Abstract
Pestiviruses, including bovine viral diarrhea virus, are important animal pathogens and are closely related to hepatitis C virus, which remains a major global health threat. They have an outer lipid envelope bearing two glycoproteins, E1 and E2, required for cell entry. They deliver their genome into the host cell cytoplasm by fusion of their envelope with a cellular membrane. The crystal structure of bovine viral diarrhea virus E2 reveals a unique protein architecture consisting of two Ig-like domains followed by an elongated β-stranded domain with a new fold. E2 forms end-to-end homodimers with a conserved C-terminal motif rich in aromatic residues at the contact. A disulfide bond across the interface explains the acid resistance of pestiviruses and their requirement for a redox activation step to initiate fusion. From the structure of E2, we propose alternative possible membrane fusion mechanisms. We expect the pestivirus fusion apparatus to be conserved in hepatitis C virus.
Keywords: domain swap, family Flaviviridae, genus hepacivirus, pH sensing, fusion motif
Viruses in the pestivirus genus are economically important animal pathogens, including bovine viral diarrhea virus (BVDV), border disease virus, and classical swine fever virus (CSFV) (1). BVDV causes both acute and chronic infections, primarily in cattle but also in sheep, pigs, goats, and other even-toed ungulates. Among the various flavivirus family members, pestiviruses are the most closely related to hepatitis C virus (HCV) of the hepacivirus genus, a serious and persistent global health threat (2).
Pestiviruses have been used as surrogates for HCV because of the many structural and functional properties shared by these viruses. Pesti- and hepaciviruses have a similar genomic organization and they both form mostly spherical particles 40–60 nm in diameter with relatively smooth surfaces. Due to variations in particle size and shape, electron microscopy studies have provided few additional structural insights and it is not yet clear whether the virions have regular icosahedral symmetry (3–5). Inhibition of cell entry by endocytic inhibitors suggests that pesti- and hepaciviruses enter the cell by clathrin-mediated endocytosis (6, 7). However, pestiviruses and hepaciviruses recognize host cells by binding to cell surface receptors—CD46 and CD81, respectively—that are not significantly internalized (8, 9), and one or more coreceptors are required for postattachment internalization (10, 11). To deliver their single-stranded positive-sense RNA genome into the cytoplasm, pestiviruses and HCV must fuse their lipid envelope with a cellular membrane. The reduced pH of endocytic compartments is required but not sufficient to initiate membrane fusion of these viruses (6, 7, 10, 12, 13). Moreover, viruses from both families are acid resistant before endocytosis, and require a poorly understood activation step to become fusogenic at endosomal pH (6, 7, 12). In pestiviruses, scission of disulfide bonds in the envelope glycoproteins is thought to activate the virion for fusion, as the combination of reducing agent and low pH allows fusion with the plasma membrane (“fusion from without”), albeit at low efficiency (6). A potential precedent is human papillomavirus, which requires a cellular protein disulfide isomerase in addition to low endosomal pH for disassembly of the disulfide-linked capsid and delivery of viral DNA into the cytoplasm (14).
Two envelope glycoproteins, E1 and E2, are required for cell entry and membrane fusion of pesti- and hepaciviruses. E2 determines cellular tropism, binds the cell-surface receptor (CD46 or CD81), and contains the major neutralizing antibody epitopes (9, 15–18). BVDV E2 forms disulfide-linked homodimers and heterodimers with E1 (19–21). Formation of BVDV E1–E2 heterodimers is essential for virus entry and depends on charged residues in the transmembrane segments of the two glycoproteins (22). Similarly, in HCV the transmembrane segment of E2 is required for correct folding and assembly of E1–E2 heterodimers (23).
In the absence of experimental structural data for E1 or E2, it has not been possible to determine the molecular basis of membrane fusion in pesti- and hepaciviruses. We have determined the crystal structure of the BVDV E2 ectodomain. Based on the structure, we propose three alternative membrane fusion mechanisms.
Results
Overall Fold of BVDV E2.
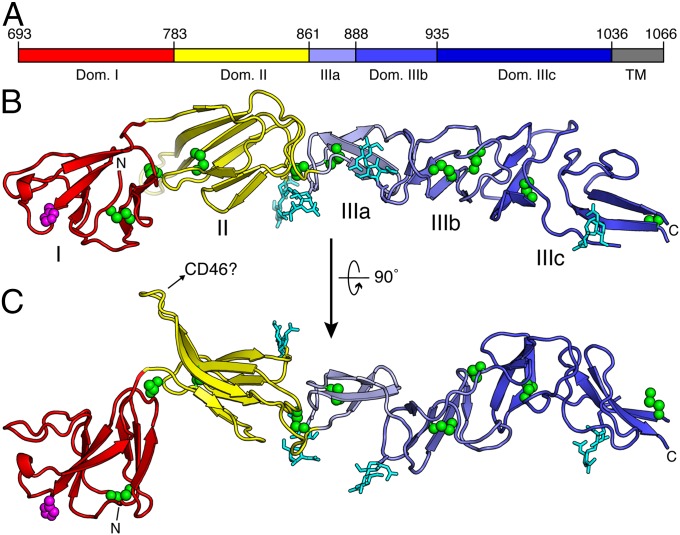
We determined two crystal structures of E2 from the American prototype cytopathic BVDV1 strain (NADL), one of the entire ectodomain, and the other of an ectodomain fragment lacking the 90 N-terminal amino acids. Both E2 constructs lacked the C-terminal transmembrane anchor and were crystallized at pH 5.5 under nonreducing conditions. The two structures were refined at 4.1 Å and 3.3 Å resolution, respectively (Table 1). The E2 ectodomain has a total span of 140 Å and forms disulfide-linked dimers. It has a unique three-domain architecture (Fig. 1 A–C). Domain I (polyprotein residues 693–782) is an Ig-like domain. Due to the lower resolution of the full-length ectodomain structure, many side chains were omitted from the atomic coordinates for domain I. The only histidine residue that is conserved across pestiviruses, His762, is exposed on the surface of domain I at the membrane-distal end of the molecule (Fig. 2A). Histidine side chains have pKa values in the range of 6–6.4 and therefore typically become protonated during endosomal acidification. The increase in positive charge resulting from histidine protonation is an important part of the pH sensing mechanism of many viruses (24–29).
