.
All liquid cultures were grown in M9 medium containing 0.3% yeast extract, 1% casamino acids, 20 mM glucose, 1 mM MgSO4, and 0.1 mM CaCl2. For enumeration of bacterial populations, we used Brain Heart Infusion agar medium containing antibiotics selective for donors [rifampicin, 200 µg/mL; tetracycline (Tet), 10 µg/mL], recipients [spectinomycin (Spec), 1,000 µg/mL], and transconjugants (Spec, Tet).
To examine the expression of conjugation genes following C induction, overnight cultures of wild-type donors were diluted 1:10 and split into different tubes containing different concentrations of cCF10 and/or iCF10. Cell samples were collected at different time points, followed by the addition of RNAprotect Bacteria Reagent (Qiagen), and subjected to RNA extraction.
To examine the dynamic response of donor cells to pheromone (C) at varying cell densities (Fig. 4 A and B), overnight cultures of donors were spun down and washed once in 1 mL PBS containing 2 mM EDTA. Cells were diluted 1:10 (high donor density) or 1:1,000 (low donor density) into 4.5 mL M9 medium. Cultures were incubated for 1 h at 37 °C before cCF10 was added at different concentrations. Thereafter, the cultures were incubated on a shaker at 37 °C. Samples of cells were spun down, washed once in 1 mL PBS containing 2 mM EDTA. Samples were treated with RNAProtect Bacteria Reagent (Qiagen) before RNA extraction.
Mathematical Modeling.
The mathematical model for conjugative transfer of pCF10 plasmid between E. faecalis donor–recipient cells consists of two main parts: a molecular model describing the regulation of conjugation of pCF10 in donor cells and a model depicting the interactions of donor and recipient cells. The first part of the model was described previously and shown to give rise to a bistable switch behavior (7). The previous model did not consider the effect of inhibitor I released by donor cells as a quorum-sensing signal. The original model was modified to allow for the consideration of the effect of donor cell density. The second part of the model considers growth of the donor and recipient population as well as the conjugation and conversion of recipient cells to donor cells. The molecular and population events depicted in the mathematical model are described in Supporting Information and shown in Fig. S1. The governing equations also are presented.
Supplementary Material
Acknowledgments
We thank Benjamin Brasseur for sample preparation, and Kristi Frank for the Δ-Eep donor strain. This work was supported, in part, by National Institutes of Health Grants GM081888 (to W.-S.H.) and GM49530 (to G.M.D.). A.C. was supported by a doctoral dissertation fellowship from The Graduate School, University of Minnesota. L.C.C.C. is the recipient of an American Academy of University Women American Fellowship (2010–2011) and Biotechnology Training Grant (2007–2009) T32GM008347.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212256110/-/DCSupplemental.
References
- 1.Paulsen IT, et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299(5615):2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 2.Dunny GM. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: Cell-cell signalling, gene transfer, complexity and evolution. Philos Trans R Soc Lond B Biol Sci. 2007;362(1483):1185–1193. doi: 10.1098/rstb.2007.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moellering RC., Jr Emergence of Enterococcus as a significant pathogen. Clin Infect Dis. 1992;14(6):1173–1176. doi: 10.1093/clinids/14.6.1173. [DOI] [PubMed] [Google Scholar]
- 4.Mori M, et al. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J Biol Chem. 1988;263(28):14574–14578. [PubMed] [Google Scholar]
- 5.Leonard BAB, Podbielski A, Hedberg PJ, Dunny GM. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci USA. 1996;93(1):260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fixen KR, et al. Analysis of the amino acid sequence specificity determinants of the enterococcal cCF10 sex pheromone in interactions with the pheromone-sensing machinery. J Bacteriol. 2007;189(4):1399–1406. doi: 10.1128/JB.01226-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee A, et al. Convergent transcription confers a bistable switch in Enterococcus faecalis conjugation. Proc Natl Acad Sci USA. 2011;108(23):9721–9726. doi: 10.1073/pnas.1101569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandler JR, Hirt H, Dunny GM. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc Natl Acad Sci USA. 2005;102(43):15617–15622. doi: 10.1073/pnas.0505545102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakayama J, Ruhfel RE, Dunny GM, Isogai A, Suzuki A. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J Bacteriol. 1994;176(23):7405–7408. doi: 10.1128/jb.176.23.7405-7408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandler JR, Dunny GM. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J Bacteriol. 2008;190(4):1172–1183. doi: 10.1128/JB.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi K, et al. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc Natl Acad Sci USA. 2005;102(51):18596–18601. doi: 10.1073/pnas.0506163102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bae T, Clerc-Bardin S, Dunny GM. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J Mol Biol. 2000;297(4):861–875. doi: 10.1006/jmbi.2000.3628. [DOI] [PubMed] [Google Scholar]
- 13.Hirt H, et al. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: Complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J Bacteriol. 2005;187(3):1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bensing BA, Meyer BJ, Dunny GM. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc Natl Acad Sci USA. 1996;93(15):7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buttaro BA, Antiporta MH, Dunny GM. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J Bacteriol. 2000;182(17):4926–4933. doi: 10.1128/jb.182.17.4926-4933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 17.Ng W-L, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson CM, et al. RNA-mediated reciprocal regulation between two bacterial operons is RNase III dependent. MBio. 2011;2(5):e00189-11. doi: 10.1128/mBio.00189-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olmsted SB, Kao SM, van Putte LJ, Gallo JC, Dunny GM. Role of the pheromone-inducible surface protein Asc10 in mating aggregate formation and conjugal transfer of the Enterococcus faecalis plasmid pCF10. J Bacteriol. 1991;173(23):7665–7672. doi: 10.1128/jb.173.23.7665-7672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnuson R, Solomon J, Grossman AD. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77(2):207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 21.Perego M, Hoch JA. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93(4):1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies DG, et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 23.Zhu J, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2002;99(5):3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins DA, et al. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450(7171):883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 25.Zhang HB, Wang LH, Zhang LH. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2002;99(7):4638–4643. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith J. Tragedy of the commons among antibiotic resistance plasmids. Evolution. 2012;66(4):1269–1274. doi: 10.1111/j.1558-5646.2011.01531.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen YQ, Staddon JH, Dunny GM. Specificity determinants of conjugative DNA processing in the Enterococcus faecalis plasmid pCF10 and the Lactococcus lactis plasmid pRS01. Mol Microbiol. 2007;63(5):1549–1564. doi: 10.1111/j.1365-2958.2007.05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourgogne A, et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9(7):R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kristich CJ, Chandler JR, Dunny GM. Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid. 2007;57(2):131–144. doi: 10.1016/j.plasmid.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.