Abstract
Rising atmospheric carbon dioxide (CO2) conditions are driving unprecedented changes in seawater chemistry, resulting in reduced pH and carbonate ion concentrations in the Earth’s oceans. This ocean acidification has negative but variable impacts on individual performance in many marine species. However, little is known about the adaptive capacity of species to respond to an acidified ocean, and, as a result, predictions regarding future ecosystem responses remain incomplete. Here we demonstrate that ocean acidification generates striking patterns of genome-wide selection in purple sea urchins (Strongylocentrotus purpuratus) cultured under different CO2 levels. We examined genetic change at 19,493 loci in larvae from seven adult populations cultured under realistic future CO2 levels. Although larval development and morphology showed little response to elevated CO2, we found substantial allelic change in 40 functional classes of proteins involving hundreds of loci. Pronounced genetic changes, including excess amino acid replacements, were detected in all populations and occurred in genes for biomineralization, lipid metabolism, and ion homeostasis—gene classes that build skeletons and interact in pH regulation. Such genetic change represents a neglected and important impact of ocean acidification that may influence populations that show few outward signs of response to acidification. Our results demonstrate the capacity for rapid evolution in the face of ocean acidification and show that standing genetic variation could be a reservoir of resilience to climate change in this coastal upwelling ecosystem. However, effective response to strong natural selection demands large population sizes and may be limited in species impacted by other environmental stressors.
Keywords: experimental evolution, population genomics, RNA sequencing, adaptation, environmental mosaic
Accelerating increases in ocean CO2 concentrations and accompanying declines in pH are expected this century (1, 2), particularly in the California Current System (3). The negative impacts of ocean acidification have been seen in a broad range of species (4–8) and are predicted to lead to future populations of individuals with low growth, reproduction, or survival. However, the capacity of marine populations to adapt to these changes is unknown (9, 10), and, as a result, there may be circumstances in which natural selection could result in populations of individuals with better-than-expected fitness under acidified conditions. Until recently, the tools to scan for standing genetic variation with adaptive potential in the face of climate change have not been broadly available. Here we combine sequencing across the transcriptome of the purple sea urchin Strongylocentrotus purpuratus, growth measurements under experimental acidification, and tests of frequency shifts in 19,493 polymorphisms during development. We detect the widespread occurrence of genetic variation to tolerate ocean acidification.
Rapid evolution in changing environments is likely to depend more on existing genetic variation than new mutations (11–13), so evolutionary response to acidification is most likely when a large population exists in a pH-variable environment. The habitat of the purple sea urchin varies across seasons and latitude in CO2 concentrations and in pH due to the upwelling of cold, high-CO2, low-pH waters from the ocean’s depths (14). This large-scale environmental mosaic within the California Current System is likely to be of ancient origin: variable upwelling conditions have existed for millions of years (15) and have led to the evolution of marine taxa dependent on these productive, cold-water conditions (16). Populations within this upwelling ecosystem are exposed episodically to elevated CO2 conditions within coastal regions of smaller spatial extent than the dispersal scales of the planktonic larvae of sea urchins (17), opening the possibility that local selection might promote and sustain alleles with greater fitness in high CO2 conditions.
Results
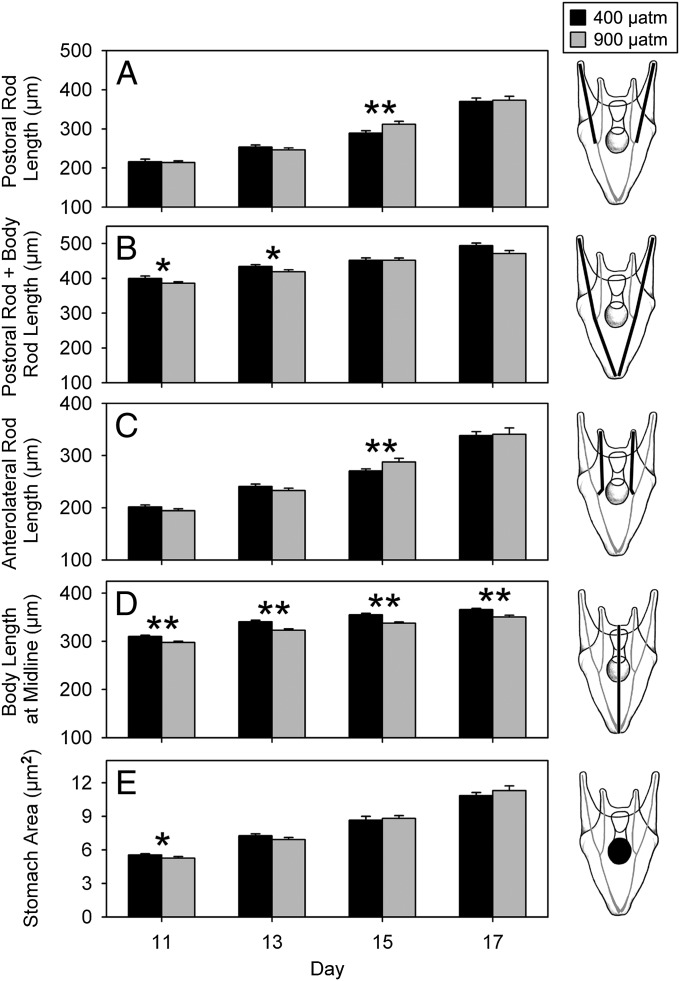
From each of seven populations along a 1,200-km mosaic of coastal upwelling-driven acidification (Table S1), we fertilized eggs from 10 females with a mixture of sperm from 10 males (40 alleles per population). Planktonic larvae were grown in replicate cultures from fertilization until metamorphosis to the benthic, juvenile stage (50 d postfertilization) under ambient (400 µatm) or elevated (900 µatm) partial pressures of CO2 (pCO2; Table S2). To quantify effects of CO2 on growth and development, we measured five morphological features during the transition period from four- to eight-arm larvae. Responses to CO2 seldom differed among populations (ANOVA, CO2 × population; P > 0.05), so we pooled across localities. Lengths of calcareous skeletal rods (Fig. 1 A–C) and stomach area (Fig. 1E) rarely differed between the two CO2 levels, although high-CO2 larvae were consistently 4–5% smaller in body length (Fig. 1D). Larvae developed at similar or slightly faster rates under high CO2 (Fig. S1A), and thus smaller body size was an effect of elevated CO2 rather than an indicator of delayed development. Moreover, at the completion of the planktonic phase, larvae under both CO2 levels were equally competent to metamorphose and settle (Fig. S1B), indicating no sustained difference in developmental trajectory.