Table 1.
Crystallographic data collection and refinement statistics
| Dataset | U ∆N90-E2-ECD | Native ∆N90-E2-ECD | E2-ECD |
| Data collection | |||
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 136.7, 54.5, 95.9 | 137.4, 56.2, 94.9 | 154.6, 67.8, 138.9 |
| α, β, γ (°) | 90, 92.2, 90 | 90, 94.9, 90 | 90, 121.9, 90 |
| Wavelength, Å | 1.2142 | 1.0711 | 1.1 |
| Resolution, Å* | 50–3.27 (3.36–3.27) | 50–3.2 (3.4–3.2) | 50–4.09 (4.34–4.09) |
| Unique reflections | 21,043 | 23,295 | 18,572 |
| Rmerge ,%* | 7.2 (50.4) | 7.2 (98.1) | 15.1 (100) |
| I/σIa | 12.1 (1.9) | 7.8 (1.2) | 4.8 (1.3) |
| Completeness, %* | 98.4 (89.1) | 97.4 (98.2) | 98.1 (98.1) |
| Redundancy* | 3.3 (1.7) | 2.12 (2.2) | 2.1 (2.1) |
| Overall figure of merit | 0.45 | ||
| Refinement | |||
| Resolution, Å* | 50–3.27 (3.36–3.27) | 50–4.09 (4.34–4.09) | |
| No. reflections | 10,539 | 9,486 | |
| Rwork/Rfree, %* | 24.6 (32.5)/28.9 (36.7) | 28.9 (44)/34.9 (62) | |
| Average B factor, Å2 | 48.9 | 141.72 | |
| Rms deviations | |||
| Bond lengths, Å | 0.010 | 0.013 | |
| Bond angles, ° | 1.89 | 1.90 | |
| Ramachandran analysis | |||
| Preferred regions, % | 86.4 | 83.4 | |
| Allowed regions, % | 12.3 | 10.8 | |
| Disallowed regions, % | 1.3 | 5.8 | |
| Synchrotron beamline | BNL X25 | APS NECAT 24ID-C | BNL X25 |
Rsym = ΣhklΣi |Ihkl,i – < I>hkl|/ΣhklΣi|Ihkl,i|, where Ihkl is the intensity of a reflection and <I>hkl is the average of all observations of the reflection. Rfree, Rwork with 10% of Fobs sequestered before refinement. Residual B-factors after TLS refinement. See PDB entry for TLS refinement parameters. rms, root mean square.
Highest resolution shell is shown in parentheses.
Fig. 1.
Topology and overall protein fold of BVDV E2. (A) The three-domain topology of E2. Domain I is in red, domain II is in yellow, and domain III is in shades of blue: light blue for module IIIa, medium blue for module IIIb, and dark blue for module IIIc. Residue numbers follow BVDV polyprotein numbering. The transmembrane domain (gray) is missing in the structure. (B and C) E2 has a unique architecture consisting of two Ig-like domains (I and II) followed by an elongated β-stranded domain with a new fold (III). The only histidine conserved in pestivirus E2 is His762 (magenta). A host cell binding sequence forms a protruding β hairpin and is a candidate for CD46 binding. Glycans (cyan) are linked to N809, N878, N922, and N990. Disulfide bonds are in green. See also Fig. S1.
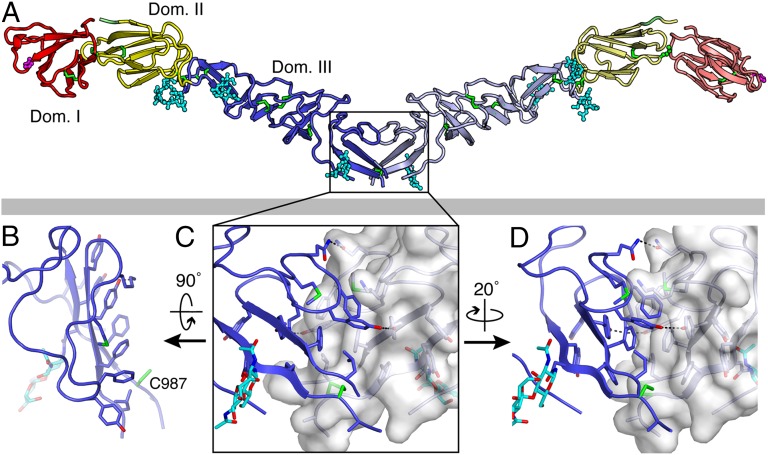
Fig. 2.
BVDV E2 forms covalently linked dimers. (A) The structure of the BVDV E2 dimer, colored as in Fig. 1. The two subunits in the end-to-end dimer are related by a dyad axis, except for domain I, whose orientation relative to domain II differs by 42° in the two subunits. The viral lipid envelope is represented by a gray rectangle (not to scale). (B) The dimer interface consists predominantly of a large cluster of aromatic residues in module IIIc. The aromatic side chains are tightly clustered in a planar array at the membrane-proximal end of E2. (C and D) Disulfide bond links each monomer across the dyad axis via Cys987. The aromatic side chains interact mostly via π stacking and hydrophobic interactions. Hydrogen bonds across the dimer interface are shown as black dashed lines.
