Abstract
Removal of introns from the precursors to messenger RNA (pre-mRNAs) requires close apposition of intron ends by the spliceosome, but when and how apposition occurs is unclear. We investigated the process by which intron ends are brought together using single-molecule fluorescence resonance energy transfer together with colocalization single-molecule spectroscopy, a combination of methods that can directly reveal how conformational transitions in macromolecular machines are coupled to specific assembly and disassembly events. The FRET measurements suggest that the 5′ splice site and branch site remain physically separated throughout spliceosome assembly, and only approach one another after the spliceosome is activated for catalysis, at which time the pre-mRNA becomes highly dynamic. Separation of the sites of chemistry until very late in the splicing pathway may be crucial for preventing splicing at incorrect sites.
Keywords: splicing mechanism, single-molecule FRET, RNA dynamics
Intron excision from precursors to messenger RNAs (pre-mRNAs) is carried out by the spliceosome, arguably the most complex macromolecular machine in the cell (1). One of the most important jobs of the spliceosome is to accurately and efficiently identify the ends of introns and bring them together to promote the chemistry of splicing. This chemistry occurs via two SN2 transesterification reactions: (i) attack by the branch site (BS) adenosine on the phosphodiester bond at the beginning of the intron (the 5′ splice site; 5′SS) and (ii) attack of the released 5′ exon on the phosphodiester bond at the end of the intron (the 3′SS) (2). The BS adenosine is internal to the intron and usually located in the vicinity of the 3′SS.
The spliceosome consists of four major subcomplexes that must assemble de novo on each new intron: the U1 and U2 small nuclear ribonucleoprotein particles (snRNPs), the U4/U6.U5 tri-snRNP, and the protein-only nineteen complex (NTC). The snRNPs each contain numerous proteins and one or more small nuclear RNAs (snRNAs). These subcomplexes assemble stepwise, with U1 and U2 recognition of the 5′SS and BS, respectively, preceding tri-snRNP and NTC recruitment (3, 4). Throughout the assembly process, numerous large-scale conformational changes occur that involve making and breaking of pre-mRNA:snRNA and snRNA:snRNA base pairing interactions. These structural transitions are necessary for both recognition of the splice sites and creation of the catalytic core in which the splice sites are juxtaposed for chemistry. The two chemical steps occur within the activated spliceosome formed after ejection of the U1 and U4 snRNPs (5).
When during spliceosome assembly are the splice sites brought into close proximity? Previous studies in human and yeast extracts using hydroxy radical cleavage or protein–RNA crosslinking led to the hypothesis that the 5′SS and BS regions are closely positioned in early complexes containing only U1 and/or U2 (6–12). However, as both of these irreversible trapping methods can capture transient excursions that are not necessarily on the pathway for splicing, when during spliceosome assembly and activation the 5′SS and BS are stably juxtaposed has remained unclear. In this work, we have used a combination of single-molecule FRET and colocalization single-molecule spectroscopy (FRET–CoSMoS) methods to monitor conformational changes in individual pre-mRNA molecules in real time and examine how these changes are coordinated with the steps of spliceosome assembly and activation.
Results
Single-Molecule FRET Suggests Dynamic Changes in SS Proximity.
