Abstract
Autophagy is a stress-induced catabolic process in which cytoplasmic components, sequestered in double-membrane autophagic vesicles (AVs) or autophagosomes, are delivered to lysosomes for degradation and recycling [Kroemer G, Mariño G, Levine B (2010) Mol Cell 40(2):280–293]. Activity of the class III phosphatidylinositol-3-OH-kinase (PI3K) vacuolar protein-sorting (Vps) 34, bound to coiled-coil moesin-like B-cell lymphoma 2 (Bcl-2)–interacting protein Beclin-1, is required for phosphoinositide generation, essential for AV formation in autophagy [Cuervo AM (2010) Nat Cell Biol 12(8):735–737]. However, how autophagy-inducing stress regulates Vps34 activity has not been fully elucidated. Our findings demonstrate that autophagy-inducing stress increases intracellular levels of acetylated inducible heat shock protein (hsp) 70, which binds to the Beclin-1–Vps34 complex. Acetylated hsp70 also recruits E3 ligase for SUMOylation, KRAB–ZFP-associated protein 1 (KAP1), inducing Lys840 SUMOylation and increasing Vps34 activity bound to Beclin 1. Knockdown of hsp70 abolished the Beclin-1–Vps34 complex formation, as well as inhibited KAP1 binding to Vps34 and AV formation. Notably, autophagy-inducing stress due to histone deacetylase inhibitor treatment induced AV formation in the wild-type but not hsp70.1/3 knockout mouse embryonic fibroblasts MEFs. These findings highlight a regulatory mechanism of Vps34 activity, which involves acetylated hsp70 and KAP1-dependent SUMOylation of Vps34 bound to Beclin 1.
Keywords: HDAC inhibitor, breast cancer
The autophagic process is initiated when at the phagophore assembly site (PAS) a cup-shaped isolation phospholipid membrane, the phagophore, derived from the endoplasmic reticulum (ER), Golgi, or the outer mitochondrial or plasma membrane, is formed through the action of a subset of autophagy related genes (1–3). During nutrient depletion and other forms of stress, induction and nucleation of the phagophore depends on the activity of the adenosine monophosphate (AMP)-activated protein kinase (AMPK) and mammalian target of rapamycin complex 1 (mTORC1)-regulated serine threonine kinase unc-51–like kinase 1/2 (ULK1/2) complex that also contains mammalian autophagy gene 13 (mATG13) and FIP200 (mammalian ortholog of Atg17) (4–7). Phagophore formation also critically depends on the production and availability of phosphatidylinositol 3-phosphate [PI(3)P] (8). This availability is controlled by the activity of the class III phosphatidylinositol-3-OH kinase (PI3K) vacuolar protein-sorting (Vps) 34, also known as PI3K catalytic subunit type 3 (PI3KC3) (8, 9). Vps34 associates with p150, the mammalian ortholog of Vps15, which anchors Vps34 to the phagophore membrane and stimulates the lipid kinase activity of Vps34 (3, 4, 9). This activity of Vps34 is required for generating PI(3)P at the PAS, which controls membrane dynamics during autophagosome formation and recruits the other autophagy regulatory proteins involved in phagophore and autophagic vesicle (AV) formation (4, 9). Thus, Vps34 is the central regulator of autophagy (4, 8, 9). Vps34 forms a complex with p150 (Vps15), Beclin 1 and ATG14L (also known as Barkor) (9–11). Beclin 1 interacting proteins or “interactome” includes Ambra1 and UV irradiation resistance-associated gene (UVRAG) (ortholog of Vps38), which binds and recruits Bif-1 (Bax-interacting factor 1) (12, 13). With its binding partners, Beclin 1 allosterically regulates Vps34 activity (1, 4, 12).
