Significance
Neuroinvasive herpesviruses cause diseases in humans ranging from cold sores to central nervous system infections. Unlike most icosahedral viruses, herpesvirus capsids are surrounded by protein layers that lack polyhedral architecture. The outer layers are critical for herpesvirus infectivity. Although the disorganized layers are visible by electron microscopy, the protein topography of these layers remains unclear. We fused fluorophores to virus proteins and pinpointed their positions within virions at nanometer resolution. The findings reveal a complex and variable asymmetric architecture, shed light on herpesvirus assembly, and provide a foundation for visualizing viral particle dynamics during herpesvirus infection.
Keywords: pseudorabies, virus, heterogeneity, asymmetry, point-spread function
Abstract
The herpesvirus virion is a multilayered structure consisting of a DNA-filled capsid, tegument, and envelope. Detailed reconstructions of the capsid are possible based on its icosahedral symmetry, but the surrounding tegument and envelope layers lack regular architecture. To circumvent limitations of symmetry-based ultrastructural reconstruction methods, a fluorescence approach was developed using single-particle imaging combined with displacement measurements at nanoscale resolution. An analysis of 11 tegument and envelope proteins defined the composition and plasticity of symmetric and asymmetric elements of the virion architecture. The resulting virion protein map ascribes molecular composition to density profiles previously acquired by traditional ultrastructural methods, and provides a way forward to examine the dynamics of the virion architecture during infection.
Herpesviruses are responsible for a broad range of diseases in humans and other animals. The herpesvirus virion consists of four components: (i) the linear dsDNA genome, (ii) a 125-nm diameter T = 16 icosahedral capsid, (iii) a tegument consisting of more than 20 proteins that surround the capsid, and (iv) a lipid bilayer envelope studded with viral glycoproteins. The fully assembled particle is ∼200–250 nm in diameter and is referred to as the heavy particle (H-particle) (1). The capsid has been solved to 8.5 Å resolution and a fraction of the tegument that is symmetrically bound to the capsid surface has been solved to 20 Å resolution by cryo-electron microscopy (cryo-EM) reconstruction (2–4). The detailed resolution afforded by cryo-EM results from the averaging of many particles that are aligned in silico, based on icosahedral symmetry. However, such studies provide an incomplete picture of herpesvirus virions because unlike some smaller enveloped viruses that project icosahedral symmetry into the envelope proteins, the herpesvirus envelope, and the majority of the tegument mass lack radial symmetry and are not “seen” by cryo-EM (5–7). Our understanding of these variable structural layers predominantly comes from single-particle imaging methods that do not use symmetry-based averaging. In particular, cryo-electron tomography (cryo-ET) has provided insight into the general structure of the outer virion layers, but has not yielded sufficient detail to resolve the organization of the constituent protein components (8).
Extracellular herpes virions are metastable structures that are triggered by interactions between virion membrane surface proteins and corresponding receptors on the cell, culminating in membrane fusion (9). Although tegument proteins are critical for postfusion steps in nuclear delivery (10–14), their perceived role before cell entry has been as rigid structural elements that bridge the capsid shell to the tails of envelope membrane proteins (15, 16). However, recent evidence indicates that the tegument may reorganize before membrane fusion (17–19). A deeper understanding of the relevance of these internal virion dynamics requires a detailed description of the herpesvirus structure, which has been limited because of its asymmetric architecture and compositional variance. Using an approach that combines advances in recombinant herpesvirus production and single-particle fluorescence imaging, we mapped the location of viral proteins to subregions within the tegument and envelope and correlated the findings to variance in protein acquisition.
Results
To study herpesvirus structure by fluorescence microscopy, an infectious clone of the alphaherpesvirus, pseudorabies virus (PRV), was modified by homologous recombination to express fluorescent protein fusions to envelope, tegument, and capsid components (20). All recombinant viruses in this study had mRFP1 (monomeric red fluorescent protein) fused to the N terminus of pUL35: a protein present on the tips of hexons, which are symmetrically distributed on the capsid (21, 22). Additionally, GFP was fused to 11 structural viral proteins, one nonstructural viral protein, one host protein, or expressed by the virus as an unfused molecule (Table S1). Although many fluorophore fusions attenuate virus production, only viruses that propagated to wild-type titers were included in this study.
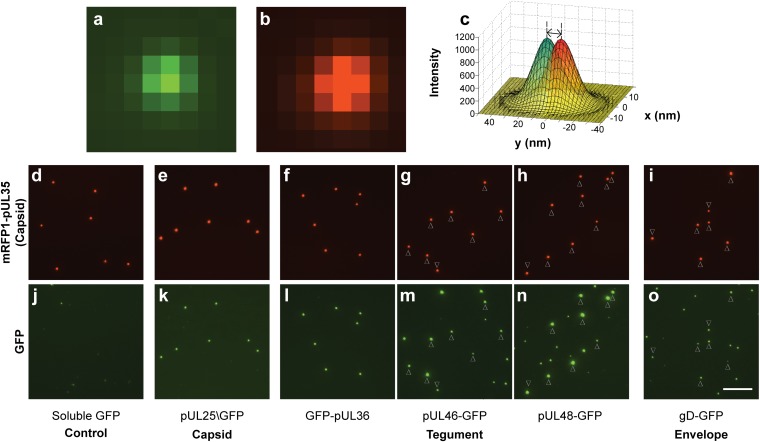
Herpesvirus particles were observed by infecting pig kidney cells (PK15) with dual-fluorescent PRV at a multiplicity of infection (MOI) of 5 pfu per cell. Eighteen hours postinfection (hpi), 10 mL of culture supernatant was collected, cleared of cell debris, and pelleted through a 10% (wt/vol) Nycodenz cushion (Accurate Chemical). Virions prepared in this method have a particle:pfu ratio below 10, indicating structural integrity is maintained (23). The viral particles were resuspended in 100 µL of PBS, further diluted 1:50 in PBS, and a 65-µL volume was applied to a plasma-cleaned cover glass. Red and green fluorescence was captured using sequential 1.5-s exposures. Fluorescently tagged H-particles produced Airy disks that were consistent with diffraction-limited emissions (Fig. 1 A and B). For each recombinant virus, three independent preparations were examined by capturing > 30 image fields each, allowing the analysis of 500–2,000 H-particles per preparation.
Fig. 1.
Acquisition of fluorescent proteins in to viral particles. (A and B) Fluorescence micrographs of a single diffraction-limited PRV particle expressing gD-GFP (A) and RFP-pUL35 (B). (C) Example of a red-green displacement measurement using simulated data: 2D Gaussian curves are shaded red and green, vertical dashed lines are the centroids, and the double-headed arrow is the measured displacement. (D–O) Micrographs of dual-fluorescent PRV particles. (D–I) mRFP1-pUL35 capsids. (J–O) corresponding GFP in matched fields. Where L-particles are abundant, arrowheads represent the position of H-particles. All exposures were 1.5 s. Images of the same color have equivalent intensity scaling. (Scale bar, 10 µm.)