Fig. 1.
Growth and morphometrics of sea urchin larvae cultured at control pCO2 (400 µatm; black bars) vs. elevated pCO2 (900 µatm; gray bars). Results are from larvae collected on days 11, 13, 15, and 17 (postfertilization) of trial 1. Responses to CO2 seldom differed among the four populations (ANOVA, CO2 × population; P > 0.05), so results shown are for responses pooled across populations (n = 12–14 culture jars per CO2 level). Drawings (based on ref. 49) illustrate the morphological features quantified. (A) Postoral rod length. (B) Postoral rod and body rod length. (C) Anterolateral rod length. (D) Body length at midline. (E) Stomach area. Error bars are +1 SE. *P < 0.05; **P < 0.005 (significant differences between CO2 levels; ANOVA).
To determine whether homeostasis in morphology and development was associated with selection at the genomic level, we measured genome-wide shifts in allele frequency across 6 critical days of development, spanning the transition from prefeeding to feeding larvae, a period of major skeletal growth. We sampled larvae at 1 d postfertilization (hatched blastulae) and again at day 7 (four-arm swimming and feeding larvae) and prepared libraries for Illumina sequencing from mRNA pools from 1,000 larvae. We sequenced the 28 samples on 28 Illumina lanes (7 populations × 2 CO2 levels × 2 time points in development) to obtain >100 billion base pairs of sequence data, an average of 80 million 50-bp reads for each sample. We mapped reads to all predicted genes from the genome sequence (18) and identified single-nucleotide polymorphisms (SNPs) across all samples. We excluded rare alleles (<5% frequency) and filtered for depth of coverage and differences in gene expression in response to CO2 to minimize the possibility of confounding changes in allele frequency with changes in allele-specific expression (see SI Text and Figs. S2 and S3 for additional analyses, results, and discussion of the small role of allele-specific expression in response to CO2 in our experiments). This process yielded 19,493 high-quality SNPs.
We found substantial change in allele frequencies through time, with greater changes in response to elevated CO2 (pCO2 = 900 µatm) than ambient [400 µatm; Kolmogorov-Smirnov (K-S) test; P = 0.0021]. We used three approaches to test for natural selection in response to elevated CO2: (i) permutation analyses to identify polymorphisms that differed in frequency between ambient and high CO2 levels more than expected by chance, (ii) protein function enrichment analyses to identify suites of genes related by biological function that responded to elevated CO2, and (iii) amino acid polymorphism analyses to contrast types of SNPs with different magnitudes of allelic response and biological functions (19, 20).
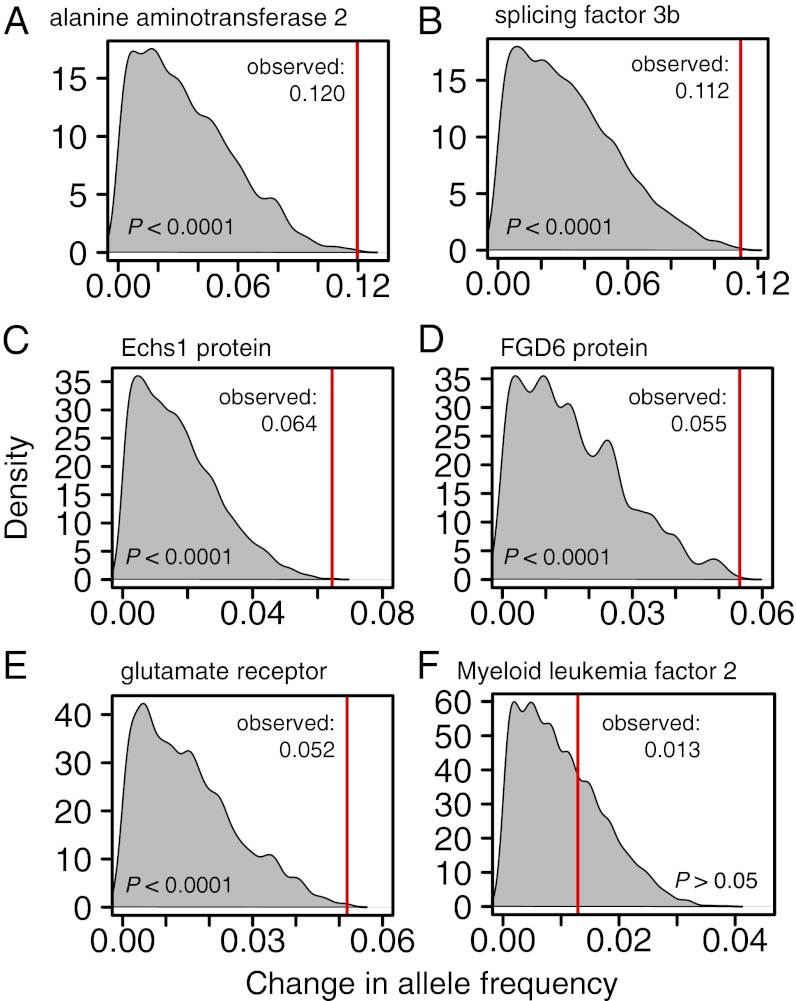
By comparing observed differences in allele frequency between control and elevated CO2 levels to empirical null distributions generated by random permutation of samples, we identified 30 outlier polymorphisms in 30 genes [false discovery rate (FDR) P < 0.05; Fig. 2 and Table S3]. Observed allele frequency changes and permuted distributions in 5 of the 30 genes are shown in Fig. 2 A–E, with an additional randomly selected gene for comparison (Fig. 2F). The alanine aminotransferase 2 enzyme and splicing factor 3b proteins have the polymorphisms with the greatest treatment effects and perform functions related to metabolism and RNA processing, respectively (Fig. 2 A and B and Table S3). The enoyl CoA hydratase (Echs1) protein is a mitochondrial enzyme involved in lipid metabolism (Fig. 2C). FYVE, RhoGEF and PH domain-containing protein 6 (FGD6) regulates the cytoskeleton and cell shape (Fig. 2D), and the glutamate receptor is involved in ion transport (Fig. 2E). Four of the 30 genes (significantly more than expected; P < 0.0001; test of percentages) are part of the biomineralization proteome and occur in adult or larval skeletal structures (see below). Other protein functions across the 30 genes include metabolism, cell structure, transcription and translation processes, cell signaling, and stress response (see Table S3 for complete list).