A recent report demonstrated that following activation by nutrient starvation, the activated Ras-like small G protein RalB and its effector Exo-84 induce the assembly of catalytically active ULK1 and Beclin-1–Vps34 complexes on the isolation membrane of the nascent AV (14). Additionally, in response to cellular stress, the heat shock protein 90 (hsp90)–cell division cycle (cdc) 37 chaperone complex was shown to stabilize ULK1 during mitochondrial autophagy or mitophagy (15). Direct phosphorylation of Vps34 by cyclin-dependent kinase 1 (CDK1) inhibits its interaction with Beclin 1 and decreases Vps34 activity, leading to reduced PI(3)P formation (16). Conversely, as a downstream effector of DAP (death associated protein) kinase, under oxidative stress protein kinase D (PKD) was shown to phosphorylate and activate Vps34 leading to PI(3)P formation at the PAS and autophagosome formation (17). In a recent report, the crystal structure of Vps34 was resolved (18). Compared with the catalytic unit of class I PI3K, Vps34 has a smaller ATP binding pocket, which allows the small molecular weight 3-methyladenine (3-MA) to fit into and inhibit the catalytic activity of Vps34. Additionally, the C-terminal helix (kα12 residues 878–886) was shown to fold over the catalytic loop in a closed autoinhibitory, off the membrane conformation (kα12-in) and an open, activating, and on-membrane conformation (kα12-out) (18). The loop between the last two helices kα11 and kα12 was proposed to act as a hinge that enables a closed to open conformation transition of the kα12. This proposed activity would suggest that other factors could potentially affect the hinge function, thereby controlling this conformational switch and catalytic lipid kinase activity of Vps34 (18). Our present studies demonstrate that autophagy-inducing stress increases intracellular levels of acetylated hsp70, which binds to the Beclin-1–Vps34 complex and recruits KRAB–ZFP-associated protein 1 (KAP1) to the complex. KAP1 mediates Lys840 SUMOylation in the loop between helices kα11 and kα12 and increases Vps34 activity bound to Beclin 1 during phagophore formation.
Results
Therapeutic Stress, e.g., Due to HDAC Inhibitor Panobinostat, Induces Vps34 and Autophagy in Breast Cancer Cells.
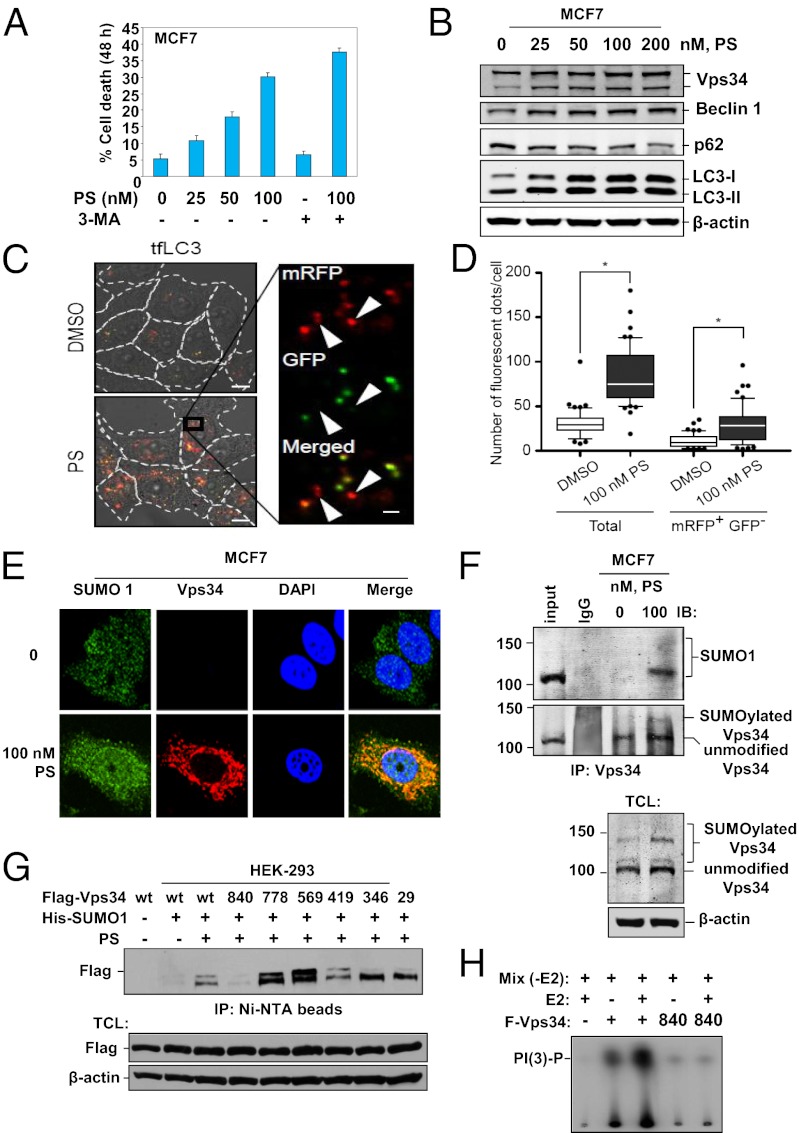
Nutrient deprivation and withdrawal of growth factors, as well as a variety of conventional and targeted therapeutic anticancer agents such as etoposide, irradiation, and histone deacetylase (HDAC) inhibitors, induce autophagy, which, depending on the context, exerts a prosurvival or prodeath activity (8, 19, 20). Consistent with this fact, pan-HDAC inhibitor panobinostat (PS) treatment not only resulted in the DNA fragmentation associated with apoptosis and in cell death (Fig. 1A and Fig. S1 A and B), but also dose-dependently depleted p62 expression and induced intracellular levels of the phosphatidylethanolamine-conjugated, microtubule-associated