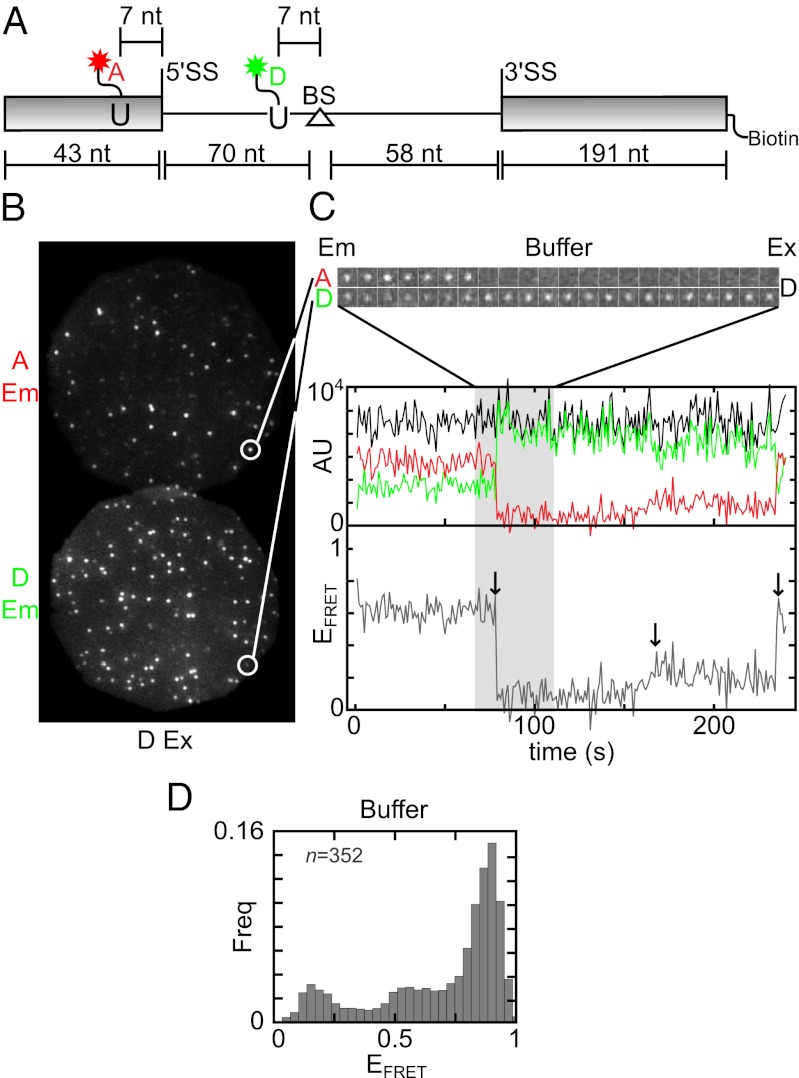
To investigate 5′SS and BS proximity, we first synthesized RP51A pre-mRNA (a well-studied splicing substrate) (13) containing a FRET donor and acceptor and a 3′ biotin to facilitate tethering to a glass surface (Fig. 1A and Fig. S1). The green-excited Cy3 donor dye (D) was 7 nt upstream of the BS adenosine and the red-excited AlexaFluor 647 or Cy5 acceptor dye (A) was 6 nt upstream of the 5′SS (i.e., in the 5′ exon). These positions were chosen to be as close as possible to the sites of chemistry without hindering access by the splicing machinery to the 5′SS and BS consensus sequences. In ensemble splicing assays in yeast whole cell extract (WCE), this modified pre-mRNA spliced with kinetics and efficiency similar to unmodified RP51A pre-mRNA (Fig. S2).
Fig. 1.
Single-molecule FRET of labeled RP51A pre-mRNA in buffer. (A) Schematic of the D, A, and biotin-labeled pre-mRNA. Rectangles, exons; line, intron; triangle, BS adenosine. Wavy lines denote multiple-atom linkers (six atoms for dyes and 16 for biotin). (B) Fluorescent spots from individual surface-tethered pre-mRNAs observed with D excitation (Ex; 532 nm). The same field of view (25 × 25 µm) was imaged at D (<635 nm) and A (>635 nm) emission (Em) wavelengths. (C) Time records of emission and EFRET from an individual pre-mRNA molecule. Image galleries show D and A emission images (1 × 1 µm; 1 s time interval) from part of the record. Plot shows the entire record D (green), A (red), and total (D + A; black) emission intensities and calculated EFRET (gray). (D) Histogram of EFRET across a population of pre-mRNA molecules. The histogram displays the relative frequencies in a dataset of five consecutive EFRET measurements on each of 352 molecules.