Fig. 2.
Change in allele frequency in 5 of 30 outlier polymorphisms with high treatment effect in response to elevated CO2 (A–E; P < 0.0001) and one randomly selected polymorphism (F; P > 0.05). (A and B) Alanine aminotransferase 2 enzyme (A) and splicing factor 3b (B) show the greatest treatment effects and perform functions related to metabolism and RNA processing, respectively. (C) Echs1 protein (enoyl CoA hydratase) is a mitochondrial enzyme involved in lipid metabolism. (D and E) FGD6 regulates the cytoskeleton and cell shape (D), and the glutamate receptor is involved in ion transport (E). Red lines mark observed change in allele frequency between control (400 µatm–days 1 and 7 averaged) and treatment (900 µatm–day 7) cultures. An empirical null distribution (gray) was generated by the random permutation of samples and recalculation of treatment effect. These five genes span the diverse protein functions and range of change in allele frequency represented across the 30 outliers. See Table S3 for the complete list of 30 outlier genes with protein function annotations.
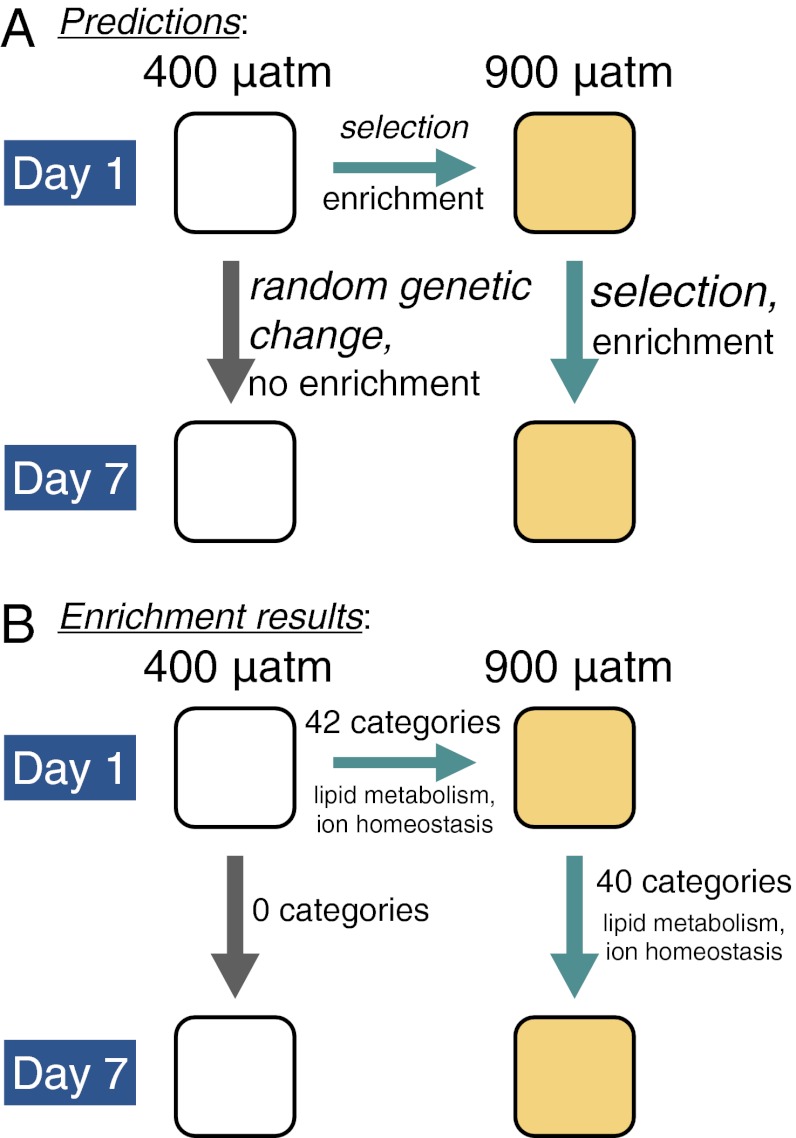
We tested for the concentration of greater changes in allele frequency in genes with specific biological functions, an approach known as functional enrichment analysis. We found that genes showing strong allele frequency changes induced by CO2 were concentrated in 40 of 1,307 functional protein classes (FDR P < 0.10; Table S4 and Figs. 3 and 4). In response to elevated CO2, we expected greater changes in allele frequency to be concentrated in functional categories that improve larval fitness in low-pH conditions (Fig. 3). Specifically, based on previous studies of physiological response to CO2, we hypothesized that genes related to biomineralization, ion homeostasis, and metabolism would show excess genetic change in response to high CO2 (21–24). In contrast, at ambient CO2, we expected changes to be random with respect to protein function. In agreement with predictions, the 40 categories enriched for greater than average allele frequency changes in high-CO2 conditions were primarily related to lipid metabolism, ion homeostasis, cell signaling, and protein modification (Table S4 and Figs. 3 and 4). There were 980 unique genes represented across these categories (Table S4). In contrast, we found no functional enrichment for temporal changes in allele frequency in larvae cultured in ambient CO2 seawater (Fig. 3B). These results provide an important negative control, validating our hypotheses regarding which classes of genes we expect to respond to ocean acidification (25).
Fig. 3.
Summary schematic of predicted evolutionary forces and enrichment results (A) and observed protein function enrichment results for greater changes in allele frequency (B) between the four day and treatment combinations (populations averaged for each day and treatment). Arrows represent comparisons of allele frequencies between day and treatment combinations. Of the genes in enriched categories, 71% were in common after 1 d at 900 µatm and after 7 d at 900 µatm (Fig. 5).