protein light chain 3 (LC3)-II in the human breast cancer MCF7 cells. This result was detected by Western blotting, immunostaining followed by confocal microscopy, and by increased acridine orange staining, which are hallmarks of AV expansion and completion (Fig. 1B and Fig. S1 C and D). Consistently, an autophagic flux assay using tfLC3 revealed that PS treatment not only induced the formation of autophagosomes (mRFP+ GFP+ dots) but also promoted the turnover of autophagolysosomes (mRFP+ GFP− dots) (Fig. 1 C and D). Cotreatment with 3-MA, which fits into the ATP-binding pocket and inhibits the catalytic activity of Vps34 (18), increased PS-induced cell death, but inhibited PS-induced apoptosis of MCF7 cells, whereas cotreatment with chloroquine enhanced PS induced apoptosis (Fig. 1A and Fig. S1 A and B). These results indicate a protective role for autophagy in this setting, and underscore that 3-MA-mediated block in autophagy promoted PS-induced nonapoptotic cell death (21, 22). We next determined whether PS treatment affected expression of the key regulators of AV formation, Vps34 and Beclin 1. Exposure to PS induced the accumulation of Vps34 and Beclin 1 levels in MCF7 cells (Fig. 1B and Fig. S1E).
Fig. 1.
HDAC inhibitor panobinostat induces autophagy in MCF7 cells. (A) Cell death of MCF7 cells induced by treatment with the indicated concentrations of PS and/or 3-MA for 48 h. For 3-MA treatment, cells were pretreated with 20 mM 3-MA for 4 h. (B) Depletion of p62 and induction of Vps34, Beclin 1, and LC3 II in MCF7 cells following 48 h of treatment with PS, as indicated. (C) MCF-7 cells were transduced with lentivirus encoding mRFP-GFP-LC3 (tfLC3) for 48 h followed by treatment with 100 nM PS or control DMSO for 24 h. Representative fluorescence images merged with DIC images are shown. Arrowheads indicate mRFP+ GFP− degradive autophagosomes. (Scale bars: 10 µm, main image; 1 µm, magnified image.) (D) The number of mRFP+ GFP+ and mRFP+ GFP− fluorescent dots per cell in C were counted using SlideBook 5.0 software (n = 50) and shown as a box plot. The lines, boxes, and error bars represent median values, 25th–75th percentiles, and 10th–90th percentiles, respectively. Statistical significance was determined using a one-way ANOVA followed by Bonferroni's multiple comparison test (GraphPad PRISM ver. 5.01). The asterisk indicates P < 0.001. (E) Immunostaining of Vps34 (red) colocalized with SUMO1 (green) in PS-treated MCF7 cells. (F) Immunoblot analysis of SUMOylation in Vps34 immunoprecipitates and total cell lysates from PS-treated MCF7 cells. (G) Lysine 840 is a major site of PS-induced SUMOylation of Vps34. Immunoblot analyses of SUMOylated Vps34 in HEK-293 cells cotransfected with wild type or K-to-R mutant Flag–Vps34, His-SUMO1 and treated with 50 nM PS for 24 h. (H) Effect of K840R mutation on in vitro lipid kinase activity of Vps34.
Vps34 SUMOylation at Lysine 840 Induces Vps34 Activity.