GFP-fusion proteins differed in abundance and distribution among H-particles and light particles (L-particles) (Fig. 1 D–O). L-particles, which lack capsids, are abundantly released from infected cells but are not themselves infectious (1). Using an image analysis algorithm, individual viral particles were identified based on the emission of diffraction-limited red and green fluorescence. The emission intensities of individual particles were measured and categorized as either H- or L-particles based on the presence or absence of red-capsid signal. This accounting was consistent with previous biochemical characterizations of H- and L-particles (1, 24, 25). The RFP-pUL35 fluorescence emissions from H-particle populations were Gaussian in distribution, consistent with the copy-controlled incorporation of the protein on the icosahedral capsid surface (Fig. 2A and Fig. S1) (22, 26). As expected, the distribution and average brightness of RFP-pUL35 did not deviate significantly among the recombinant viruses studied with the exception of the pUL47 sample, which was distinct statistically from pUL25 sample (Fig. 2B).
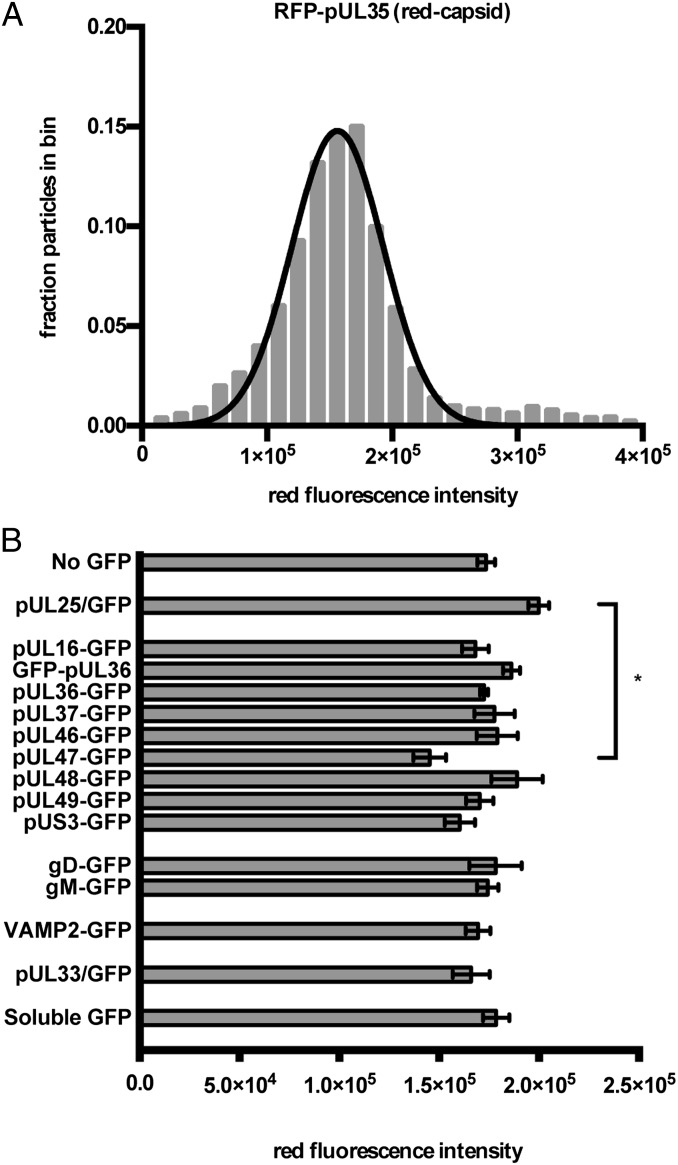
Fig. 2.
Capsid fluorescence properties of recombinant H-particles. (A) Representative frequency distribution of red capsid fluorescence. Red fluorescence intensity (x axis) is expressed in arbitrary units. Nonlinear regression was used to fit a normal distribution to the histogram. For all strains, the goodness of fit (R2) of the data to a normal distribution was greater than 0.95. (B) The average red fluorescence particle intensity for each recombinant virus. The x axis indicates intensity values expressed as arbitrary fluorescence units. Error bars represent the SEM for n ≥ 3 independent experiments. *P < 0.05, as determined by one-way ANOVA and a post hoc Tukey test.
Although all H-particles contain tegument and an envelope, we set out to determine how frequently and to what degree specific protein species contribute to these layers. GFP emissions were detected from ≥ 85% of H-particles when fused to any of the 11 structural proteins. In contrast, the majority of H-particles isolated from cells infected with PRV encoding GFP fused to a nonstructural viral protein, GFP-pUL33, were not green fluorescent (Fig. 3A). The copy number of GFP-tagged proteins in each H-particle was measured as a proportion of the fluorescence intensity of the capsid protein, pUL25 (24, 27, 28). Acquisition of pUL25 during assembly is copy-controlled, and estimates of pUL25 copy number were previously obtained by cryo-EM (60 copies; 5 per vertex) and immunoblot (estimated within the range of 75 ± 15 to 82 ± 19 copies) (29–31). We refined the measurement of pUL25 by comparing the fluorescence of pUL25/GFP to rotavirus (RV) virus-like particles (VLPs), the latter of which invariantly contain 120 copies of GFP (Fig. S2) (32). GFP emission intensities from RV VLPs and PRV H-particles indicated 77 ± 3 copies of pUL25/GFP in monofluorescent (green only) virions or 64 ± 2 copies of pUL25/GFP in dual-fluorescent (red capsid) virions. Thus, using a mean value of 70.5 copies of pUL25/GFP as a standard, the specific H-particle incorporation of other observed GFP-tagged viral proteins ranged between 30 and 458 copies with 9.2% uncertainty. pUL35 was not used as the basis of protein estimates because of previous evidence that GFP-tagged pUL35 is present in reduced copy numbers in virions (33).
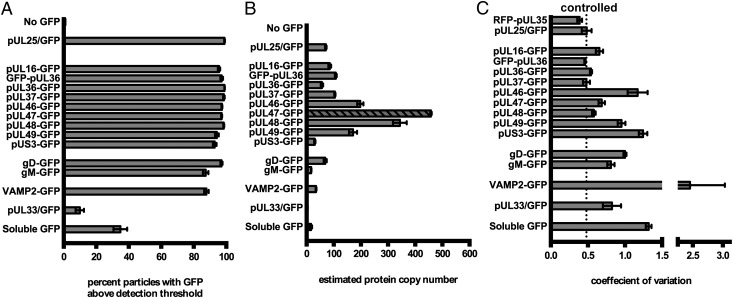
Fig. 3.