Apparent FRET efficiencies (EFRET) (SI Materials and Methods) of surface-tethered RNAs were monitored using a micromirror total internal reflection fluorescence (TIRF) microscope equipped for simultaneous multiwavelength excitation and detection (14). We excited the D and recorded emission from both D and A (Fig. 1B) to measure time-dependent EFRET of individual molecules (e.g., Fig. 1C). In all cases, we limited our analysis to molecules in which A photobleaching had not occurred, as confirmed by direct excitation of the A at the end of the recording.
When the labeled pre-mRNA was recorded in buffer only, the majority of single molecules exhibited EFRET > 0.7 (60%; 210/352), with smaller populations at EFRET = ∼0.2 (13%; 46/352 at EFRET < 0.3) and ∼0.6 (17%; 61/352 at 0.5 < EFRET < 0.7) (Fig. 1D). Some individual molecules switched between different discrete EFRET values (e.g., Fig. 1C, arrows), indicating that a single molecule can assume multiple conformations. This is consistent with the variety of secondary structures calculated for this RNA (Fig. S3); in the most stable structure, the dye attachment sites are constrained in comparative proximity, consistent with the majority high EFRET population.
U1 Binding Is Accompanied by a Decrease in EFRET.
To determine the FRET state(s) associated with each stage of spliceosome assembly, we next performed FRET–CoSMoS experiments in which we followed both EFRET of and subcomplex binding to individual pre-mRNA molecules in WCE + 2 mM ATP (e.g., Fig. 2A). For these experiments, we prepared WCE in which two constituents of a single spliceosomal subcomplex (U1, U2, U4/U6.U5, or NTC) were SNAP-tagged and labeled with the blue-excited Atto488 dye (15). These tagged and labeled WCEs were active for splicing in ensemble assays (Fig. S4). In single-molecule experiments, individual-labeled subcomplexes (S in Fig. 2 A–C) could be clearly seen and did not significantly quench D or A fluorescence when bound to the pre-mRNA (Figs. S5 and S6).
Fig. 2.
Simultaneous observation of 5′SS and BS proximity and U1 snRNP binding by FRET–CoSMoS. (A) Experimental design; same symbols as Fig. 1A. U1 snRNP (circle) was labeled with dye moieties (S) on two subunits. (B) Fluorescent spots from individual surface-tethered molecules from the same field of view (65 × 65 µm). (Left) A excitation (633 nm) and emission (>635 nm). (Center) As in Fig. 1B. (Right) S excitation (488 nm) and emission (500–550 nm). White circle indicates the same molecule in all images. (C) Example images (0.2 s duration) of the single pre-mRNA molecule circled in B exhibiting binding of a U1 snRNP. Key shows which dye was excited and from which dye emission was monitored in each image. Each set of 12 images (acquired over 2 s total) was separated by a 78 s delay (wavy lines) during which there was no excitation. (D) EFRET from 56 pre-mRNA molecules before (Buffer; left column) and after adding U1-labeled WCE plus 2 mM ATP (remaining 14 columns). The time interval in which U1 binding to each pre-mRNA molecule was first detected (tU1 = 0) was used to align the records relative to one another. Images were acquired as in C. Bar graph shows mean EFRET (± SEM, based on number of molecules) for each acquisition interval. ND, no data. (E–G) 2D histograms showing how EFRET of each molecule changed from one acquisition interval to another. Peaks on the diagonal (dashed line) indicate no change; off-diagonal peaks indicate significant changes in EFRET between acquisition intervals. Bar graphs indicate the distributions of EFRET values within each acquisition interval. (E) EFRET in buffer versus EFRET 0–80 s before U1 binding was detected. (F) EFRET 0–80 s before versus 0–80 s after U1 binding was detected. (G) EFRET 0–80 s after versus 80 s to 160 s after U1 binding was detected.
We first conducted time-lapse FRET–CoSMoS experiments in WCE containing labeled U1 snRNP and 2 mM ATP. At each observation time, we verified the presence of A fluorescence; made five successive measurements of EFRET, which also verified the presence of D; and then determined if labeled U1 was bound to the pre-mRNA (Fig. 2C). This sequence of observations was repeated eight times at 80 s intervals. To assess EFRET changes that accompanied U1 binding, we selected records that showed no U1 fluorescence in the first observation after WCE addition but did exhibit a well-defined spot of U1 fluorescence (Fig. 2C, arrows; Fig. S7) in at least one of the next seven observations. For each molecule, the time of U1 arrival (tU1 = 0) was estimated to be the midpoint of the 80 s interval before U1 was first observed. We then aligned the experimental records according to tU1 to examine EFRET changes coupled to U1 binding (Fig. 2D).