We also observed that the molecular weight of PS-induced Vps34 was about 20 kDa larger than previously reported (Fig. 1B), which approximated an addition of one small ubiquitin-related modifier (SUMO) 1 (23). This finding prompted us to check whether PS treatment induced Vps34 SUMOylation. First, the in vitro translated Flag-tagged Vps34 was applied to an in vitro SUMOylation reaction mixture, containing E1, E2, or SUMO1 alone or their combinations (Fig. S2A). Slower migrating bands of Flag–Vps34 were observed, when complete components for the in vitro SUMOylation were available (Fig. S2A). PS treatment resulted in SUMO1 (green) colocalizing with the induced Vps34 (red) (Fig. 1E). PS also induced binding of the endogenous Vps34 with SUMO1 in MCF7 cells (Fig. 1F). PS treatment also dose-dependently increased the binding of ectopically expressed Flag-tagged Vps34 to endogenous SUMO1, or to HA-tagged SUMO1 but not SUMO2, in HEK-293 cells (Figs. S1F and S2B). Indicative of the covalent linkage, nickel beads precipitated Flag–Vps34 from the extracts of HEK-293 cells expressing Flag–Vps34 and His-SUMO1 (Fig. S2C). Ectopically expressed, cytoplasmic, wild-type but not mutant de-SUMOylase SENP5 abolished the higher molecular weight species of Vps34 (Fig. S2C) (23). Using nickel beads pull-down, Lys to Arg (K to R) mutation at the putative SUMOylation sites on Vps34 did not prevent the SUMOylation of Vps34, except K to R substitution at Lys-840 (Fig. 1G). Located in the hinge loop between kα11 and kα12 in the catalytic domain of Vps34, Lys-840 modifications may be essential for maintaining the open, on-the-membrane, activating conformation (kα12-out) of the C-terminal kα12 helix (residues 878–886), thereby supporting the catalytic lipid kinase activity of Vps34 (18). Using phosphatidylinositol phosphate (PIP) as a substrate, M2-immunoaffinity purified Vps34 was first applied to an in vitro SUMOylation mix followed by determination of its activity in an in vitro lipid kinase assay. Results showed that mutation of Vps34 at Lys-840 to Arg markedly impaired the lipid kinase activity of Vps34 (Fig. 1H).
Hsp70 Regulates Vps34 SUMOylation and Beclin-1–Vps34 Interaction During Autophagosome Formation.
Next, we speculated that PS treatment may directly modify factors involved in Vps34-specific SUMOylation and activity of Vps34 in MCF7 cells. Using immunoprecipitation and proteomic analysis by mass spectrometry (24), 10 proteins were identified in the PS-induced Vps34 complex (Fig. 2A). Among these were stress-inducible hsp70 (also called HSP72, HSP70-1 or HSPA1A), hsp90 and glucose-regulated protein 78 (GRP78, also known as Ig heavy chain binding protein BiP), which may form a complex with Vps34 (Fig. 2A). Knockdown of GRP78 was previously shown not to disrupt binding of VPS34 and Beclin 1 during ER stress-induced AV formation (25). Because treatment with the hsp90 inhibitor AUY922 induced hsp70 and promoted autophagy in MCF7 cells (21, 24), we focused on the potential role of hsp70 in regulating Vps34 function in the AV formation. Although hsp70 is known to exhibit tumor-specific localization to lysosomes and promote autophagy (26, 27), the effects of hsp70 on Vps34 and the upstream AV nucleation have not been determined. Although hsp70 was noted to be present in a complex with Vps34 and Beclin 1, PS treatment further increased hsp70 levels in the complex with both the endogenous and ectopically expressed Beclin 1 and Vps34 (Fig. 2B and C). This finding prompted us to explore whether hsp70 is an important regulator of Vps34 SUMOylation. Presence of SUMO1 augmented the interaction of Flag–hsp70 with endogenous Vps34 and induced SUMOylation of Vps34 in MCF7 cells (Fig. 2D). Although hsp70 has been shown as one of the components of the SUMO1 conjugates (28), so far there is no direct evidence that hsp70 is a substrate for SUMOylation. Recently, hsp70 was demonstrated to act as an adaptor protein enhancing “apoptosome” formation (29). Therefore, we speculated that hsp70 may also act as an adaptor for assembly of Beclin-1–Vps34 complex during autophagosome formation. This was confirmed by knocking down hsp70 expression in MCF7 cells. Abrogation of hsp70 by anti-sense mRNA to hsp70 attenuated PS-induced Beclin-1–Vps34 complex formation and reduced the intracellular accumulation of LC3-II, suggesting decreased AV formation in MCF7 cells (Fig. 2E). Consistent with this finding, compared with wild-type MEFs, in hsp70.1/3−/− MEFs PS treatment neither induced SUMOylation and expression of Vps34, or expression of Beclin 1, nor increased LC3 II (Fig. 2 F and G) levels. Therefore, hsp70 is required for PS-triggered in vivo AV formation.
Fig. 2.