Incorporation of GFP-fusion proteins into viral particles. (A) Viral particles were scored positive for GFP-RFP colocalization if GFP was detectable in RFP+ particles after background subtraction. (B) Estimated copy number of GFP-fused proteins in PRV virions. Copy number was determined by dividing the mean green fluorescence of triplicate experiments by the mean fluorescence of pUL25/GFP and multiplied by 70.5 (see text). For pUL47, the exposure time was reduced from 1.5 s to 0.5 s, and the diagonal fill represents the normalized value based on linear extrapolation. (C) CV of GFP fluorescence for each strain. CV was obtained for each sample by dividing the SD of each sample by its mean. Error bars represent SEM for n ≥ 3 independent experiments.
Unlike Western blot analysis, which yields average protein copy numbers in virion populations, single-particle fluorescence analysis allowed for quantification of protein variance. We used the coefficient of variation (CV, σ/μ) as an indicator of the heterogeneity of protein acquisition, where σ is the sample SD and µ is the sample mean fluorescence intensity. The CV for pUL25 was 0.48, which provided a baseline for copy-controlled incorporation (Fig. 3C). In contrast, the amount of unfused GFP incorporated into H-particles was highly variable (CV = 1.3), which may be explained in part by heterogeneity in virion size (34). Because GFP does not contain motifs that direct assembly into virions, the GFP CV provided a baseline for stochastic incorporation. The CV for tegument proteins ranged between 0.46 (pUL36) and 1.24 (pUS3) and approached, but did not exceed, that of unfused GFP. On the high end, the pUS3 and pUL46 tegument proteins had CV approximating unfused GFP indicating there was no copy control of these proteins despite, at least in the case of pUL46, a high average copy number. The low CV of pUL36 and pUL37 was equivalent to the copy-controlled acquisition of capsid protein pUL25, and was consistent with findings of an immunoblot study of pUL37 incorporation (35). These findings indicated a degree of flexibility in virion assembly that varied among the tegument proteins.
Nonselective incorporation of GFP in the virion envelope was modeled by fusing GFP to the cellular vesicle-associated membrane protein 2 (VAMP2). As with incorporation of unfused GFP into the tegument, VAMP2 was incorporated into the envelope with greater variance than virally encoded proteins (CV = 2.5), which was suggestive of random incorporation. Of the two envelope glycoproteins that were tagged with GFP, gD and gM, incorporation was more uniform than VAMP2 but lacked the copy control seen with capsid proteins and a subset of the tegument.
In addition to examining H-particles, we quantified the number and intensity of L-particles that were observed from each fluorescent virus. L:H particle ratios were determined by quantifying all green fluorescent puncta that lacked corresponding red capsid fluorescence (Fig. S3). We could not determine the total percentage of L-particles that contained a given tegument or envelope protein, because L-particles lacking a GFP-tagged protein would be nonfluorescent. Instead, the relative incorporation of specific tegument protein species into L-particles was normalized to H-particle count. The L:H particle ratio was highly variable between the different dual-fluorescent viruses. Tegument proteins had the highest disparities in L:H-particle ratios. pUL36 and pUL37 were observed in approximately half as many L-particles as H-particles, whereas pUS3, pUL16, pUL46, pUL48, and pUL49 were observed in L-particles more frequently than H-particles.
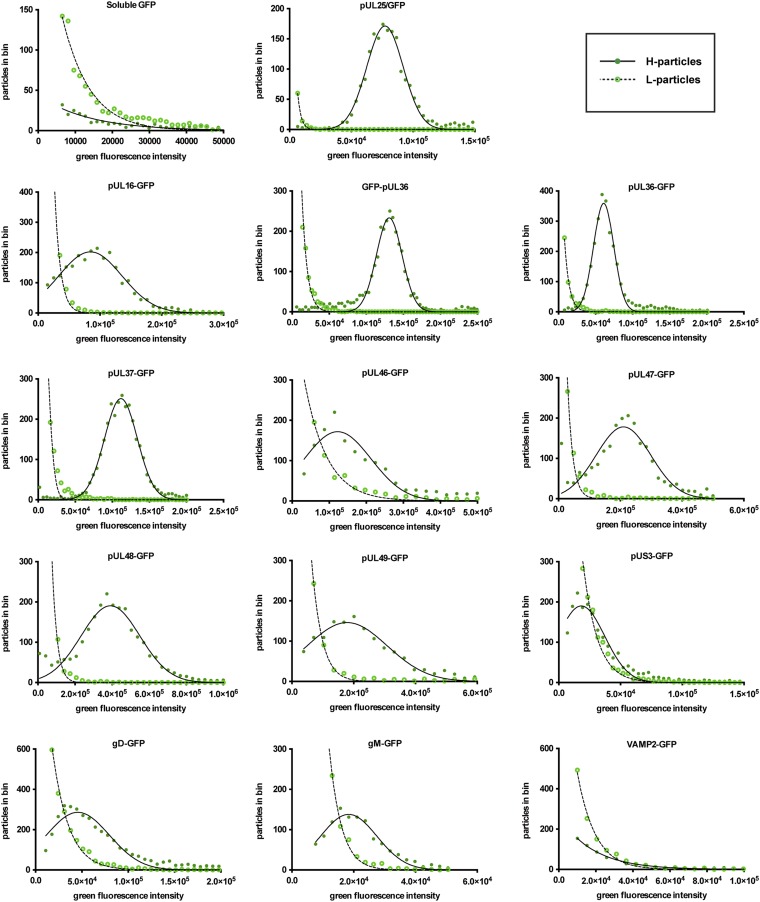
A comparison of the brightness of green fluorescence in H- and L-particles revealed that the presence of a capsid enhanced the acquisition of a subset of proteins into viral particles (Fig. 4). This enhancement was most notable for the pUL36, pUL37, pUL47, and pUL48 tegument proteins, which displayed profiles similar to the H-particle–restricted pUL25 capsid protein (24). In contrast, pUS3, pUL16, pUL46, pUL49, and gD had overlapping levels of protein incorporation between L- and H-particles, indicating that capsids played a smaller role in their acquisition. In stark contrast to all viral proteins, capsids did not enhance incorporation of unfused GFP and VAMP2.
Fig. 4.
Comparison of L- and H-particle protein incorporation. The x axis indicates intensity values expressed as arbitrary fluorescence units. H-particle fluorescence (filled circles) was fit to a normal distribution (solid line), and L-particle fluorescence (empty circles) was fit to a decaying exponential distribution (dashed line). All L-particle decaying exponentials had a goodness of fit R2 > 0.98. Normal distributions fit H-particle data with R2 > 0.95, with the exception pUS3-GFP (R2 = 0.89), pUL46-GFP (R2 = 0.88), and UL47-GFP (R2 = 0.92), pUL48-GFP (R2 = 0.92), and gD-GFP (R2 = 0.93).