Both before and after U1 arrival, different pre-mRNA molecules in the population displayed a variety of EFRET values, and there was some fluctuation of EFRET for individual molecules within each 5 × 0.2 s set of measurements (Fig. 2D). Just before U1 binding both the EFRET population average <EFRET> = 0.60 ± 0.03 (S.E.) and its extent of fluctuation were essentially unchanged from those measured in buffer alone (<EFRET> = 0.64 ± 0.03; compare buffer data to data just before U1 arrival in Fig. 2D). Although discrete changes in EFRET were detected in a small number of molecules, most molecules that had been at EFRET > 0.7 in buffer were also at EFRET > 0.7 just before U1 arrival (Fig. 2E). These data suggest that proteins interacting with the pre-mRNA before U1 arrival most often do not significantly diminish 5′SS and BS proximity. In contrast, in the subpopulation of molecules in which U1 binding was observed, we saw upon U1 binding a clear shift to lower EFRET values (Fig. 2D). Most or all of this change occurred between the EFRET measurements immediately before and immediately after U1 arrival (Fig. 2F). The <EFRET> immediately after U1 binding, 0.38 ± 0.03, was significantly different (P = 2.8 × 10−4) from that immediately before (Fig. 2D). Furthermore, molecules in this low EFRET state after U1 binding tended to remain in that state and exhibited reduced EFRET fluctuation (Fig. 2 D and G). The decrease in EFRET upon U1 binding could not be explained by quenching of D or A by the snRNP or its dye label (Figs. S5 and S6). Assuming free mobility of D and A tethered by their aliphatic linkers (Fig. S1), these data suggest that binding of U1 is associated with stable separation of the 5′SS and BS to a greater distance than was present before U1 binding. A likely explanation is that U1 binding competed away structures (e.g., secondary structures shown in Fig. S3) that maintained 5′SS and BS proximity in the uncomplexed pre-mRNA.
Low EFRET Is Maintained Throughout Spliceosome Assembly.
We next proceeded to determine whether the EFRET changed in subsequent assembly steps. Following U1 acquisition, spliceosome assembly on RP51A pre-mRNA proceeds via U2 binding, followed by U4/U6.U5 binding, and then U1 and U4 release and NTC binding (Fig. 3A) (15). Spliceosome assembly can readily be blocked before U2 addition (16), before U4/U6.U5 addition (17), or before U4 release (18) (Fig. S8). In reactions in which U2 addition was blocked, we examined pre-mRNAs with bound fluorescent U1. Most of these molecules exhibited EFRET = ∼0.2 (Fig. 3B; 57%; 13/23 with EFRET < 0.3; Fig. 3C; 61%; 19/31 with EFRET < 0.3). These EFRET distributions were essentially indistinguishable from those seen <80 s after U1 binding in nonblocked reactions (Fig. 2G, Upper). Similarly, RNAs with bound fluorescent U2 in WCE in which U4/U6.U5 binding was blocked (Fig. 3D) and RNAs with bound fluorescent U5 in WCE in which U4 departure was blocked (Fig. 3E) both also predominantly exhibited EFRET = ∼0.2 (Fig. 3D; 69%; 24/35 with EFRET < 0.3; Fig. 3E; 91%; 32/35 with EFRET < 0.3). Taken together, these data suggest that the 5′SS and BS do not become closely juxtaposed in any stable assembly intermediate up to and including the U4/U6.U5 association step.
Fig. 3.