Domain II (residues 783–860) is also a seven-stranded Ig-like domain, with similar overall shape and size as domain I. A 12-residue sequence between the last two β strands in domain II forms a highly exposed β hairpin that protrudes into the solvent (Fig. 1C). A peptide containing the analogous sequence from CSFV E2, which is not identical to the one in BVDV, was shown to interact with a host cell receptor (15), presumably CD46. (Overall, the amino acid sequences of BVDV and CSFV E2 are 65% identical.) The most similar structure to domain II is the first Ig domain of receptor for advanced glycation end products (RAGE), Protein Data Bank entry 3O3U, with a Z score of 6.1 in Dali (30). It is tempting to speculate that E2, like RAGE, binds carbohydrates, but the extensive positively charged carbohydrate binding surface in RAGE Ig-1 is not conserved in E2.
Domain III consists of a series of three small β-sheet modules (IIIa–c), which together form an elongated domain. The overall fold and topology of domain III bears no significant similarity to previously determined protein structures (Fig. S1). There are no internal or terminal fusion motifs in E2 with an obvious resemblance to those of other viral fusion proteins. A hydrophobic surface at the dimer interface in domain IIIc containing a large cluster of conserved aromatic side chains could, however, serve as an anchor in either the cellular or viral membrane (see below). The structures of domains II and III are essentially identical in the full-length and truncated E2 ectodomain structures.
E2 contains extensive posttranslational modifications. Four glycans are distributed over the molecule, one in domain II and three in domain III (Fig. 1 B and C). All 17 cysteine residues in the ectodomain are involved in disulfide bonds. Domains I, II, and III contain one, two, and four internal disulfides, respectively; one disulfide bridges domains I and II; and the 17th cysteine forms an intermolecular bond with the dimer partner molecule (Fig. 2). The cysteines are in different positions in pestivirus and HCV E1 and E2 proteins. The disulfide-bonding pattern is similar in HCV E2, however, with a total of nine disulfide bonds linking cysteines that are mostly close to each other in the amino acid sequence (31), consistent with a linear domain architecture as in BVDV E2. In contrast, class II viral membrane fusion proteins have a nonlinear topology that allows cross-links between cysteines far from each other in the amino acid sequence, especially in the N-terminal domain. Although E2 does not form cross-linked homodimers in HCV, E1 and E2 form disulfide-linked complexes in HCV virions (32).
Disulfide-Linked Dimer Interface of E2.
The ectodomain of BVDV E2 forms disulfide-linked dimers in solution, as determined by hydrodynamic and multiangle scattering analysis, and by nonreducing SDS/PAGE (Fig. S2). Domain I is not required for dimerization and dimers are stable at low pH and in the presence of reducing agent. The E2 dimers dissociated in SDS/PAGE, however, even when the samples were not boiled (Fig. S2). The two E2 crystal forms each contain one end-to-end dimer per asymmetric unit (Fig. 2A). The dimer interface is made up predominantly of a large cluster of aromatic residues in module IIIc (Phe989, Tyr991, Tyr1006, Phe1007, Tyr1016, Tyr1018, and Phe1020). The side chains are tightly clustered in a planar array at the membrane-proximal extremity of E2 (Fig. 2B). The aromatic side chains interact mostly via π stacking and hydrophobic interactions and via a single hydrogen bond between the hydroxyl groups of Tyr1006 and Tyr1018 (Fig. 2 C and D). The intersubunit disulfide bond extends across the twofold axis, between Cys987 in each monomer. Dimer formation buries a total surface area of ∼1,200 Å2. The two subunits in the dimer are related by the dyad, except for domain I, whose orientation relative to domain II differs by 58° in the two subunits, despite the disulfide bond between Cys751 and Cys798, which links the two domains (Fig. S3). Thus, the domain I–II interface appears to be flexible and to allow large hinge motions of the type observed during the fusogenic conformational change in class II fusion proteins (33). E2 dimers elute from a size-exclusion column slightly earlier at pH 7.5 than at pH 5.5, indicating that the dimers have a larger hydrodynamic radius at pH 7.5 (Fig. S2). This is consistent with the E2 domains maintaining an elongated configuration at pH 7.5 and adopting a more compact configuration, or range of configurations, possibly due to increased flexibility of the domain I–II interface as the pH is lowered.
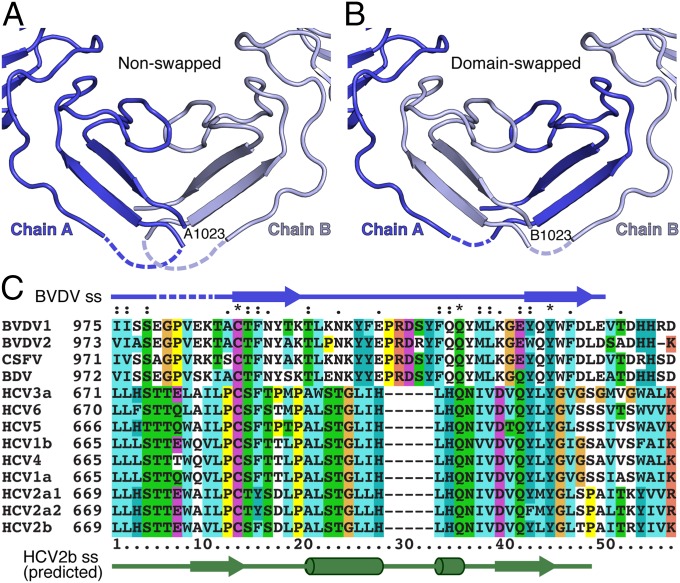
A loop in module IIIc (residues 979–984) is disordered in the E2 structure. Because this loop is close to the corresponding loop in the dimer partner molecule, the connectivity of the protein is ambiguous. It is possible that the polypeptide chain of each subunit crosses the dyad into the dimer partner (Fig. 3). This “domain swap” would approximately double the buried surface area within the dimer interface, which would then include three β-sheet hydrogen bonds (between residues 976–978 and 989–991). A domain swap would greatly increase the energetic cost of dissociating E2 dimers into monomers, consistent with the stability of the dimers in solution. Electron density features near the twofold axis in one subunit of the full-length E2 ectodomain structure are also consistent with a domain-swapped configuration. We note that module IIIc is the most conserved region of E2 in pestiviruses and that its sequence has a significant level of similarity to the C-terminal region of HCV E2 proteins (Fig. 3C). Threading of the HCV E2 sequence into the BVDV E2 structure with Phyre2 reveals a good match between the predicted secondary structure of HCV E2 and the known secondary structure of BVDV E2 (Fig. S4).