The single-particle analysis used to examine virion heterogeneity was next extended to examine protein localization within viral particles. Herpesviruses cannot be spatially resolved by conventional light microscopy. Using a technique derived from fluorescence imaging with one nanometer accuracy (FIONA), we overcame the resolution limit to pinpoint sources of diffraction-limited fluorescence (36). Displacements between diffraction-limited red (capsid; pUL35) and green emissions provided a first indication that some structural proteins were asymmetrically distributed around the capsid surface. To refine this analysis, H-particles were fit to point-spread functions (PSF) to determine the centroids of fluorescence (Fig. 1C). The analysis was restricted to images containing five or more H-particles, which allowed for correction of red-green fluorescence alignment resulting from light path aberrations. The corrected distance between the projected red and green fluorescence centroids was converted to a 3D vector to take into account the random orientation of the viral particles on the cover glass.
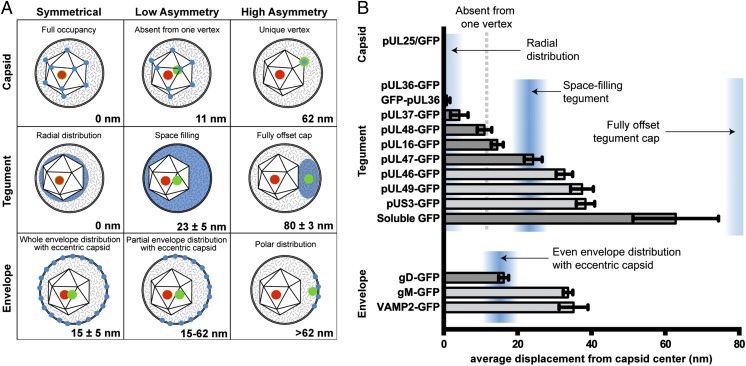
The displacement of capsid protein pUL25 from the red capsid centroid was below a calculated optical noise threshold of 7 nm (Methods), consistent with the reported radial symmetry of both proteins (Fig. 5) (37, 38). Two tegument proteins, pUL36 and pUL37, similarly exhibited no measurable displacement from the capsid, indicative of radial symmetry and consistent with a recent report of the pUL36-capsid interface (3). Other tegument proteins, including pUS3, pUL46, and pUL49, were displaced on average by 30–40 nm from the capsid center. Because the capsid is itself 125 nm in diameter, displacements of this magnitude indicated a biased, but not exclusive, localization on one side of the capsid. Based on these displacement values, we conclude that pUS3, pUL46, and pUL49 are components of the tegument cap that was previously observed by cryo-ET reconstruction (8). Surprisingly, the asymmetric architecture of the herpesvirus H-particle was not limited to the tegument. Although gD and gM are both envelope proteins, they projected strikingly different average displacements from the capsid center.
Fig. 5.
Subvirion protein distributions. (A) Models of predicted displacement values for green fluorescent proteins (blue: postulated distribution; green: centroid of fluorescence emission) from the capsid (red: centroid of capsid fluorescence). Models were derived by determining the centroids of icosahedra (Top), and spheres, which were solid (Middle) or hollow (Bottom). A capsid eccentricity of 15 nm was modeled based on measurements of HSV-1 by Grünewald et al. (8). (B) Displacements of green fluorescence from the red fluorescent capsid. As illustrated in Figs. 1C and 2D, Gaussian curves were used to determine the point source of Airy disks in red and green channels. The bar graph shows the average displacement of the green fluorescence centroid from the red fluorescence centroid. Blue shaded regions indicate likely locations of proteins within the virion based on the models in A. The center of the shaded regions represents the mean expected value, and width represents 1 SD to either side of the mean. The five bars shaded in light gray are not significantly different from one another (see text). Error bars represent the SEM for n ≥ 3 independent experiments.
Discussion
Unlike the structures of many viruses, components of herpesvirus particles have irregular architecture and possess compositional plasticity (39). For some herpesviruses, this flexibility allows for cell-type–dependent changes in particle composition that specify tropism (40, 41). Aspects of herpesvirus structural variance also manifest between particles released from the same cell. H-particles (which include infectious virions) and L-particles (which lack the capsid and viral genome) are readily observed examples of particle variation, but H-particles also display intrinsic heterogeneity (8, 42, 43). Although there were initial concerns that variations in H-particle appearance could be an artifact of electron microscopy preparative techniques (34), cryo-ET has confirmed that the icosahedral capsid is positioned eccentrically within the surrounding envelope (8, 44) and monitoring the occupancy of specific viral proteins using fluorescent protein fusions has confirmed that herpesvirus H-particles consist of mixed populations (26, 45, 46).
By imaging single-particle fluorescence, we examined a collection of recombinant viruses derived from the neuroinvasive herpesvirus, PRV. All viruses were designed to encode a RFP tag on the outer capsid shell (mRFP1-VP26) to produce a radially symmetric emission source to mark the center of the capsids within H-particles. In addition, each virus-encoded GFP fused to a protein of interest. A panel of 15 dual-fluorescent viruses that propagated to wild-type titers were examined for variance in particle-to-particle protein acquisition and intravirion localization by superresolution positioning relative to the RFP-capsid. The PSF mapping used for superresolution imaging was similar to applications of FIONA and STORM (stochastic optical reconstruction microscopy), but unlike the latter methods red and green PSFs were fit and the displacement of the centroids was measured. This approach did not require stochastic activation of a sparse fluorophore subpopulation in reiterative cycles nor did it yield image reconstruction, unlike STORM. The 7-nm spatial resolution afforded by this method, coupled with the collection of dual-fluorescent recombinant viruses, made probing the asymmetric protein distributions in the 200- to 250-nm viral particles possible. The accuracy of the analysis was strengthened by automated analysis of large datasets. Although viral particles are often analyzed en masse for protein composition, our analysis allowed the traits of individual particles to be compared (47).
The results reveal how specific protein constituents contribute to particle structure. For example, the pUL36 protein is the only component of the tegument that is known to bind directly to the icosahedral capsid surface in PRV and other alphaherpesviruses (48). In this study, pUL36 fulfilled all expectations of being a tegument protein located proximal to the capsid; the protein was highly enriched in H-particles, copy-controlled, and symmetrically distributed around the capsid. Multiple isoforms of pUL36 are expressed in infected cells, but only full-length protein is readily incorporated into PRV particles (49, 50). Consistent with this, placing GFP at either end of pUL36 did not substantially alter the results. Recently, cryo-EM revealed a portion of pUL36 on the capsid surface and showed a symmetric distribution of pUL36 around capsid vertices (3). The results obtained here are consistent with these observations and further indicate that pUL36 is present at all 12 capsid vertices, including the portal vertex. The latter point was debated because the unique portal vertex lacks the major capsid protein, VP5, which is juxtaposed to pUL36 in reconstructions (3, 51). The pUL37 tegument protein is a binding partner of pUL36 and possessed properties indistinguishable from pUL25 and pUL36 (52, 53).