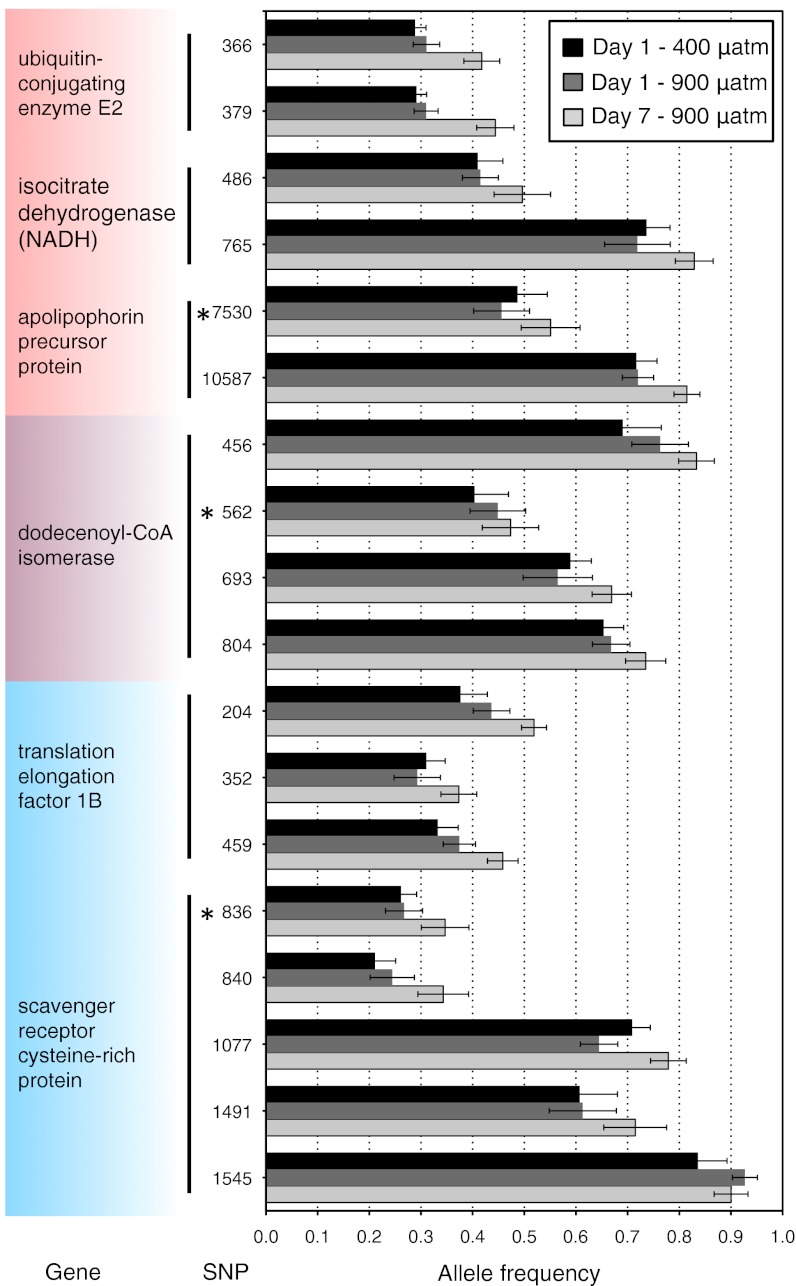
Fig. 4.
Consistent changes in allele frequencies associated with elevated CO2 in multiple SNPs within single genes. Genes are in functionally enriched categories related to lipid metabolism (highlighted in red), ion homeostasis (blue), or both (purple); these six genes represent many in these categories that have multiple SNPs per gene showing greater than average change in allele frequency and are enriched for response to high CO2 after 1 and 7 d. For 13 of 18 SNPs shown, changes in allele frequency are in the same direction after 1 d in elevated CO2 conditions (day 1–900 µatm) as after 7 d in elevated CO2 conditions (day 7–900 µatm) relative to allele frequencies in the wild (day 1–400 µatm). See Fig. 5 for data from SNPs in genes enriched for change after both 1 and 7 d in high CO2 culture. Numbers indicate base position of the polymorphism in the coding region of the gene. Bars are mean frequencies (±SE) in seven populations. Asterisks mark amino acid changing polymorphisms.
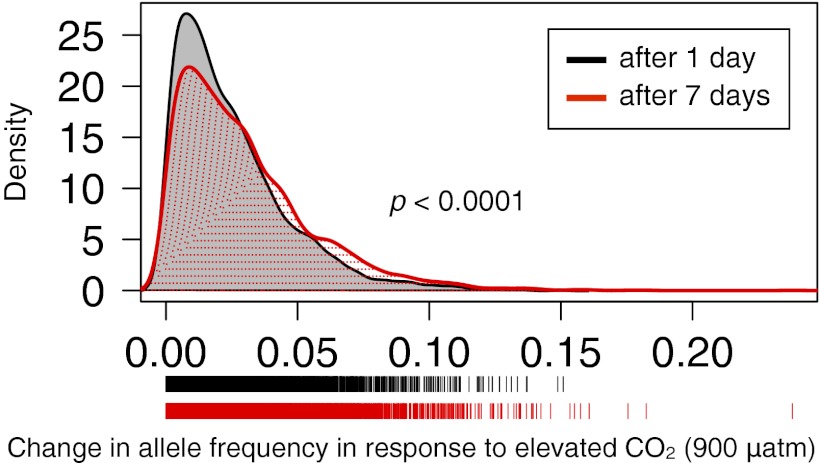
In addition, we found significant functional enrichment for changes in allele frequency after a single day at elevated CO2 in 42 of 1,307 functional protein classes (FDR P < 0.10; comparing day 1–400 µatm pCO2 to day 1–900 µatm pCO2; Table S5 and Fig. 3B). Again, as predicted, the greatest changes in allele frequency were concentrated in metabolism, particularly lipid metabolism, and ion homeostasis genes (Table S5; 846 unique genes represented). As expected, if changes were due to natural selection rather than random effects, there was a high degree of overlap (71%) in the genes in the enriched functional categories after 1 d at 900 µatm pCO2 and after 7 d at 900 µatm pCO2. Further, the magnitude of change in allele frequency was greater after 7 d compared with 1 d (Fig. 5; K-S test; P < 0.0001). These results suggest the continued action of selection on a similar suite of genes through developmental time.
Fig. 5.
Selection in action. There is a greater magnitude of change in allele frequency after 7 d than 1 d in elevated CO2 conditions (red and black, respectively) in the same suite of genes. Change in allele frequency for the same set of 4,782 SNPs comparing day 1–400 µatm with day 1–900 µatm (black) and day 1–900 µatm with day 7–900 µatm (red) are represented in each distribution. These SNPs are in the 601 genes in common (71% overlap) in functional categories enriched for change in allele frequency in response to high CO2 conditions after 1 and 7 d (Tables S4 and S5). Distributions are significantly different (K-S test; P < 0.0001).