Hsp70 regulates Beclin-1–Vps34 interaction and autophagosome formation. (A) Identification of proteins in a PS-induced Vps34 complex. Vps34 immunoprecipitates from PS-treated MCF7 cells were separated by SDS/PAGE and silver stained. Distinct bands were excised and analyzed by mass spectrometry. (B) Hsp70 forms a complex with Vps34 and Beclin 1 in PS-treated cells. MCF7 cells were treated with 100 nM PS for 24 h. Immunoblot analyses were performed on immunoprecipitates, as indicated. (C) Hsp70 facilitates the interaction of Vps34 with Beclin 1. Immunoblot analyses of Beclin 1 in Vps34 immunoprecipitates from HEK-293 cells transfected, as indicated. (D) Vps34 SUMOylation enhances binding to hsp70. Immunoblot analyses of SUMO1 and Vps34 in hsp70 immunoprecipitates from MCF7 cells transfected as indicated, and treated with PS (100 nM) for 24 h. (E) Hsp70 is essential for PS-induced Vps34 and Beclin 1 interaction and autophagosome formation. Immunoblot analyses of Vps34 immunoprecipitates from MCF7 cells stably transfected as indicated, then treated with PS (100 nM) for 24 h. (F and G) Dysregulation of autophagosome formation in PS-treated hsp70.1/3−/− MEFs. Hsp70 wt and hsp70.1/3−/− MEFs were treated with 100 nM PS for 24 h. Immunoprecipitation of Vps34 and immunoblot analyses were performed on total cell lysates. (H) PS treatment induces hsp70 acetylation. Immunoblot analyses of acetylated lysine in hsp70 immunoprecipitates or Ac K159 hsp70, hsp70, and β-actin in cell lysates of PS-treated MCF7 cells.
Acetylation of hsp70 Regulates SUMOylation of Vps34 and Its Association with Beclin 1 in Breast Cancer Cells.
In a previous study, we demonstrated that by inhibiting HDAC6 activity, PS treatment induced hsp90 acetylation (24). Here, we determined that PS also dose-dependently induced hsp70 acetylation in MCF7 cells, with slight induction of hsp70 expression (Fig. 2H). Knockdown of HDAC6 by siRNA also induced hsp70 acetylation (Fig. S2D). This acetylation was shown in vitro to be mediated by acetyltransferase p300 and confirmed in vivo by demonstrating acetylation of ectopically expressed Flag–hsp70, coexpressed with HA-tagged p300 in HEK-293 cells (Fig. S2 E and F). Consistent with this finding, acetylation of hsp70 by p300 was decreased by the add-back of HDAC6 in a dose-dependent manner (Fig. S3A). Moreover, in vitro deacetylation of acetylated hsp70 by in vitro translated HDAC6 was inhibited by HDAC inhibitors PS and trichostatin A (TSA) (Fig. S3B). In HEK-293 cells, PS, a more potent HDAC6 inhibitor, induced stronger acetylation of the ectopically expressed Flag–hsp70 than sodium butyrate, a weak inhibitor of HDAC6 (Fig. S3C). This result suggested that HDAC6 is the major deacetylase for hsp70, but is not the sole one. We identified, by a previously described mass spectrometry method (24), six lysine residues which could potentially be the acetylation sites on hsp70 (Fig. S3D). Because we had observed increased binding affinity of hsp70 with Vps34 in the presence of SUMO1 and PS (Fig. 2D), this finding prompted us to test whether SUMO1 modification of Vps34 is promoted by acetylated hsp70. Ectopically expressed acetylation mimics (K to Q) of hsp70 exhibited differential ability to enhance V5–Vps34 SUMOylation by HA–SUMO1 in HEK-293 cells (Fig. S3E). Although K558Q and K560Q did not increase SUMOylation of V5–Vps34, K88Q, K126Q, K159Q, and K523Q did increase SUMOylation of V5–Vps34. We next generated an anti-acetylated hsp70 (Ac K159) antibody and confirmed its specificity by demonstrating that it specifically detected WT, but not the acetylation-deficient K159R mutant of Flag–hsp70 by ELISA. Moreover, treatment with PS robustly increased acetylation signal intensity of the endogenous hsp70, as detected by Ac K159 antibody, which was also seen by immunoblotting the immunoprecipitates of hsp70 with an anti-acetyl lysine antibody (Fig. 2H). To address whether Lys159-acetylated hsp70 facilitates Beclin-1–Vps34 complex formation, MCF7 cells were transfected with either empty vector or Flag–hsp70 vector encoding wild-type (wt), K159Q (Q) or K159R (R) mutants (Fig. S4A). K159Q (Q) mutant of hsp70 dramatically increased the association of endogenous Vps34 with Beclin 1. This finding was further confirmed in HEK-293 cells expressing Myc–Beclin-1, V5–Vps34, or Flag–hsp70 (wt, Q, or R) (Fig. S4B). Notably, the ability of hsp70 K159Q to increase the association of Vps34 with Beclin 1 was comparable to treatment with PS (Fig. S4B).