Before this study, the tegument was inferred to have an “inner” and “outer” layer based on proximity to membrane or capsid surfaces in infected cells before the final envelopment stage of virion assembly, and these designations were bolstered by results obtained by differential virion extraction (54, 55). Our findings further support that proteins such as pUL36 and pUL37 fit the traditional inner tegument definition in that the proteins are symmetrically present on capsids and are scarce in L-particles. However, the findings also indicate that, in the strictest sense, inner and outer tegument is an inexact dichotomy. Three tegument proteins (pUL16, pUL48, and pUL47) had medial asymmetries that fell between the radially distributed tegument proteins and the tegument cap group. The pUL16 and pUL48 displacements were consistent with occupation at all but one vertex of the capsid, which could signify absence of these proteins from the unique portal vertex (28, 51, 56, 57). A vertex assignment for pUL16 is attractive because it is a capsid-associated protein, although the molecular interaction linking it to the capsid is not defined (17, 58). The pUL48 tegument protein is the herpesvirus transactivating factor (α-TIF; VP16), and binds pUL36 (59, 60). pUL48 was proposed as a candidate for linking tegument proteins proximal to the capsid with those more closely associated with the envelope, which is supported by the current findings (61). The pUL47 displacement was accurately modeled as space filling, and there is evidence for its contribution to both inner and outer tegument layers (61–63).
Several tegument and envelope proteins had similar virion properties, which included: pUL46, pUL49, pUS3, gM, and VAMP2. Each protein of this group was heterogeneously acquired into H-particles and abundant in L-particles. In addition, capsid displacements within H-particles were statistically indistinguishable (Fig. 5B, light gray bars). The common displacement of these proteins indicated an intravirion distribution that was not predicted by simple mathematical predictions but was none-the-less consistent with localization to a previously described asymmetric tegument cap (8). The putative tegument cap proteins are accurately predicted by the space-filling model if the capsid-proximal tegument proteins displace the surrounding tegument cap proteins by ∼11 nm from the capsid surface. This model is reasonable given that pUL36 and pUL37 are the largest tegument proteins and collectively amount to an estimated 30 MDa per H-particle.
We were surprised to see that envelope proteins gD and gM had significantly different displacements from the capsid, suggestive of partitioning in the viral envelope. This finding was predicted by the authors of a cryo-ET study of herpesvirus entry into cells, who noted that virions orient at cell surfaces with the tegument dense pole facing away from the cell (18). The authors suggested that the gD entry receptor is exposed at the entry pole but enmeshed in other proteins elsewhere in the envelope. This interpretation is consistent with our observation. Also of note, the gM protein antagonizes gD-triggered fusion in cell fusion assays, which may indicate that the juxtaposition of gD and gM in the envelope can regulate virion entry into cells (64, 65). The concept that partitioning of envelope components may modulate virion entry was recently established for HIV using superresolution imaging (66). In this instance, the envelope of HIV was determined to remodel during virion maturation by clustering the Env protein to one pole, which was essential for efficient HIV entry into cells. Dynamic virion rearrangements are also documented for herpesviruses, and future work will focus on whether maturation or triggering events contribute to herpes entry into cells (17, 19).
Methods
BAC Mutagenesis.
All recombinant PRV strains were derived from the infectious clone pBecker3 as described in Table S1 (20). The viral genome was engineered to express GFP-fusion proteins by either a BAC-flp or En Passant two-step recombination procedure (46, 67). The GFP coding sequence was most frequently inserted into a pBecker3 derivative that encodes a mRFP1-pUL35 (RFP-capsid) fusion (68). Primers used to insert the GFP coding sequence into the PRV genome are listed in Table S2. Soluble GFP and VAMP2-GFP were inserted as an expression cassette driven by the CMV IE promoter that disrupted the gene encoding the nonessential protein pUS4, as previously described (33, 69).
Cells and Viruses.
Infectious clone DNA was transfected into PK15 cells as described previously (46). The harvested virus stock was passaged once more through PK15 cells to create a working stock. Titers of working stocks were determined by plaque assay on PK15 cells as previously described (20). PK15 cells were maintained in DMEM (Gibco) supplemented with 10% (vol/vol) bovine growth serum (BGS; HyClone). Sf9 (Spodoptera frugiperda) cells were grown in Sf-900 II serum free media (Gibco) at 28 °C.
Sample Preparation.
Extracellular viral particle fluorescence was analyzed by collecting supernatants from PRV-infected PK15 cells as previously described (33). Briefly, PK15 cells in 10 cm dishes were infected with PRV strains at an MOI of 5 pfu/cell and incubated for 18 h in phenol red-free DMEM/F12 media (Gibco) supplemented with 2% (vol/vol) BGS (HyClone). Supernatants were cleared of cell debris with a 10 min 3,000 × g centrifugation. Next, 8 mL of cleared supernatant was underlaid with a 1-mL cushion of 10% (wt/vol) Nycodenz (Accurate Chemical) in PBS in a SW41 centrifuge tube. Samples were centrifuged at 38,500 × g for 60 min. Media and the Nycodenz cushion were aspirated from pelleted virus, which was resuspended in 0.1 mL PBS. For light microscopy, resuspended virus was diluted 1:50 in PBS and 0.07 mL was transferred into a sealed microscope slide chamber with a plasma-cleaned No. 1.5, 22 × 22-mm coverslip.
Light Microscopy.
Particles were imaged on a Nikon Eclipse TE2000 U wide-field fluorescence microscope with a Cascade II camera (Roper Scientific), an apochromat 100× 1.49 numerical aperture (NA) objective, and a Lumen PRO Fluorescence Illumination System (Prior Scientific). GFP was imaged using a 470/40-nm excitation filter and 525/50-nm emission filter; mRFP1 was imaged using a 572/35 nm excitation filter and 632/60-nm emission filter (Chroma Technology Corporation). Both fluorophores were imaged with the same Chroma ET GFP/mCherry polychroic beamsplitter. Green and red fluorescence emissions were sequentially captured using 1.5-s exposures. The digitizer of the Cascade II camera was set to 1 MHz with zero electron-multiplying (EM) gain.
Incorporation of GFP into PRV Virions and RV VLPs.
Extracellular PRV pUL25/GFP with no capsid tag (PRV-GS4379) and PRV pUL25/GFP + RFP-pUL35 (PRV-GS3171) were isolated from infected PK15 cells at 18 hpi, as described above. To make RV VLPs, Sf9 cells were coinfected with two baculoviruses (a generous gift from Annie Charpilienne, Laboratory of Molecular and Structural Virology, Gif-sur-Yvette, France), RV VP2-GFP, and RV VP6, at MOI = 10. Expression of RV VP2-GFP and RV VP6 was sufficient for VLP (GFP-VLP2/6) formation (32). Sf9 cell supernatants were collected after baculovirus-induced lysis at 3–4 d after infection. RV VLPs were pelleted and imaged identically to PRV. Next, 100 nm TetraSpeck Beads were sonicated, diluted 1:1,000 in PBS, and imaged identically to PRV.