We also tested for enrichment of amino acid-changing SNPs as a function of increased experimental treatment effect. These tests parallel studies of human genetic variation that use the fraction of replacement SNPs as a signal of environmental selection (20). First, we ranked SNPs by the average degree of allele frequency difference between control (average of 400-µatm pCO2 samples) and treatment (day 7–900 µatm pCO2) samples. We identified amino acid-changing SNPs across the entire dataset and compared them to amino acid-changing SNPs that occur in the 5% and 1% upper tail of the distribution of ranks (as in ref. 20). Across our entire dataset, only 10.8% of all SNPs are amino acid replacements. Of these, 25.6% involve charge-changing amino acids most likely to be functionally significant. By contrast, among SNPs with the 5% greatest treatment effects, 14.5% are amino acid-replacing SNPs, with 32.7% changing charge (χ2 = 8.2; P = 0.004; Fig. S2). The 1% tail of the treatment distribution shows 17.3% amino acid-changing SNPs, and 42.9% of these change amino acid charge (χ2 = 2.7; P = 0.10; Fig. S2). The replacement enrichment over the complete dataset (60.2%: 17.3% vs. 10.8%) is similar to that observed in SNPs the human genome that show the greatest association with environmental variables (table 1 in ref. 20). These patterns suggest major changes in protein function occurring in polymorphisms with the greatest change in allele frequency.
Elevated CO2 can impact calcification in many marine taxa (4). However, we found the formation of calcareous skeletal rods by larval purple sea urchins to be resilient to elevated CO2 (Fig. 1 A–C). Thus, it is particularly interesting to assay biomineralization genes for experimental CO2 effects. Because sea urchin biomineralization genes are not included in current Gene Ontology categories, we compiled a list based on the protein components of major skeletal elements (26) and regulatory genes that generate larval skeletons in the primary mesenchyme cells (27, 28). In total, the list included 350 genes known to be involved in sea urchin skeletal formation, and here we report data on 884 SNPs from these genes.
As mentioned above, 4 of these biomineral genes appear in our list of 30 outlier loci (Table S3). Among 84 biomineral SNPs with the strongest treatment effects (the 5% tail of the biomineral distribution), there is an excess of amino acid-changing SNPs (14.5%; χ2 = 8.2; P < 0.01; Fig. S2) compared with background (10.8%). These 84 high-treatment-effect SNPs occur in only 37 biomineralization genes, including two previously identified outlier genes (a glucose-regulated structural protein in spicules and a GTP binding protein; four SNPs each), an extracellular matrix protein (three SNPs) found in larval skeletons, a protein similar to a TRAP-family protein (TFP250, six SNPs, including an amino acid replacement), a thioester-containing protein of unknown function (three SNPs), and two chaperonins (three SNPs) associated with adult skeletons. TFP250, similar to a calcium-binding growth factor, had the largest number of polymorphic SNPs (57), amino acid-changing SNPs (12), and high-treatment-effect SNPs (6) among the biomineralization genes.
Other gene categories with large CO2 effects included those involving lipid metabolism and ion homeostasis. As described above, these categories have excess SNPs with high magnitude change in allele frequency. In addition, these categories have an excess number of amino acid-changing polymorphisms, a signal of selection independent of allele frequency estimates (Fig. S2). Lipid metabolism genes showed 17.8% amino acid-changing SNPs vs. a background level of 10.8% (χ2 = 23.8; P < 0.0001), whereas ion homeostasis genes were slightly, but not significantly, enhanced for amino acid-changing SNPs (12.5%; P > 0.05; Fig. S2).
Such results may reflect strong selection directly on some SNPs along with associated changes in nearby polymorphisms through hitchhiking, a pattern often observed in studies of genes associated with environmental variation, selective sweeps, and positive selection (29). Silent SNPs could alternatively be linked with amino acid-changing SNPs elsewhere or with non-amino-acid-changing polymorphisms selected for more efficient transcription or translation (30).
Discussion
Our results indicate that normal morphological and developmental progress in sea urchins raised under low-pH conditions may arise in part from natural selection for larvae with specific alleles that improve performance under these conditions. As hypothesized, allelic shifts were most pronounced in genes related to membrane composition and ion transport, categories that both critically influence ion homeostasis (31). Sterols specifically enhance mechanical stability and create lipid rafts where large transport proteins can be organized and concentrated (32). Ion and sterol categories together suggest selective survival of larvae with alleles that allow them to regulate internal pH in the face of elevated CO2. These results are in strong contrast to the absence of functional enrichment in control cultures (25). It remains to be determined whether these selected alleles have unmeasured future tradeoffs or costs (e.g., in terms of reduced larval tolerance of other stressors or altered juvenile performance).
Effects of Acidification on Larval Growth and Development.
Previous ocean-acidification experiments on purple sea urchin and other echinoderm larvae have often found stronger negative effects of CO2 on growth and development than those documented here (23, 24, 33, 34). The primary difference between our experimental design and previous studies is that our larval cultures were >10 times less dense, reflecting a more ecologically relevant condition (35), with perhaps less laboratory-generated stress. We hypothesize that the higher larval densities used in other studies may have interacted with elevated CO2 to exacerbate the negative effects on developing larvae. At the lower larval densities used in our experiments, effects of elevated CO2 on purple sea urchin development and morphology appear to be minimal. The most consistent negative response to acidified conditions in our cultures was a 4–5% reduction in body length, which may reflect increased energetic costs of maintenance and skeletal rod formation under elevated CO2 (24). However, these minor effects did not influence the timing of settlement or the proportion of larvae that were competent to metamorphose.
Allele-Specific Expression.
Because we estimate allele frequency from mRNA from pools of larvae, it is conceivable that a change in allele frequency at a given locus is due to a change in allele-specific expression of one haplotype over another. To reduce this potential confounding effect, we excluded the SNPs in the few (23) genes that showed differential expression in response to CO2 treatment. However, there could also be differences in allele-specific expression in genes with constant transcript abundances. To explore the potential effect of allele-specific expression on our allele-frequency estimates, we compared SNPs from our dataset with SNPs at exactly the same positions using (i) PCR-based allele resequencing for SNPs at two high-treatment-effect loci and (ii) across the genome with Restriction Site Tiling Analysis arrays (36). Allele frequency estimates were within 0.7% and 1.6% accurate comparing estimates from RNA and DNA in the two high treatment effect loci (Fig. S3), and we found high correspondence between genome-wide datasets (Fig. S4A; R2 = 0.75; P < 0.0001), suggesting that allele frequencies inferred from RNA-sequencing data were representative of true genomic allele frequencies.