Fig. 3.
Potential domain swap at the conserved dimer interface. A loop in module IIIc (residues 979–984) is disordered in the structure of E2 (dashed lines). Because the ends connecting to the loop are close to the dyad, residues on the C-terminal end of the loop (985–1023) may cross the dyad into the dimer partner. (A) The nonswapped structure, as deposited. (B) The domain-swapped structure, which doubles the buried surface area within the dimer interface. (C) Module IIIc is the most conserved region of E2 in pestiviruses. The sequence of module IIIc has similarity to the C-terminal region of HCV E2 proteins. Residue numbers (Left) follow polyprotein numbering. The secondary structure elements of BVDV E2 (blue arrows) and predicted secondary structure elements of HCV2b E2 (green arrows, cylinders) are shown.
Antibody Epitope Mapping.
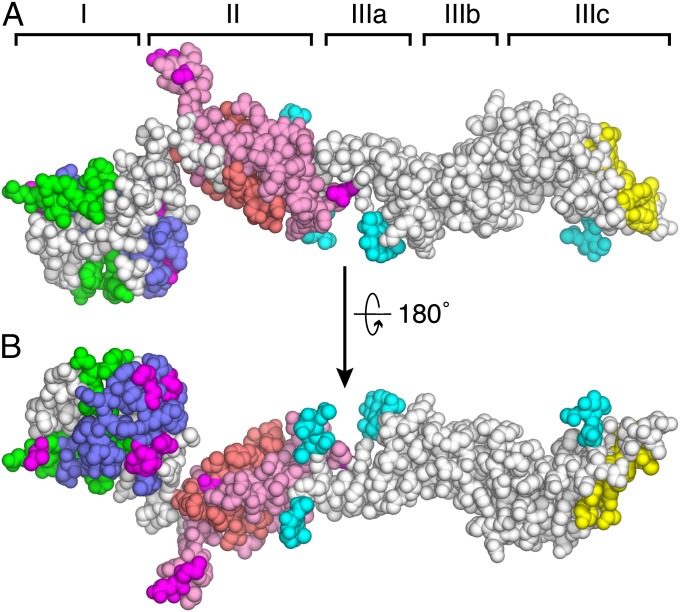
The antibody reactivity of CSFV E2 has been studied in more detail than that of BVDV E2. Epitope mapping based on competitive antibody binding assays and neutralization-escape mutations has identified four distinct antigenic domains (A–D) (34). Domains A and D map to domain II in the BVDV E2 crystal structure; domains B and C correspond to domain I (Fig. 4). Domain III does not contain any antibody epitopes, suggesting that it is not exposed on the viral surface. Most of the neutralization-escape mutations and glycans are clustered on one face of E2 (Fig. 4B), suggesting that this face is solvent exposed. The opposite face (Fig. 4A) is likely to define the interface with E1.
Fig. 4.
Mapping of antigenic regions. (A) Location of the four antigenic domains (A–D) of CSFV E2 in the structure of BVDV E2 viewed in the same orientation as in Fig. 1C in space-filling representation. Antigenic domains B (green) and C (blue) are in domain I. Domains A (pink and salmon) and D (salmon) map to domain II. Most of the neutralization-escape mutations (magenta) and glycans (cyan) are exposed on the opposite face of E2 (B), suggesting that this face is exposed on the viral surface. Residues forming the E2 dimer interface are in yellow.
Discussion
Although HCV E1 and E2 have both been predicted to resemble the class II fusion proteins found in alphaviruses and flaviviruses (35), several strands of evidence suggest that the HCV glycoproteins are instead more similar to their pestivirus homologs. Viruses from the two genera have similar morphologies in electron micrographs and contain only two envelope glycoproteins, E1 and E2 (3–5). Moreover, formation of E1–E2 heterodimers through interactions involving the transmembrane segments of the two proteins is required for cell entry of both pesti- and hepaciviruses (22, 23). In HCV, as in pestiviruses, E1 is half the size of E2, which is the immunodominant protein and binds a cellular receptor that is not efficiently internalized (8, 9). HCV and pestiviruses both appear to require one or more coreceptors for postattachment internalization and membrane fusion (10, 11). HCV and pestiviruses are also both unusually resistant to acid outside the cell yet depend on low pH and an additional activation step for fusion (6, 7, 12). Like BVDV, hepaciviruses have a conserved histidine residue in the N-terminal region of E2 that may contribute to pH sensing, and the two genera share some sequence similarity in the functionally important C-terminal region of E2. Together, these observations suggest that the structure of BVDV E2 should provide a useful model for HCV E2.