Quantification of Extracellular Particle Fluorescence and Statistical Analysis.
A custom algorithm for the MetaMorph software package (Molecular Devices) was used to quantify fluorescence intensity of individual extracellular virus particles. Briefly, the algorithm identifies multiple diffraction-limited fluorescent puncta consistent with individual virions. Each spot is measured for total fluorescence intensity using a defined region of interest that is constant for all particles and experiments. The algorithm excludes particles with spatial overlap so that the analysis is restricted to single diffraction-limited particles. Automation of the algorithm allowed for quantification of hundreds of particles per sample.
Intensity measurements (red and green) were plotted as bar graphs, which summarize the results of independent experiments performed in triplicate, or as histograms, where the Freedman–Diaconis rule was applied to green H-particle intensity values to independently determine bin sizes for each graph (70). Nonlinear Gaussian regression was performed in GraphPad Prism 6.0 (GraphPad Software). For L:H-particle ratios, a threshold value of 5,000 gray levels was set for minimum L-particle fluorescence, corresponding with approximately four copies of GFP. Where indicated, one-way ANOVA followed by a post hoc Tukey Test was used to determined statistical differences.
Quantification of Particle Asymmetry.
Asymmetry in the distribution of proteins within individual H-particles was attained by measuring the distance between corresponding GFP and RFP emissions. Because H-particles are physically smaller than the spatial resolution afforded by light microscopy, the centroids of GFP and RFP diffraction-limited fluorescence were determined by 2D Gaussian fitting. GFP/RFP image pairs used for heterogeneity analysis were also the source for asymmetry analysis. First, a 512 × 512-pixel image was cropped to isolate the center 300 × 300-pixel region. The cropped image was convolved with a 3 × 3 Gaussian matrix (low-pass filter) and noise in the image was removed with a band-pass filter (71). Otsu’s method was next used to select a threshold and create a binary mask of light and dark sections of each image (72). A second binary matrix was made from local maxima from the original image, which were represented with the value of one. Punctae representing viral particles were analyzed if a single pixel in both matrices had a value of 1. To obtain subpixel spatial positioning of the GFP and RFP emissions, a 7 × 7 matrix was centered at the local maxima of each virus punctae and the centroid of each spectral distribution was identified by 2D Gaussian fitting. The form of the Gaussian was:
Here, 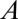 is the peak intensity,
is the peak intensity, 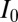 is the background intensity,
is the background intensity, 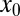 and
and 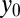 are the positions, and
are the positions, and 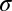 is the radius of each puncta. The analysis was repeated with the corresponding GFP image. From this procedure the locations of each punctum were identified at subpixel resolution. H-particles were defined as punctae that had intensity peaks of RFP and GFP emissions within five pixels.
is the radius of each puncta. The analysis was repeated with the corresponding GFP image. From this procedure the locations of each punctum were identified at subpixel resolution. H-particles were defined as punctae that had intensity peaks of RFP and GFP emissions within five pixels.
The displacements of GFP and RFP puncta were nonuniform across the field of view and varied slightly between image pairs, likely as a result of imperfections in the light path. Nonuniformity was corrected based on multiple H-particle displacements in each image pair. Because the random orientation of H-particles on the coverslip results in a zero average displacement between the GFP and RFP centroids, displacements were corrected independently in both the x and y directions with the linear function: 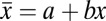 . The value was subtracted from the measured displacement (
. The value was subtracted from the measured displacement (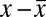 ) for each particle. Any RFP-GFP pairs with displacements larger than three times the SD were not considered H-particles and discarded.
) for each particle. Any RFP-GFP pairs with displacements larger than three times the SD were not considered H-particles and discarded.
We applied a 3D correction to account for the projection of fluorescence emission from randomly oriented 3D viral particles to a 2D image field. We ran a computer simulation in which two points with displacement r were projected randomly in 3D Cartesian space. The individual x, y, and z components that contributed to displacement r after 100,000 random orientations were determined. Gaussian error to x- and y-component projections was set at 7 nm, based on the displacement of red and green point sources of PRV-GS4848, which expresses mRFP1 and GFP fused in tandem on the N terminus of pUL35. Empirical observations were matched to the output of simulated r values (every 0.1 nm between 0 and 60 nm) and plotted in Fig. 5B. The corrected value never differed by more than 10 nm from the empirical value.
Modeling of Virion Substructure Displacement Measurements.
We modeled the capsid as an icosahedron centered at (0,0) with vertices of (0, ±1, ±1.62), (±1.62, 0, ±1), and (±1, ±1.62, 0). To model displacement with GFP absent from one unique vertex, the barycenter shift was determined after deleting 1 of 12 vertices. The displacement of protein at a unique vertex was modeled as the maximum capsid radius, 62.5 nm.
Models of tegument and envelope asymmetry are based on measurements made by Grünewald et al. in cryo-ET of HSV-1, where the capsid center is displaced 15 ± ≅5 nm from the center of the entire virion on average (8). The centroid of protein filling the space between the capsid and the envelope in an eccentric capsid was modeled by finding the barycenter of a large sphere with a smaller, eccentric inner sphere removed:
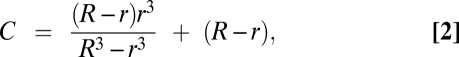 |
where R is the experimental virion radius (88 nm) and r the experimental capsid radius (62.5 nm). To approximate a tegument distribution in a fully offset cap from the capsid, the tegument was modeled as a 35-nm sphere adjacent to the capsid, resulting in a projected displacement of 80 nm, the distance between the center of the two adjacent spheres. An H-particle with homogenously distributed GFP in the envelope will project a 15-nm centroid displacement from the capsid center. However, polar distributions of GFP within the envelope could be modeled by any value up to 114, or the diameter of a virion minus the capsid radius.
Supplementary Material
Acknowledgments
We thank Annie Charpilienne and Didier Poncet at the Laboratory of Molecular and Structural Virology in Gif-sur-Yvette, France, for the gift of baculoviruses and rotavirus virus-like particles; Jenifer Klabis, Oana Maier, and Sarah Haverlock for assisting with BAC mutagenesis; and Massimo DiPierro and Marcus Schaefer from the School of Computing at DePaul University for their assistance with mathematical modeling. This work was funded by National Institutes of Health Grants R01 AI080658 (to G.A.S.) and R01 GM64624 (to S.P.G.). K.P.B. was supported by the training program in Immunology and Molecular Pathogenesis from the National Institutes of Health (T32AI07476).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1221896110/-/DCSupplemental.