Overall, our data suggest that allele-specific expression is rare in purple sea urchins. Significant allele-specific expression at high CO2 should result in significant shifts in expression value because a chromosome that produces few transcripts of a gene in high CO2 will reduce the cell-wide abundance of those transcripts (see SI Text for a discussion of potential exceptions). However, only 32 genes across our dataset showed significant expression differences between CO2 levels, suggesting that the potential for allele-specific expression is limited. We also saw no excess in treatment effect among these 32 differentially expressed genes [unlike a previous study in Drosophila hybrids (37); SI Text]. Lastly, CO2 treatment had little impact on developmental programming of gene expression because day 1 to 7 expression changes were the same for ambient and high CO2 levels (Fig. S4B; R2 = 0.92, P < 0.0001) showing that even under conditions of strong gene expression regulation, CO2 levels have little effect on expression levels, leaving a very small role for CO2-based allele-specific expression.
Evidence from the published literature on allele-specific expression suggests a small role for allelic differences among genetically related individuals of the same species (38–40). As a result, our analyses (see additional examples in SI Text) cannot preclude allele-specific expression at a few loci. However, our gene function results cannot be solely driven by such rare loci. In addition, the identification of amino acid-changing polymorphisms does not rely on estimates of allele frequency; therefore, our results that show positive tests for excess amino acid substitutions in the three functional classes of genes tested provide additional evidence of selection that is independent of allele frequency estimates (Fig. S2).
Any rare changes that could be due to allele-specific expression would point to environmentally sensitive cis-regulatory regions. This finding would be an equally interesting result that we cannot completely exclude at a few loci. Although evidence from our data and the literature suggest a small role for allele-specific expression in the present study, further investigations are needed to learn of its role in response to acute stress within a population.
Selection Across Environmental Mosaics.
Our growth results, taken alone, would have signaled relatively low impact of acidified conditions on sea urchin larvae. However, our investigation of the genetics of these populations shows that underneath the observed homeostasis of larval growth and development, there was significant genetic change in classes of genes involved in growth and biomineralization. Complementing studies of negative fitness consequences of acidification (4–8) and plasticity in gene regulation (23, 41, 42), our results demonstrate another important impact of ocean acidification: population genetic change.
Factors that favor adaptive genetic variation in purple sea urchins—and perhaps other marine taxa—include a spatial and temporal mosaic in carbonate chemistry, along with high genetic variability, high fecundity, and high dispersal potential. Purple sea urchins have evolved in the California Current System where coastal taxa are exposed seasonally to higher CO2 and reduced pH and carbonate ion concentrations. Indeed, during upwelling events, urchin larvae in some regions are likely already exposed to elevated seawater CO2 levels similar to those used in these experiments (900 µatm pCO2) (14, 34, 43).
Our results suggest that this long-term environmental mosaic has led to a reservoir of genetic variation in purple sea urchins that can respond to acidification during larval development and buffer some of the negative consequences of low pH (24). We saw small differences in many genes, suggesting that the genetic basis of adaptation is highly multigenic. However, the concentration of genetic effects in biomineralization, ion regulation, and membrane genes suggests that some of the major adaptive mechanisms reside in regulation of skeletal growth. In support of this hypothesis, results from another study of common-garden-acclimated adult purple sea urchins originally from Boiler Bay, OR, and San Diego showed consistent differences in the regulation of growth and biomineralization genes (28), potentially due to adaptation to differences in pH and temperature between the Oregon coast, a region of strong upwelling and low pH, and the Southern California Bight, a region of warmer, high-pH waters (14, 43).
Link Between Acidification and Population Growth.
Our results suggest a conceptual model in which acidification experienced by a population of developing larvae results in impaired growth or mortality in some individuals, but natural selection across this fitness differential results in a population that is better adapted to acidified conditions. For this process to occur, two conditions must be met. First, there must be genetic variation within the population for response to low pH. In addition to our data, three recently published studies using breeding and growth experiments in two sea urchin species and a bryozoan highlight the importance of standing genetic variation for an organism’s ability to respond to ocean acidification (44–46). Second, there must be robust populations with excess reproductive capacity.
This second condition is important because adaptive capacity is a double-edged sword. Populations experiencing selection pay a selective cost in terms of reduced demographic growth and reproductive potential (12, 47) because individuals that are less fit experience lower growth or survival. Populations impacted by climate change, overfishing, and other anthropogenic impacts may have low population growth and impaired capacity to absorb these selective impacts of acidification. Thus, although marine populations in upwelling systems may possess the capacity for adaptation, maintenance of robust populations in the future is an important part of any climate response strategy. Future studies are needed to empirically test for loss of genetic diversity through rapid evolution in response to ocean acidification.
Materials and Methods
Larval Culture of Sea Urchins.
Adult urchins were collected from seven populations from Central Oregon to Southern California, spanning 1,200 km of the S. purpuratus species range, and were shipped to Bodega Marine Laboratory (see SI Text for complete experimental details). Adults were spawned in the laboratory, and for each population gametes were mixed from 10 females and 10 males. Fertilized embryos were cultured under present-day global-mean pCO2 (partial pressure of CO2∼400 µatm) and a fossil-fuel intensive projection (∼900 µatm) (48). After 24 h, hatched blastulae were transferred to four replicate jars per population and CO2 level and were maintained at 14 °C in jars with oscillating paddles (SI Text). Seawater was exchanged and sampled for pH and alkalinity, and larvae were fed and sampled every other day during 50-d trials, from 3 d postfertilization to metamorphosis.
Population Genomics.