Specific features within the E2 structure provide insight into the fusion mechanism. Histidine protonation during endosomal acidification is a common mechanism for pH sensing in viral fusion proteins. The position of the only conserved histidine residue (His762) on the surface of domain I at the membrane-distal end of the molecule indicates that His762 protonation does not destabilize the structure of E2. Thus, if His762 contributes to pH sensing it may be by destabilizing an E1–E2 interface that is present only in the prefusion conformation. Protonation of His762 may also promote membrane fusion by stabilizing the postfusion conformation of E2. Consistent with a possible role for His762 in pH sensing, a conserved histidine near the N terminus of HCV E2 (His445) was recently shown to be important for pH sensing (36).
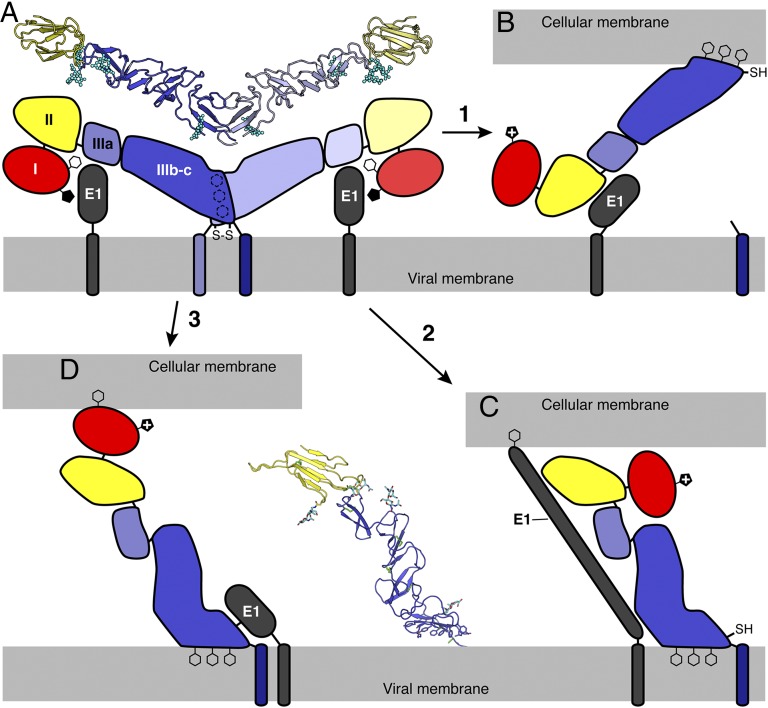
Viral fusion proteins respond to the reduced pH of endocytic compartments (or to other environmental cues) with a conformational change that exposes a hydrophobic fusion motif allowing it to insert into the endosomal membrane. The proteins then fold back on themselves, forcing the cell membrane (held by the fusion loop) and the viral membrane (held by a transmembrane anchor) against each other, resulting in fusion of the viral and endosomal membranes. In the absence of an obvious fusion motif, a key question regarding the pestivirus fusion mechanism is how the viral fusion apparatus anchors itself in the host cell membrane. In HCV, a hydrophobic sequence in E1 (residues 276–286) has been proposed to function as a fusion motif (37). If this is the case and E1 is the fusion protein, E2 is likely to function as a coeffector of fusion providing structural integrity to the fusion complex (Fig. 5). Protonation of His762 in BVDV E2 could still control exposure of the fusion motif in E1, for example by destabilizing an interaction with the fusion motif region.
Fig. 5.
Possible pestivirus membrane fusion mechanisms. Three alternative mechanisms are proposed for insertion of a fusion motif into the target host cell membrane. (A) E2 forms a disulfide-linked dimer and is associated with E1 on the surface of the infectious virion. The black pentagon represents the unprotonated side chain of His762. (B) Activation of E2 by the reduced pH of the endosome, disulfide isomerase activity and proteolytic cleavage of E2 near the C-terminal transmembrane domain causes E2 dimers to dissociate. The cluster of aromatic residues in domain IIIc functions as a fusion motif. E2 remains associated to the viral membrane via E1. Both glycoproteins contribute to the subsequent fusogenic conformational change. (C) E1 contains the fusion motif. E2 functions as a coeffector of fusion providing structural integrity to the fusion complex. Protonation of His762 controls exposure of the fusion motif in E1 by destabilizing an interaction between E2 and the fusion motif. (D) Domain I contains an as yet unidentified fusion motif, which becomes exposed under specific conditions, for example in the presence of a lipid bilayer. This topology would place the fusion motif of the opposite end of E2 from the viral membrane, as would be expected in a fusion protein. (C and D) If domain IIIc does not function as the fusion motif, the array of aromatic side chains in domain IIIc could insert into cellular membrane, positioning the fusion motif toward the target membrane. See also Fig. S5.
Alternative cellular membrane-anchoring mechanisms are also possible. The cluster of aromatic residues at the dimer interface in domain IIIc, could function as a highly effective membrane anchor. This would require disruption of the E2 dimer interface and proteolytic cleavage of E2 at the C-terminal end of its ectodomain, by a cathepsin for example, while maintaining indirect association of E2 to the viral membrane via E1 (Fig. 5B). BVDV E2 dimers dissociate in Laemmli buffer without heating (Fig. S2). Low pH and reducing agent support only low levels of fusion activity (6). There is, however, no direct evidence that E2 requires proteolytic activation.
As a third possible membrane anchoring mechanism we propose that domain I may contain an as yet unidentified fusion motif, which may only become exposed under specific conditions, for example in the presence of a lipid bilayer (Fig. 5D). This topology would place the fusion motif on the opposite end of E2 from the viral transmembrane domain, as would be expected in a fusion protein.