Schematic of spliceosome assembly (A) and EFRET distributions for different stages of spliceosome assembly (B–E). (B and C) Spliceosome assembly was blocked before U2 addition in U1-labeled WCE by ATP depletion (B) or by RNase H ablation of U2 snRNA (C). EFRET of pre-mRNAs exhibiting U1 fluorescence was measured 10 min after WCE addition. (D) Assembly was blocked before U4/U6.U5 addition in U2-labeled WCE by RNase H ablation of U6. EFRET of pre-mRNAs exhibiting U2 fluorescence was measured 15 min after WCE addition. (E) Activation was blocked before U4 release in U5-labeled WCE by limiting the concentration of ATP to 50 µM. EFRET of pre-mRNAs exhibiting U5 fluorescence was measured 45 min after WCE addition. The histograms (B–E) include five EFRET values (0.2 s each) for each of the n molecules measured.
EFRET Increases in Multiple Steps After NTC Arrival.
Do the 5′SS and BS come together upon NTC acquisition or in some subsequent structural rearrangement of the fully assembled spliceosome? To investigate this we performed FRET–CoSMoS experiments to measure pre-mRNA EFRET in splicing reactions containing labeled NTC. Of the pre-mRNA molecules observed to acquire NTC, 81% (90/111) were subsequently observed to lose both NTC (S) and intron (D) fluorescence simultaneously (within the experimental time resolution of 0.75 min; Fig. 4A). This likely reflected lariat intron and spliceosome release from the surface-tethered mRNA upon completion of exon ligation. We selected this subset of molecules for further analysis, as doing so allowed us to specifically characterize the changes in EFRET occurring in fully assembled, catalytically active spliceosomes (e.g., the single-molecule record in Fig. 4B). Aligning the EFRET records according to the time of NTC binding (Fig. 4C) revealed <EFRET> = 0.2 ± 0.1 (S.D.) immediately before and <EFRET> = 0.2 ± 0.1 (S.D.) immediately after NTC acquisition (Fig. 4D, Upper). However, an exponential transition (apparent first-order rate constant 0.75 min−1) to <EFRET> = 0.40 ± 0.16 was observed subsequently (Fig. 4D). Thus, NTC binding did not in itself alter EFRET, but subsequent step(s) did.
Fig. 4.
EFRET changes between NTC binding and intron release. (A) Example images (0.2 s duration) of a single pre-mRNA molecule exhibiting binding of labeled NTC (S; at tNTC = 0) and subsequent release of D-labeled intron and NTC (at trelease = 0). Key as in Fig. 2C. Each set of four images (acquired over 1 s total) was separated by a 45 s delay (wavy lines) during which there was no excitation. (B) Time record of EFRET from the same molecule shown in A. (C–F) Analysis of EFRET for pre-mRNA molecules (n = 90) observed to bind NTC, and subsequently lose both NTC and intron fluorescence in the same acquisition interval. For C and D, the records of 85 molecules were aligned so that the time of the last detected NTC binding was positioned at tNTC = 0; for E and F, the records from 56 molecules were aligned so that the time of disappearance of NTC and intron fluorescence was positioned at trelease = 0. Both the evolution of the EFRET distribution over the molecular population (C and E) and the <EFRET> ± SD (D and F, Upper) are shown. Also shown (D and F, Lower) are the relative populations of three species with EFRET = ∼0.2, EFRET = ∼0.3, and EFRET > 0.5 derived assuming a three EFRET-state model (SI Materials and Methods and Figs. S9 and S10). (G) Schematic working model of reaction pathway from NTC addition to product release and hypothesized EFRET values for the reaction intermediates. The complex shown at top is the same as that shown at the bottom of Fig. 3A.
Examination of the EFRET records for individual molecules showed that the <EFRET> = 0.4 was caused by fluctuation on the time scale of tens of seconds between a highly populated state with EFRET = ∼0.3 and less populated states of EFRET > 0.5 (Fig. 4 B and C). The latter nearly always did not occur immediately but appeared to lag behind the former; this observation was confirmed by kinetic modeling (Fig. 4D, Lower; Fig. S9; lag time ∼2 min).