For genomic analyses, we sampled ∼1,000 larvae at 1 and 7 d postfertilization from each population and CO2 level. Total RNA was extracted from pooled samples of larvae, and mRNA was isolated and prepared for sequencing by using Illumina’s TruSeq kit. Each of the 28 samples was sequenced on a single Illumina HiSeq lane, yielding ∼80 million 50-bp reads per sample. Sequence data were processed for length and quality and aligned to all predicted genes of the purple sea urchin (downloaded from www.spbase.org). SNPs were detected and allele frequencies were calculated for each sample at each SNP position as the number of reads with the reference allele divided by the total number of quality reads mapped at that position (see SI Text for more details). A detailed pipeline of all data processing steps and parameters used can be found at http://sfg.stanford.edu (50). We used permutation analyses to identify outlier polymorphisms and enrichment analyses to identify suites of functionally related genes enriched for high changes in allele frequency (see SI Text for more details).
Supplementary Material
Acknowledgments
We thank B. Menge, G. Hofmann, and our other collaborators in the Ocean Margin Ecosystems Group for Acidification Studies (OMEGAS) for assistance with urchin collections; N. Rose for assistance in generating PCR-based sequence data; J. Hodin, K. Montooth, and M. Hahn for helpful discussions; and anonymous reviewers for thoughtful and constructive critiques of the manuscript. This work was supported by National Science Foundation (NSF) Collaborative Grants OCE-1041222 (to S.R.P), OCE-1041089 (to E.S., B.G., T.M.H., and A.D.R.), and OCE-0927255 (to B.G., T.M.H., A.D.R., and E.S.). M.H.P. was supported by NSF predoctoral and postdoctoral fellowships.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.S. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Dryad Repository, http://dx.doi.org/10.5061/dryad.6j51n.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220673110/-/DCSupplemental.
References
- 1.Gattuso JP, Buddemeier RW. Ocean biogeochemistry. Calcification and CO2. Nature. 2000;407(6802):311–313, 313. doi: 10.1038/35030280. [DOI] [PubMed] [Google Scholar]
- 2.Hönisch B, et al. The geological record of ocean acidification. Science. 2012;335(6072):1058–1063. doi: 10.1126/science.1208277. [DOI] [PubMed] [Google Scholar]
- 3.Gruber N, et al. Rapid progression of ocean acidification in the California Current System. Science. 2012;337(6091):220–223. doi: 10.1126/science.1216773. [DOI] [PubMed] [Google Scholar]
- 4.Kroeker KJ, Kordas RL, Crim RN, Singh GG. Meta-analysis reveals negative yet variable effects of ocean acidification on marine organisms. Ecol Lett. 2010;13(11):1419–1434. doi: 10.1111/j.1461-0248.2010.01518.x. [DOI] [PubMed] [Google Scholar]
- 5.Ries JB, Cohen AL, McCorkle DC. Marine calcifiers exhibit mixed responses to CO2-induced ocean acidification. Geology. 2009;37(12):1131–1134. [Google Scholar]
- 6.Dupont S, Ortega-Martínez O, Thorndyke M. Impact of near-future ocean acidification on echinoderms. Ecotoxicology. 2010;19(3):449–462. doi: 10.1007/s10646-010-0463-6. [DOI] [PubMed] [Google Scholar]
- 7.Gaylord B, et al. Functional impacts of ocean acidification in an ecologically critical foundation species. J Exp Biol. 2011;214(Pt 15):2586–2594. doi: 10.1242/jeb.055939. [DOI] [PubMed] [Google Scholar]
- 8.Hoegh-Guldberg O, et al. Coral reefs under rapid climate change and ocean acidification. Science. 2007;318(5857):1737–1742. doi: 10.1126/science.1152509. [DOI] [PubMed] [Google Scholar]
- 9.Doney SC, Fabry VJ, Feely RA, Kleypas JA. Ocean acidification: The other CO2 problem. Annu Rev Mar Sci. 2009;1:169–192. doi: 10.1146/annurev.marine.010908.163834. [DOI] [PubMed] [Google Scholar]
- 10.Harley CDG, et al. The impacts of climate change in coastal marine systems. Ecol Lett. 2006;9(2):228–241. doi: 10.1111/j.1461-0248.2005.00871.x. [DOI] [PubMed] [Google Scholar]
- 11.Kirkpatrick M, Barton NH. Evolution of a species’ range. Am Nat. 1997;150(1):1–23. doi: 10.1086/286054. [DOI] [PubMed] [Google Scholar]
- 12.Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169(4):2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lande R, Shannon S. The role of genetic variation in adaptation and population persistence in a changing environment. Evolution. 1996;50(1):434–437. doi: 10.1111/j.1558-5646.1996.tb04504.x. [DOI] [PubMed] [Google Scholar]
- 14.Feely RA, Sabine CL, Hernandez-Ayon JM, Ianson D, Hales B. Evidence for upwelling of corrosive “acidified” water onto the continental shelf. Science. 2008;320(5882):1490–1492. doi: 10.1126/science.1155676. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs DK, Haney TA, Louie KD. Genes, diversity, and geologic process on the Pacific Coast. Annu Rev Earth Planet Sci. 2004;32(1):601–652. [Google Scholar]
- 16.Estes JA, Steinberg PD. Predation, herbivory, and kelp evolution. Paleobiology. 1988;14(1):19–36. [Google Scholar]
- 17.Strathmann R. Length of pelagic period in echinoderms with feeding larvae from the Northeast Pacific. J Exp Mar Biol Ecol. 1978;34(1):23–28. [Google Scholar]
- 18.Sodergren E, et al. Sea Urchin Genome Sequencing Consortium The genome of the sea urchin Strongylocentrotus purpuratus. Science. 2006;314(5801):941–952. doi: 10.1126/science.1133609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slotte T, et al. Genomic determinants of protein evolution and polymorphism in Arabidopsis. Genome Biol Evol. 2011;3:1210–1219. doi: 10.1093/gbe/evr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancock AM, et al. Colloquium paper: Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc Natl Acad Sci USA. 2010;107(Suppl 2):8924–8930. doi: 10.1073/pnas.0914625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pörtner HO, Langenbuch M, Michaelidis B. Synergistic effects of temperature extremes, hypoxia, and increases in CO2 on marine animals: From Earth history to global change. J Geophys Res. 2005;110(C9):C09S10. [Google Scholar]
- 22.Todgham AE, Hofmann GE. Transcriptomic response of sea urchin larvae Strongylocentrotus purpuratus to CO2-driven seawater acidification. J Exp Biol. 2009;212(Pt 16):2579–2594. doi: 10.1242/jeb.032540. [DOI] [PubMed] [Google Scholar]