Even if domain IIIc does not function as the fusion motif, the array of aromatic side chains in domain IIIc could insert into viral membrane, without the requirement for proteolytic cleavage. Membrane insertion of the motif would position E2 in an orientation approximately perpendicular to the viral and host cell membranes, with the fusion motif positioned toward the target membrane (Fig. 5 C and D). If the E2 dimers do not dissociate, the hydrophobic contacts between side chains in the aromatic array may serve as a hinge (Fig. S5). A role for domain IIIc as a membrane anchor or hinge would be consistent with the high degree of conservation of its sequence across pestiviruses. Any significant molecular motions involving domain IIIc would require the breakage or isomerization of the disulfide bond at Cys987, which would explain the requirement for reducing agent in addition to low pH for fusion-from-without by pestiviruses. Reports of BVDV E1–E2 disulfide cross-links (19–21) suggest that an unpaired cysteine in E1 catalyzes E2 dimer dissociation by exchange with the disulfide at Cys987. Retroviruses provide a precedent for such a mechanism. A disulfide bond between the two subunits of the cleaved retroviral Env protein inhibits fusion until receptor binding exposes an unpaired cysteine thiol, which isomerizes the disulfide bond, allowing the regulatory subunit to dissociate (38).
After we completed this study, another article reporting the structure of BVDV1 E2 (strain Pe515) was published independently (39). The sequence of the Pe515 strain is 85% identical to the American prototype cytopathic BVDV1 NADL strain used in our study. The structure of BVDV1 Pe515 E2 determined at neutral pH is similar to the structures described here, except that domain IIIc is in the domain-swapped configuration. The root-mean square deviation of main chain atoms in domain I, the least conserved domain in the Pe515 and NADL structures (79% sequence identity), is 0.95 Å. In a second structure of Pe515 E2 determined at low pH, domain I was completely disordered (39), in contrast to our structure. David I. Stuart and colleagues reached the conclusion, consistent with our own, that E1 contains the fusion motif and E2 acts as a structural scaffold for E1. Additional parallels are identified is this study between the pestiviruses and HCV. Their many common structural and functional properties suggest that the structure of BVDV E2 will be useful as a model for HCV E2, and that the membrane fusion mechanisms discussed here are likely to be conserved in HCV.
Materials and Methods
Protein Expression and Purification.
Genes encoding residues 693–1030 (E2-ECD) or residues 783–1030 (∆N90-E2-ECD) of E2 from bovine viral diarrhea virus 1 strain NADL (GenBank accession code 9626650), preceded by an N-terminal eight-histidine purification tag followed by the linker sequence Ser-Ser-Gly and a tobacco etch virus (TEV) protease cleavage site (ENLYFQGP), were subcloned into the pAcGP67 vector (BD Biosciences) in frame with the baculovirus gp67 signal sequence. Spodoptera frugiperda (Sf9) insect cells (Invitrogen) were cotransfected with one of the E2 expression constructs and DiamondBac baculovirus genomic DNA (Sigma-Aldrich) to produce a recombinant baculovirus expressing E2-ECD or ∆N90-E2-ECD. Virus stocks were amplified with three sequential infections of Sf9 cells. For E2 expression, Trichoplusia ni (Tni) insect cells (Expression Systems) grown at 27 °C were infected at a density of 2 × 106 cells/mL with 1.0% (vol/vol) of third-passage (P3) baculovirus stock. After culture in suspension for 96–105 h at 20 °C the culture media was collected and its pH was adjusted with 10 mM triethanolamine (TEA) pH 7.5.
E2 was purified by nickel affinity chromatography with a HisTrap HP column (GE Healthcare) and then treated with TEV protease and Peptide N-glycosidase (PNGase) F (New England Biolabs) at 25 °C for 12 h to remove the histidine tag and glycans. Glycan removal was incomplete and PNGase treatment was not necessary to obtain crystals; however, the treatment improved the diffraction properties of the crystals. Uncleaved E2 and N-terminal TEV cleavage fragments were removed with a second round of nickel affinity chromatography. Untagged E2 was purified to homogeneity by size-exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare). The size-exclusion buffer was 10 mM TEA, pH 7.5, 0.3 mM DTT, 0.5 mM glutathione disulfide, 50 mM NaCl, and 2 mM β-mercaptoethanol. Protein samples were concentrated to 4.5–6 g/L, frozen in liquid nitrogen, and stored at −80 °C in 10 mM TEA, pH 7.5, 50 mM NaCl, and 2 mM β-mercaptoethanol.
Hydrodynamic and Multiangle Scattering Analysis.
Analytical size-exclusion chromatography and multiangle light scattering (MALS) experiments were performed in 50 mM Bis-Tris pH 5.0 or TEA pH 7.5 and 50 mM NaCl with or without 10 mM DTT or Tris(2-carboxyethyl)phosphine (TCEP). A total of 0.1 mL of E2-ECD or E2-ECD (1-3 g/L), pretreated with 10 mM DTT at pH 5 or 7.5, was loaded onto a Superdex 200 10/300 column coupled to a DAWN EOS spectrometer and OPTILAB DSP interferometric refractometer (Wyatt Technology) at a flow rate of 0.5 mL/min. E2-ECD was detected as it eluted from the column with a UV detector at 280 nm, a light scattering detector at 690 nm, and a refractive index detector. The molar mass of E2-ECD was determined from the Debye plot of light scattering intensity versus scattering angle. Data processing was performed with ASTRA software (Wyatt Technology).
Crystallization and Structure Determination of E2.