When we analyzed the records by aligning them to the time of intron and NTC release (trelease = 0), we observed a progressive shift from EFRET = ∼0.3 states to EFRET > 0.5 states (Fig. 4 E and F and Fig. S10). Only in the last 1–2 min before intron release does the population in the highest EFRET state in the model exceed those in the lower EFRET states. Thus, the data are consistent with reversible passage through a sequence of two or more states in which the 5′SS and BS become closer after NTC binding and before intron release.
Discussion
By simultaneously monitoring the energy transfer efficiency of a FRET pair in the pre-mRNA and the binding of spliceosomal subcomplexes, this work defines by direct observation the temporal relationship between pre-mRNA conformational changes and specific steps in the assembly/activation process. Importantly, in some experiments we could restrict our analysis to catalytically active spliceosomes, allowing us to disregard the significant fraction of pre-mRNA molecules that assemble in vitro into dead-end complexes not on the pathway to splicing (4). In general, changes in EFRET can be caused by changes in D or A quantum efficiency, changes in the orientation of D relative to A, or changes in distance between D and A. Our measurements exclude significant changes in quantum efficiency. Changes in relative orientation are possible, but we view these as unlikely to cause the large EFRET changes we observe: orientation effects are likely to be averaged out given that the dyes are attached to the pre-mRNA through long linkers that likely allow for high mobility. Thus, we hypothesize that the EFRET changes predominantly reflect distance changes (19). If so, our EFRET data suggest that the 5′SS and BS are held apart from the earliest stage of spliceosome assembly and remain separated until after the very last assembly step, NTC arrival. After NTC arrival we detect multiple structural transitions in which the 5′SS and BS appear to more closely approach one another before spliced exon release.
Our results are superficially at odds with previous crosslinking and hydroxyl radical cleavage studies suggesting close apposition of the 5′SS and BS at early states of spliceosome assembly (6–11). Using FRET–CoSMoS, we see no evidence for a stable close approach of these sites in early complexes. Nonetheless, we did observe transient excursions into higher EFRET (e.g., the single-molecule record in Fig. 2D); these excursions might correspond to the close approach detected in trapping studies. Transient fluctuations in single-molecule EFRET were also observed in a previous splicing study (20). However, the relationship between those observations and ours is unclear because the D and A positions were different, and the earlier experiments were conducted with higher time resolution but shorter data record durations.
The most straightforward interpretation of our data are that in WCE, the 5′SS and BS of RP51A pre-mRNA are close together until U1 binds, at which point they are held apart and remain so for the rest of the spliceosome assembly process. Only after binding of NTC do the sites likely come into closer proximity, as evidenced by a transition to a state with EFRET = ∼0.3 (Fig. 4 C and D). These conclusions are consistent with the NTC providing essential structural support for the activated spliceosome (21) by stabilizing interactions between the U5 and U6 snRNAs and the pre-mRNA (22). Furthermore, the NTC component Cwc2 has recently been proposed to tether the 5′SS:U6 snRNA duplex to the spliceosome’s catalytic center including the essential U2:U6 snRNA duplex (helix I) immediately adjacent to the U2:BS duplex. UV-crosslinking and structural probing experiments have shown that Cwc2 interactions with these components increase in catalytic spliceosomes relative to spliceosomes stalled at earlier stages (23). Our observation of a FRET transition with an apparent rate of 0.75 min−1 may reflect the formation of these interactions and suggests that these conformational changes are not rate-limiting for the overall splicing reaction.
Once in this conformation, the complex can transiently and reversibly form states with EFRET > 0.5, suggesting large-scale pre-mRNA conformational fluctuations within the activated spliceosome. These fluctuations are reminiscent of those observed in earlier single-molecule FRET studies of protein-free U2 and U6 snRNAs (24). In the spliceosome, these structural alterations could well be events mediated by ATPases [Prp2 (25) and Prp16 and Prp22 (26, 27)] or other first or second step splicing factors [e.g., Yju2 (28) or Cwc25 (29)]. In the high EFRET states, the distance between the 5′SS and BS is probably greatly reduced, suggesting that one or more of these states may be the catalytic state in which the first chemical step of splicing occurs (Fig. 4G).