- 23.O'Donnell MJ, et al. Ocean acidification alters skeletogenesis and gene expression in larval sea urchins. Mar Ecol Prog Ser. 2010;398:157–171. [Google Scholar]
- 24.Stumpp M, Wren J, Melzner F, Thorndyke MC, Dupont ST. CO2 induced seawater acidification impacts sea urchin larval development I: Elevated metabolic rates decrease scope for growth and induce developmental delay. Comp Biochem Physiol A Mol Integr Physiol. 2011;160(3):331–340. doi: 10.1016/j.cbpa.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 25.Pavlidis P, Jensen JD, Stephan W, Stamatakis A. A critical assessment of storytelling: gene ontology categories and the importance of validating genomic scans. Mol Biol Evol. 2012;29(10):3237–3248. doi: 10.1093/molbev/mss136. [DOI] [PubMed] [Google Scholar]
- 26.Livingston BT, et al. A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 2006;300(1):335–348. doi: 10.1016/j.ydbio.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 27.Wilt FH. Biomineralization of the spicules of sea urchin embryos. Zoolog Sci. 2002;19(3):253–261. doi: 10.2108/zsj.19.253. [DOI] [PubMed] [Google Scholar]
- 28.Pespeni MH, Barney BT, Palumbi SR. Differences in the regulation of growth and biomineralization genes revealed through long-term common garden acclimation and experimental genomics in the purple sea urchin. Evolution. 2013 doi: 10.1111/evo.12036. [DOI] [PubMed] [Google Scholar]
- 29.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12(12):1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plotkin JB, Kudla G. Synonymous but not the same: The causes and consequences of codon bias. Nat Rev Genet. 2011;12(1):32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forte M, Satow Y, Nelson D, Kung C. Mutational alteration of membrane phospholipid composition and voltage-sensitive ion channel function in Paramecium. Proc Natl Acad Sci USA. 1981;78(11):7195–7199. doi: 10.1073/pnas.78.11.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alberts B, et al. Molecular Biology of the Cell. 4th Ed. New York: Garland Science; 2002. [Google Scholar]
- 33.Dupont S, Havenhand J, Thorndyke W, Peck L, Thorndyke M. Near-future level of CO2-driven ocean acidification radically affects larval survival and development in the brittlestar Ophiothrix fragilis. Mar Ecol Prog Ser. 2008;373:285–294. [Google Scholar]
- 34.Yu PC, Matson PG, Martz TR, Hofmann GE. The ocean acidification seascape and its relationship to the performance of calcifying marine invertebrates: Laboratory experiments on the development of urchin larvae framed by environmentally-relevant pCO2/pH. J Exp Mar Biol Ecol. 2011;400(1-2):288–295. [Google Scholar]
- 35.Strathmann MF. Reproduction and Development of Marine Invertebrates of the Northern Pacific Coast: Data and Methods for the Study of Eggs, Embryos, and Larvae. Seattle: Univ of Washington Press; 1987. pp. 511–534. [Google Scholar]
- 36.Pespeni MH, Oliver TA, Manier MK, Palumbi SR. Restriction Site Tiling Analysis: Accurate discovery and quantitative genotyping of genome-wide polymorphisms using nucleotide arrays. Genome Biol. 2010;11(4):R44. doi: 10.1186/gb-2010-11-4-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graze RM, et al. Allelic imbalance in Drosophila hybrid heads: Exons, isoforms, and evolution. Mol Biol Evol. 2012;29(6):1521–1532. doi: 10.1093/molbev/msr318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K, et al. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat Methods. 2009;6(8):613–618. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang HY, et al. Complex genetic interactions underlying expression differences between Drosophila races: Analysis of chromosome substitutions. Proc Natl Acad Sci USA. 2008;105(17):6362–6367. doi: 10.1073/pnas.0711774105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heap GA, et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum Mol Genet. 2010;19(1):122–134. doi: 10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stumpp M, Dupont S, Thorndyke MC, Melzner F. CO2 induced seawater acidification impacts sea urchin larval development II: Gene expression patterns in pluteus larvae. Comp Biochem Physiol A Mol Integr Physiol. 2011;160(3):320–330. doi: 10.1016/j.cbpa.2011.06.023. [DOI] [PubMed] [Google Scholar]
- 42.Martin S, et al. Early development and molecular plasticity in the Mediterranean sea urchin Paracentrotus lividus exposed to CO2-driven acidification. J Exp Biol. 2011;214(Pt 8):1357–1368. doi: 10.1242/jeb.051169. [DOI] [PubMed] [Google Scholar]
- 43.Hofmann GE, et al. High-frequency dynamics of ocean pH: A multi-ecosystem comparison. PLoS ONE. 2011;6(12):e28983. doi: 10.1371/journal.pone.0028983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunday JM, Crim RN, Harley CDG, Hart MW. Quantifying rates of evolutionary adaptation in response to ocean acidification. PLoS ONE. 2011;6(8):e22881. doi: 10.1371/journal.pone.0022881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pistevos JCA, Calosi P, Widdicombe S, Bishop JDD. Will variation among genetic individuals influence species responses to global climate change? Oikos. 2011;120(5):675–689. [Google Scholar]
- 46.Foo SA, Dworjanyn SA, Poore AGB, Byrne M. Adaptive capacity of the habitat modifying sea urchin Centrostephanus rodgersii to ocean warming and ocean acidification: Performance of early embryos. PLoS ONE. 2012;7(8):e42497. doi: 10.1371/journal.pone.0042497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haldane JBS. The cost of natural selection. J Genet. 1957;55(3):511–524. [Google Scholar]
- 48.Moss RH, et al. The next generation of scenarios for climate change research and assessment. Nature. 2010;463(7282):747–756. doi: 10.1038/nature08823. [DOI] [PubMed] [Google Scholar]
- 49.Smith MM, Cruz Smith L, Cameron RA, Urry LA. The larval stages of the sea urchin, Strongylocentrotus purpuratus. J Morphol. 2008;269(6):713–733. doi: 10.1002/jmor.10618. [DOI] [PubMed] [Google Scholar]
- 50.De Wit P, et al. The Simple Fool's Guide to population genomics via RNA-Seq: An introduction to high-throughput sequencing data analysis. Molecular Ecology Resources. 2012;12:1058–1067. doi: 10.1111/1755-0998.12003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.