References
- 1.Szilágyi JF, Cunningham C. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol. 1991;72(Pt 3):661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 2.Zhou ZH, Chen DH, Jakana J, Rixon FJ, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73(4):3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardone G, et al. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J Virol. 2012;86(8):4058–4064. doi: 10.1128/JVI.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou ZH, et al. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288(5467):877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- 5.Paredes AM, et al. Three-dimensional structure of a membrane-containing virus. Proc Natl Acad Sci USA. 1993;90(19):9095–9099. doi: 10.1073/pnas.90.19.9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, et al. Placement of the structural proteins in Sindbis virus. J Virol. 2002;76(22):11645–11658. doi: 10.1128/JVI.76.22.11645-11658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R, et al. 4.4 Å cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J. 2011;30(18):3854–3863. doi: 10.1038/emboj.2011.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grünewald K, et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science. 2003;302(5649):1396–1398. doi: 10.1126/science.1090284. [DOI] [PubMed] [Google Scholar]
- 9.Spear PG, Longnecker R. Herpesvirus entry: An update. J Virol. 2003;77(19):10179–10185. doi: 10.1128/JVI.77.19.10179-10185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batterson W, Roizman B. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J Virol. 1983;46(2):371–377. doi: 10.1128/jvi.46.2.371-377.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ace CI, McKee TA, Ryan JM, Cameron JM, Preston CM. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J Virol. 1989;63(5):2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krautwald M, Fuchs W, Klupp BG, Mettenleiter TC. Translocation of incoming pseudorabies virus capsids to the cell nucleus is delayed in the absence of tegument protein pUL37. J Virol. 2009;83(7):3389–3396. doi: 10.1128/JVI.02090-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schipke J, et al. The C terminus of the large tegument protein pUL36 contains multiple capsid binding sites that function differently during assembly and cell entry of herpes simplex virus. J Virol. 2012;86(7):3682–3700. doi: 10.1128/JVI.06432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abaitua F, Hollinshead M, Bolstad M, Crump CM, O’Hare P. A Nuclear localization signal in herpesvirus protein VP1-2 is essential for infection via capsid routing to the nuclear pore. J Virol. 2012;86(17):8998–9014. doi: 10.1128/JVI.01209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: An update. Virus Res. 2009;143(2):222–234. doi: 10.1016/j.virusres.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Kelly BJ, Fraefel C, Cunningham AL, Diefenbach RJ. Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 2009;145(2):173–186. doi: 10.1016/j.virusres.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Meckes DG, Jr, Wills JW. Structural rearrangement within an enveloped virus upon binding to the host cell. J Virol. 2008;82(21):10429–10435. doi: 10.1128/JVI.01223-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurer UE, Sodeik B, Grünewald K. Native 3D intermediates of membrane fusion in herpes simplex virus 1 entry. Proc Natl Acad Sci USA. 2008;105(30):10559–10564. doi: 10.1073/pnas.0801674105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newcomb WW, Brown JC. Time-dependent transformation of the herpesvirus tegument. J Virol. 2009;83(16):8082–8089. doi: 10.1128/JVI.00777-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith GA, Enquist LW. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc Natl Acad Sci USA. 2000;97(9):4873–4878. doi: 10.1073/pnas.080502497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trus BL, et al. Herpes simplex virus capsids assembled in insect cells infected with recombinant baculoviruses: structural authenticity and localization of VP26. J Virol. 1995;69(11):7362–7366. doi: 10.1128/jvi.69.11.7362-7366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou ZH, et al. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat Struct Biol. 1995;2(11):1026–1030. doi: 10.1038/nsb1195-1026. [DOI] [PubMed] [Google Scholar]
- 23.Zaichick SV, et al. The Herpesvirus VP1/2 protein is an effector of dynein-mediated capsid transport and neuroinvasion. Cell Host Microbe. 2013;13(2):193–203. doi: 10.1016/j.chom.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thurlow JK, et al. The herpes simplex virus type 1 DNA packaging protein UL17 is a virion protein that is present in both the capsid and the tegument compartments. J Virol. 2005;79(1):150–158. doi: 10.1128/JVI.79.1.150-158.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuchs W, Klupp BG, Granzow H, Mettenleiter TC. Essential function of the pseudorabies virus UL36 gene product is independent of its interaction with the UL37 protein. J Virol. 2004;78(21):11879–11889. doi: 10.1128/JVI.78.21.11879-11889.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.del Rio T, Ch’ng TH, Flood EA, Gross SP, Enquist LW. Heterogeneity of a fluorescent tegument component in single pseudorabies virus virions and enveloped axonal assemblies. J Virol. 2005;79(7):3903–3919. doi: 10.1128/JVI.79.7.3903-3919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaelin K, Dezélée S, Masse MJ, Bras F, Flamand A. The UL25 protein of pseudorabies virus associates with capsids and localizes to the nucleus and to microtubules. J Virol. 2000;74(1):474–482. doi: 10.1128/jvi.74.1.474-482.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newcomb WW, et al. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J Virol. 2001;75(22):10923–10932. doi: 10.1128/JVI.75.22.10923-10932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toropova K, Huffman JB, Homa FL, Conway JF. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J Virol. 2011;85(15):7513–7522. doi: 10.1128/JVI.00837-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cockrell SK, Huffman JB, Toropova K, Conway JF, Homa FL. Residues of the UL25 protein of herpes simplex virus that are required for its stable interaction with capsids. J Virol. 2011;85(10):4875–4887. doi: 10.1128/JVI.00242-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway JF, et al. Labeling and localization of the herpes simplex virus capsid protein UL25 and its interaction with the two triplexes closest to the penton. J Mol Biol. 2010;397(2):575–586. doi: 10.1016/j.jmb.2010.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charpilienne A, et al. Individual rotavirus-like particles containing 120 molecules of fluorescent protein are visible in living cells. J Biol Chem. 2001;276(31):29361–29367. doi: 10.1074/jbc.M101935200. [DOI] [PubMed] [Google Scholar]
- 33.Bohannon KP, Sollars PJ, Pickard GE, Smith GA. Fusion of a fluorescent protein to the pUL25 minor capsid protein of pseudorabies virus allows live-cell capsid imaging with negligible impact on infection. J Gen Virol. 2012;93(Pt 1):124–129. doi: 10.1099/vir.0.036145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schrag JDJ, Prasad BVBV, Rixon FJF, Chiu WW. Three-dimensional structure of the HSV1 nucleocapsid. Cell. 1989;56(4):651–660. doi: 10.1016/0092-8674(89)90587-4. [DOI] [PubMed] [Google Scholar]
- 35.McLauchlan J. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J Gen Virol. 1997;78(Pt 1):189–194. doi: 10.1099/0022-1317-78-1-189. [DOI] [PubMed] [Google Scholar]