One of the most important jobs of the spliceosome is to ensure that chemistry occurs only at appropriate splice sites. To ensure high fidelity of splice site selection, multiple steps of spliceosome assembly are reversible and/or are subject to proofreading processes that remove inappropriately formed complexes (4, 26). Our finding that potential sites of chemistry are likely to be kept physically separate until the spliceosome is correctly assembled for catalysis is a previously unknown feature of the splicing mechanism. Such enforced separation may be essential to prevent splicing at incorrect sites.
Materials and Methods
Materials.
The 5′-GpppG capped RP51A pre-mRNAs containing indicated FRET D and A dye pairs and a 3′ biotin tag were assembled from six separate chemically or enzymatically generated RNA fragments by enzymatic ligation. For single-molecule experiments, pre-mRNAs were attached to biotin-conjugated polyethylene glycol fused silica coverslips via a streptavidin sandwich. Haploid yeast strains containing C-terminally SNAP-tagged proteins expressed from their endogenous loci were generated by homologous recombination. WCE for ensemble and single-molecule splicing reactions was prepared by grinding frozen yeast cells in a ball mill and then subjecting the clarified lysate to size exclusion chromatography. SNAP-tagged WCEs were Atto488 dye-labeled by incubation with 1–2 µM SNAP–Surface 488 (New England Biolabs 124S) for 30 min at 25 °C, followed by size exclusion chromatography to remove excess dye. U2 and U6 snRNA ablation was accomplished by adding a cDNA oligo to the appropriate WCE, followed by 10 min incubation at 25 °C. See SI Materials and Methods for more details.
Splicing and Spliceosome Assembly Reactions.
All reactions contained 40% (vol/vol) WCE in splicing buffer consisting of 100 mM potassium phosphate pH 7.3, 2.5 mM MgCl2, 3% PEG 8000 (wt/wt), 1 mM DTT, 400 U/mL RNasin+ (Promega), and containing the protocatechuate 3,4 dioxygenase/protocatechuic acid oxygen scavenging system and triplet state quenchers (Trolox, n-propyl gallate, 4-nitrobenzyl alcohol) (4), and were carried out at 21–23 °C (single-molecule experiments) or 25 °C (ensemble experiments) for the times indicated. See SI Materials and Methods for more details.
Microscopy and Data Analysis.
Single-molecule FRET and FRET–CoSMoS image sequences were recorded using a previously described multiwavelength single-molecule fluorescence microscope (14). A 488, 532, or 633 nm laser was used for dye excitation, and the emission optics produced a spectrally discriminated dual view of a sample region: fluorescence emissions at wavelengths <635 nm formed one image, while those with wavelengths >635 nm formed a second image of the same sample region. Data were analyzed with custom MATLAB (The Mathworks) image-processing software. A three EFRET-state model was fit with program vbFRET (30). See SI Materials and Methods for more details.
Supplementary Material
Acknowledgments
We thank E. Anderson, I. Shcherbakova, J. Yan, J. Chung, M. Fairman-Williams, and B. Smith for discussions and technical assistance. This work was supported by National Institutes of Health (NIH) RO1s GM053007 (to M.J.M.), GM43369 and GM81648 (to J.G.), NIH Training Grant GM759628 (to D.J.C.), National Research Service Award Fellowship GM079971 (to A.A.H.) and K99/R00 GM086471 (to A.A.H.). M.J.M. is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219305110/-/DCSupplemental.