Crystals of E2-ECD or ∆N90-E2-ECD were grown by hanging drop vapor diffusion at 20 °C. E2 at 4–6 g/L was mixed with a half-volume of reservoir solution: 10% (wt/vol) polyethylene glycol 3350 (PEG 3350), 0.1 M Bis-Tris pH 5.5, 50 mM cesium chloride, 40 mM calcium acetate, and 10% (vol/vol) glycerol. Plate-shaped crystals reached a size of 100 × 50 × 10 µm in 2–3 wk. After 3 mo, the crystals were dehydrated for 24 h by replacing the well solution with 30% (wt/vol) PEG 3350, 0.1 M Bis-Tris pH 5.5, 50 mM cesium chloride, 41 mM calcium acetate, and 10% (vol/vol) glycerol. ∆N90-E2-ECD crystals were derivatized by soaking in the reservoir solution plus 5.0 mM UO2(CH3COO)2 for 48 h and then were frozen in liquid nitrogen. Data were collected at 100 K on a PILATUS detector (Dectris) and processed with XDS (40). Initial experimental electron density maps with interpretable solvent boundary features were obtained using a merged U-soaked single-anomalous scattering (SAS) ∆N90-E2-ECD dataset at 3.5 Å resolution and a back-soaked native ∆N90-E2-ECD dataset at 3.8 Å resolution followed by density-modification procedures with PHENIX AutoSolve (41). These maps were interpretable for domain II and domains IIIa–b of both molecules in the asymmetric unit, but not for domain IIIc. Using the initial density-modified experimental phases as external phases as described (42), we refined individual heavy atom structures for each of three U-soaked datasets and calculated improved experimental phases. Before density modification, experimental maps generated using three independently refined U-SAS structures followed by phase combination showed a much clearer solvent boundary than the corresponding maps after merging three U-SAS datasets. In parallel, these datasets were treated as three isomorphous pairs to yield three single-isomorphous replacement (SIR) phases, phase recombination as described above also yielded a clear solvent boundary. All SAS and SIR phases were then combined, producing a greatly improved solvent boundary for domains II and III (including IIIc). Twofold domain averaging was performed for density modification as implemented in CCP4 DMMULTI (43). The atomic model was completed with COOT (44) and refined to an Rfree of 28.9% with PHENIX and REFMAC (45). The structure of E2-ECD was determined by molecular replacement with PHENIX using ∆N90-E2-ECD as the search model. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID codes 4ILD and 4JNT). See Table 1 for data collection and refinement statistics. See also SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Charles M. Rice (The Rockefeller University) for the generous gift of cDNA of BVDV E2, William Eliason for guidance on MALS experiments, Wuyi Meng for advice on data processing, and the staff of beamline 24-ID-C of the Northeastern Collaborative Access Team at the Advanced Photon Source (Argonne National Laboratory) and the staff of beamline X25 at the National Synchrotron Light Source (Brookhaven National Laboratory). Both beamlines are supported by the US Department of Energy. This work was supported by a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease Award and National Institutes of Health Grants P01 GM022778 and R01 GM102869 (to Y.M.). J.W. was supported by the Steitz Center for Structural Biology, Gwangju Institute of Science and Technology, Republic of Korea.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4ILD and 4JNT).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300524110/-/DCSupplemental.
References
- 1.Lindenbach BD, Thiel H-J, Rice CM. 2007. Flaviviridae: The viruses and their replication. Fields Virology, eds Knipe DM, Howley PM (Lippincott Williams and Wilkins, Philadelphia), 5th Ed, pp 1102–1152.
- 2.Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5(9):558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 3.Gastaminza P, et al. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J Virol. 2010;84(21):10999–11009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wegelt A, Reimann I, Granzow H, Beer M. Characterization and purification of recombinant bovine viral diarrhea virus particles with epitope-tagged envelope proteins. J Gen Virol. 2011;92(Pt 6):1352–1357. doi: 10.1099/vir.0.029330-0. [DOI] [PubMed] [Google Scholar]
- 5.Yu X, et al. Cryo-electron microscopy and three-dimensional reconstructions of hepatitis C virus particles. Virology. 2007;367(1):126–134. doi: 10.1016/j.virol.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 6.Krey T, Thiel HJ, Rümenapf T. Acid-resistant bovine pestivirus requires activation for pH-triggered fusion during entry. J Virol. 2005;79(7):4191–4200. doi: 10.1128/JVI.79.7.4191-4200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meertens L, Bertaux C, Dragic T. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J Virol. 2006;80(23):11571–11578. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krey T, et al. Function of bovine CD46 as a cellular receptor for bovine viral diarrhea virus is determined by complement control protein 1. J Virol. 2006;80(8):3912–3922. doi: 10.1128/JVI.80.8.3912-3922.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petracca R, et al. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J Virol. 2000;74(10):4824–4830. doi: 10.1128/jvi.74.10.4824-4830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tscherne DM, Evans MJ, Macdonald MR, Rice CM. Transdominant inhibition of bovine viral diarrhea virus entry. J Virol. 2008;82(5):2427–2436. doi: 10.1128/JVI.02158-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Hahn T, Rice CM. Hepatitis C virus entry. J Biol Chem. 2008;283(7):3689–3693. doi: 10.1074/jbc.R700024200. [DOI] [PubMed] [Google Scholar]
- 12.Tscherne DM, et al. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J Virol. 2006;80(4):1734–1741. doi: 10.1128/JVI.80.4.1734-1741.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathapati BS, et al. Entry of bovine viral diarrhea virus into ovine cells occurs through clathrin-dependent endocytosis and low pH-dependent fusion. In Vitro Cell Dev Biol Anim. 2010;46(5):403–407. doi: 10.1007/s11626-009-9263-9. [DOI] [PubMed] [Google Scholar]