- 36.Hoffman MT, Sheung J, Selvin PR. Fluorescence imaging with one nanometer accuracy: In vitro and in vivo studies of molecular motors. Methods Mol Biol. 2011;778:33–56. doi: 10.1007/978-1-61779-261-8_4. [DOI] [PubMed] [Google Scholar]
- 37.Booy FP, et al. Finding a needle in a haystack: Detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91(12):5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trus BL, et al. Allosteric signaling and a nuclear exit strategy: Binding of UL25/UL17 heterodimers to DNA-Filled HSV-1 capsids. Mol Cell. 2007;26(4):479–489. doi: 10.1016/j.molcel.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spear PG, Roizman B. Buoyant density of herpes simplex virus in solutions of caesium chloride. Nature. 1967;214(5089):713–714. doi: 10.1038/214713a0. [DOI] [PubMed] [Google Scholar]
- 40.Scrivano L, Sinzger C, Nitschko H, Koszinowski UH, Adler B. HCMV spread and cell tropism are determined by distinct virus populations. PLoS Pathog. 2011;7(1):e1001256. doi: 10.1371/journal.ppat.1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borza CM, Hutt-Fletcher LM. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat Med. 2002;8(6):594–599. doi: 10.1038/nm0602-594. [DOI] [PubMed] [Google Scholar]
- 42.Morgan C, Rose HM, Mednis B. Electron microscopy of herpes simplex virus. I. Entry. J Virol. 1968;2(5):507–516. doi: 10.1128/jvi.2.5.507-516.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vernon SK, Lawrence WC, Long CA, Rubin BA, Sheffield JB. Morphological components of herpesvirus. IV. Ultrastructural features of the envelope and tegument. J Ultrastruct Res. 1982;81(2):163–171. doi: 10.1016/s0022-5320(82)90072-7. [DOI] [PubMed] [Google Scholar]
- 44.Schmid MF, et al. A tail-like assembly at the portal vertex in intact herpes simplex type-1 virions. PLoS Pathog. 2012;8(10):e1002961. doi: 10.1371/journal.ppat.1002961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clarke RW, et al. Two-color fluorescence analysis of individual virions determines the distribution of the copy number of proteins in herpes simplex virus particles. Biophys J. 2007;93(4):1329–1337. doi: 10.1529/biophysj.107.106351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luxton GWG, et al. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc Natl Acad Sci USA. 2005;102(16):5832–5837. doi: 10.1073/pnas.0500803102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Handler CG, Eisenberg RJ, Cohen GH. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J Virol. 1996;70(9):6067–6070. doi: 10.1128/jvi.70.9.6067-6070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coller KE, Lee JI-H, Ueda A, Smith GA. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J Virol. 2007;81(21):11790–11797. doi: 10.1128/JVI.01113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leelawong M, Lee JI, Smith GA. Nuclear egress of pseudorabies virus capsids is enhanced by a subspecies of the large tegument protein that is lost upon cytoplasmic maturation. J Virol. 2012;86(11):6303–6314. doi: 10.1128/JVI.07051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michael K, Böttcher S, Klupp BG, Karger A, Mettenleiter TC. Pseudorabies virus particles lacking tegument proteins pUL11 or pUL16 incorporate less full-length pUL36 than wild-type virus, but specifically accumulate a pUL36 N-terminal fragment. J Gen Virol. 2006;87(Pt 12):3503–3507. doi: 10.1099/vir.0.82168-0. [DOI] [PubMed] [Google Scholar]
- 51.Chang JT, Schmid MF, Rixon FJ, Chiu W. Electron cryotomography reveals the portal in the herpesvirus capsid. J Virol. 2007;81(4):2065–2068. doi: 10.1128/JVI.02053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klupp BGB, Fuchs WW, Granzow HH, Nixdorf RR, Mettenleiter TCT. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J Virol. 2002;76(6):3065–3071. doi: 10.1128/JVI.76.6.3065-3071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vittone V, et al. Determination of interactions between tegument proteins of herpes simplex virus type 1. J Virol. 2005;79(15):9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolfstein A, et al. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic. 2006;7(2):227–237. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- 55.Mettenleiter TC. Budding events in herpesvirus morphogenesis. Virus Res. 2004;106(2):167–180. doi: 10.1016/j.virusres.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 56.Cardone G, et al. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology. 2007;361(2):426–434. doi: 10.1016/j.virol.2006.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng B, O’Connor CM, Kedes DH, Zhou ZH. Direct visualization of the putative portal in the Kaposi’s sarcoma-associated herpesvirus capsid by cryoelectron tomography. J Virol. 2007;81(7):3640–3644. doi: 10.1128/JVI.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meckes DG, Jr, Wills JW. Dynamic interactions of the UL16 tegument protein with the capsid of herpes simplex virus. J Virol. 2007;81(23):13028–13036. doi: 10.1128/JVI.01306-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Svobodova S, Bell S, Crump CM. Analysis of the interaction between the essential herpes simplex virus 1 tegument proteins VP16 and VP1/2. J Virol. 2012;86(1):473–483. doi: 10.1128/JVI.05981-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ko DH, Cunningham AL, Diefenbach RJ. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2) J Virol. 2010;84(3):1397–1405. doi: 10.1128/JVI.01721-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuchs W, Granzow H, Klupp BG, Kopp M, Mettenleiter TC. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J Virol. 2002;76(13):6729–6742. doi: 10.1128/JVI.76.13.6729-6742.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scholtes LD, Yang K, Li LX, Baines JD. The capsid protein encoded by U(L)17 of herpes simplex virus 1 interacts with tegument protein VP13/14. J Virol. 2010;84(15):7642–7650. doi: 10.1128/JVI.00277-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radtke K, et al. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 2010;6(7):e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klupp BG, Nixdorf R, Mettenleiter TC. Pseudorabies virus glycoprotein M inhibits membrane fusion. J Virol. 2000;74(15):6760–6768. doi: 10.1128/jvi.74.15.6760-6768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koyano S, Mar E-C, Stamey FR, Inoue N. Glycoproteins M and N of human herpesvirus 8 form a complex and inhibit cell fusion. J Gen Virol. 2003;84(Pt 6):1485–1491. doi: 10.1099/vir.0.18941-0. [DOI] [PubMed] [Google Scholar]
- 66.Chojnacki J, et al. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science. 2012;338(6106):524–528. doi: 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 67.Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40(2):191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 68.Smith GA, Gross SP, Enquist LW. Herpesviruses use bidirectional fast-axonal transport to spread in sensory neurons. Proc Natl Acad Sci USA. 2001;98(6):3466–3470. doi: 10.1073/pnas.061029798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Antinone SE, Zaichick SV, Smith GA. Resolving the assembly state of herpes simplex virus during axon transport by live-cell imaging. J Virol. 2010;84(24):13019–13030. doi: 10.1128/JVI.01296-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wand MP. Data-based choice of histogram bin width. Am Stat. 1997;51(1):59–64. [Google Scholar]
- 71.Ruusuvuori P, et al. Evaluation of methods for detection of fluorescence labeled subcellular objects in microscope images. BMC Bioinformatics. 2010;11:248. doi: 10.1186/1471-2105-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Otsu N. A threshold selection method form gray-level histograms. IEEE T Syst Man Cyb. 1979;9(1):62–66. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.