References
- 1.Nilsen TW. The spliceosome: The most complex macromolecular machine in the cell? Bioessays. 2003;25(12):1147–1149. doi: 10.1002/bies.10394. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ, Sharp PA. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993;365(6444):364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- 3.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 4.Hoskins AA, Moore MJ. The spliceosome: A flexible, reversible macromolecular machine. Trends Biochem Sci. 2012;37(5):179–188. doi: 10.1016/j.tibs.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 6.Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 1998;18(8):4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valcárcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected] Science. 1996;273(5282):1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 8.Kent OA, MacMillan AM. Early organization of pre-mRNA during spliceosome assembly. Nat Struct Biol. 2002;9(8):576–581. doi: 10.1038/nsb822. [DOI] [PubMed] [Google Scholar]
- 9.MacMillan AM, et al. Dynamic association of proteins with the pre-mRNA branch region. Genes Dev. 1994;8(24):3008–3020. doi: 10.1101/gad.8.24.3008. [DOI] [PubMed] [Google Scholar]
- 10.McPheeters DS, Muhlenkamp P. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol Cell Biol. 2003;23(12):4174–4186. doi: 10.1128/MCB.23.12.4174-4186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dönmez G, Hartmuth K, Kastner B, Will CL, Lührmann R. The 5′ end of U2 snRNA is in close proximity to U1 and functional sites of the pre-mRNA in early spliceosomal complexes. Mol Cell. 2007;25(3):399–411. doi: 10.1016/j.molcel.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 12.Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8(7):843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 13.Séraphin B, Rosbash M. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA-pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991;10(5):1209–1216. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedman LJ, Chung J, Gelles J. Viewing dynamic assembly of molecular complexes by multi-wavelength single-molecule fluorescence. Biophys J. 2006;91(3):1023–1031. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331(6022):1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McPheeters DS, Fabrizio P, Abelson J. In vitro reconstitution of functional yeast U2 snRNPs. Genes Dev. 1989;3(12B):2124–2136. doi: 10.1101/gad.3.12b.2124. [DOI] [PubMed] [Google Scholar]
- 17.Fabrizio P, McPheeters DS, Abelson J. In vitro assembly of yeast U6 snRNP: A functional assay. Genes Dev. 1989;3(12B):2137–2150. doi: 10.1101/gad.3.12b.2137. [DOI] [PubMed] [Google Scholar]
- 18.Fabrizio P, et al. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell. 2009;36(4):593–608. doi: 10.1016/j.molcel.2009.09.040. [DOI] [PubMed] [Google Scholar]
- 19.dos Remedios CG, Moens PD. Fluorescence resonance energy transfer spectroscopy is a reliable “ruler” for measuring structural changes in proteins. Dispelling the problem of the unknown orientation factor. J Struct Biol. 1995;115(2):175–185. doi: 10.1006/jsbi.1995.1042. [DOI] [PubMed] [Google Scholar]
- 20.Abelson J, et al. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat Struct Mol Biol. 2010;17(4):504–512. doi: 10.1038/nsmb.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hogg R, McGrail JC, O’Keefe RT. The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing. Biochem Soc Trans. 2010;38(4):1110–1115. doi: 10.1042/BST0381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan S-P, Cheng S-C. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem. 2005;280(35):31190–31199. doi: 10.1074/jbc.M505060200. [DOI] [PubMed] [Google Scholar]
- 23.Rasche N, et al. Cwc2 and its human homologue RBM22 promote an active conformation of the spliceosome catalytic centre. EMBO J. 2012;31(6):1591–1604. doi: 10.1038/emboj.2011.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Z, Karunatilaka KS, Rueda D. Single-molecule analysis of protein-free U2-U6 snRNAs. Nat Struct Mol Biol. 2009;16(11):1154–1159. doi: 10.1038/nsmb.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohrt T, et al. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA. 2012;18(6):1244–1256. doi: 10.1261/rna.033316.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semlow DR, Staley JP. Staying on message: Ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci. 2012;37(7):263–273. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng C-K, Liu H-L, Cheng S-C. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011;17(1):145–154. doi: 10.1261/rna.2459611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y-C, Chen H-C, Wu N-Y, Cheng S-C. A novel splicing factor, Yju2, is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol Cell Biol. 2007;27(15):5403–5413. doi: 10.1128/MCB.00346-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warkocki Z, et al. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol. 2009;16(12):1237–1243. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 30.Bronson JE, Fei J, Hofman JM, Gonzalez RL, Jr, Wiggins CH. Learning rates and states from biophysical time series: A Bayesian approach to model selection and single-molecule FRET data. Biophys J. 2009;97(12):3196–3205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.