- 14.Campos SK, Chapman JA, Deymier MJ, Bronnimann MP, Ozbun MA. Opposing effects of bacitracin on human papillomavirus type 16 infection: Enhancement of binding and entry and inhibition of endosomal penetration. J Virol. 2012;86(8):4169–4181. doi: 10.1128/JVI.05493-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, et al. Identification of host cell binding peptide from an overlapping peptide library for inhibition of classical swine fever virus infection. Virus Genes. 2011;43(1):33–40. doi: 10.1007/s11262-011-0595-7. [DOI] [PubMed] [Google Scholar]
- 16.Liang D, et al. The envelope glycoprotein E2 is a determinant of cell culture tropism in ruminant pestiviruses. J Gen Virol. 2003;84(Pt 5):1269–1274. doi: 10.1099/vir.0.18557-0. [DOI] [PubMed] [Google Scholar]
- 17.Pileri P, et al. Binding of hepatitis C virus to CD81. Science. 1998;282(5390):938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 18.Hadlock KG, et al. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J Virol. 2000;74(22):10407–10416. doi: 10.1128/jvi.74.22.10407-10416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiland E, et al. Pestivirus glycoprotein which induces neutralizing antibodies forms part of a disulfide-linked heterodimer. J Virol. 1990;64(8):3563–3569. doi: 10.1128/jvi.64.8.3563-3569.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rümenapf T, Unger G, Strauss JH, Thiel HJ. Processing of the envelope glycoproteins of pestiviruses. J Virol. 1993;67(6):3288–3294. doi: 10.1128/jvi.67.6.3288-3294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiel HJ, Stark R, Weiland E, Rümenapf T, Meyers G. Hog cholera virus: Molecular composition of virions from a pestivirus. J Virol. 1991;65(9):4705–4712. doi: 10.1128/jvi.65.9.4705-4712.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronecker S, Zimmer G, Herrler G, Greiser-Wilke I, Grummer B. Formation of bovine viral diarrhea virus E1-E2 heterodimers is essential for virus entry and depends on charged residues in the transmembrane domains. J Gen Virol. 2008;89(Pt 9):2114–2121. doi: 10.1099/vir.0.2008/001792-0. [DOI] [PubMed] [Google Scholar]
- 23.Patel J, Patel AH, McLauchlan J. The transmembrane domain of the hepatitis C virus E2 glycoprotein is required for correct folding of the E1 glycoprotein and native complex formation. Virology. 2001;279(1):58–68. doi: 10.1006/viro.2000.0693. [DOI] [PubMed] [Google Scholar]
- 24.Fritz R, Stiasny K, Heinz FX. Identification of specific histidines as pH sensors in flavivirus membrane fusion. J Cell Biol. 2008;183(2):353–361. doi: 10.1083/jcb.200806081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nayak V, et al. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J Virol. 2009;83(9):4338–4344. doi: 10.1128/JVI.02574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin ZL, Zheng Y, Kielian M. Role of conserved histidine residues in the low-pH dependence of the Semliki Forest virus fusion protein. J Virol. 2009;83(9):4670–4677. doi: 10.1128/JVI.02646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, Sánchez-San Martín C, Qin ZL, Kielian M. The domain I-domain III linker plays an important role in the fusogenic conformational change of the alphavirus membrane fusion protein. J Virol. 2011;85(13):6334–6342. doi: 10.1128/JVI.00596-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Boer SM, et al. Acid-activated structural reorganization of the Rift Valley fever virus Gc fusion protein. J Virol. 2012;86(24):13642–13652. doi: 10.1128/JVI.01973-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dessau M, Modis Y. Crystal structure of glycoprotein C from Rift Valley fever virus. Proc Natl Acad Sci USA. 2013;110(5):1696–1701. doi: 10.1073/pnas.1217780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545-9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krey T, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6(2):e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vieyres G, et al. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J Virol. 2010;84:10159–10168. doi: 10.1128/JVI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427(6972):313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 34.van Rijn PA, Miedema GK, Wensvoort G, van Gennip HG, Moormann RJ. Antigenic structure of envelope glycoprotein E1 of hog cholera virus. J Virol. 1994;68(6):3934–3942. doi: 10.1128/jvi.68.6.3934-3942.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garry RF, Dash S. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology. 2003;307(2):255–265. doi: 10.1016/s0042-6822(02)00065-x. [DOI] [PubMed] [Google Scholar]
- 36.Boo I, et al. Distinct roles in folding, CD81 receptor binding and viral entry for conserved histidine residues of hepatitis C virus glycoprotein E1 and E2. Biochem J. 2012;443(1):85–94. doi: 10.1042/BJ20110868. [DOI] [PubMed] [Google Scholar]
- 37.Drummer HE, Boo I, Poumbourios P. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J Gen Virol. 2007;88(Pt 4):1144–1148. doi: 10.1099/vir.0.82567-0. [DOI] [PubMed] [Google Scholar]
- 38.Wallin M, Ekström M, Garoff H. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 2004;23(1):54–65. doi: 10.1038/sj.emboj.7600012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.El Omari K, Iourin O, Harlos K, Grimes JM, Stuart DI. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013;3(1):30–35. doi: 10.1016/j.celrep.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55(1):94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rould MA, Perona JJ, Steitz TA. Improving multiple isomorphous replacement phasing by heavy-atom refinement using solvent-flattened phases. Acta Crystallogr A. 1992;48(Pt 5):751–756. doi: 10.1107/s0108767392003404. [DOI] [PubMed] [Google Scholar]
- 43.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.