KAP1 Is an Acetylated hsp70-Dependent SUMO E3 Ligase for Vps34.
One of the components of the Vps34 complex detected by mass spectrometry was KAP1 (Fig. 2A), previously described as a corepressor of gene transcription (30). KAP1 has also been reported to function as a SUMO E3 ligase to direct intramolecular SUMOylation (31). Therefore, we tested the hypothesis that KAP1 is a SUMO E3 ligase for Vps34. We first observed that PS treatment induced KAP1 levels and caused its cytoplasmic accumulation in MCF7 cells (Fig. 3A). In HEK-293 cells expressing Flag–Vps34, PS treatment stimulated His-SUMO1 conjugation to Flag–Vps34, which was further augmented by ectopic expression of HA–KAP1 (Fig. 3B). The C651 residue in the PHD domain of KAP1 is the determinant of KAP1 binding with E2 SUMO-conjugating enzyme Ubc9 and of the SUMOylation of specific Lys residues in the KAP1 bromodomain (31). Therefore, we introduced the C651A mutation into HA–KAP1 to abolish its ability to bind with E2, and tested whether the mutation negatively affected KAP1 in causing SUMO modification of Vps34. HA–KAP1 increased the His-SUMO–conjugated Flag–Vps34 in HEK-293 cells in a dose-dependent manner, but C651A HA–KAP1 (mt) abolished His-SUMO conjugation to Flag–Vps34 (Fig. S4C). His-SUMO conjugation of Flag–Vps34 was drastically reduced by knockdown of KAP1 by two separate siRNAs in MCF7 cells (Fig. 3C). Because we had observed enhancement of Vps34 SUMOylation by acetylated hsp70 (Fig. S3E), we next determined whether acetylated hsp70 is required for KAP1-mediated Vps34 SUMOylation. To address this possibility, we first showed by immunostaining that following PS treatment, hsp70 associates with KAP1 in MCF7 cells (Fig. 3D). Knockdown of hsp70 by siRNA resulted in a dramatic decrease in Vps34 binding with KAP1 (Fig. 3E). Notably, the C651A mutation in KAP1 dramatically decreased the binding of KAP1 with Vps34 in HEK-293 cells, even following exposure to PS (Fig. 3E). Compared with Flag–hsp70 (wt), acetylation mimic hsp70 (Q) increased, whereas acetylation-deficient hsp70 (R) reduced Vps34 binding to KAP1 (Fig. 3F). Taken together, our results demonstrate that KAP1 is the acetylated hsp70-dependent E3 SUMO ligase for Vps34 SUMOylation.
Fig. 3.
KAP1 is the SUMO E3 ligase for Vps34. (A) PS induces cytoplasmic accumulation of KAP1. MCF7 cells were treated with PS for 24 h, as indicated. (Upper) Immunoblot analyses of KAP1 and β-actin in cytosolic extracts. (Lower) Immunostaining of KAP1 (green) in the cytosol. (B) PS enhances KAP1-dependent SUMOylation of Vps34. Immunoblot analyses of Vps34 in His-SUMO1 immunoprecipitates from HEK-293 cells transfected as indicated, then treated with PS for 24 h. (C) KAP1 depletion attenuates PS-induced SUMOylation of Vps34. Immunoblot analyses of His-SUMO1-conjugated Vps34 from MCF7 cells transfected as indicated for 48 h, and treated with PS for 24 h. (D) PS induces colocalization of KAP1 with hsp70 in the cytosol of breast cancer cells. Immunostaining of KAP1 (green) and hsp70 (red) in MCF7 cells treated with 50 nM PS for 24 h. (E) Hsp70 is essential for the interaction of Vps34 with KAP1. Immunoblot analyses of HA-tagged KAP1 in Flag–Vps34 immunoprecipitates from HEK-293 cells transfected as indicated for 24 h, and then treated with 100 nM PS for 24 h. (F) Acetylated hsp70 recruits KAP1 to Vps34. Immunoblot analyses of V5 immunoprecipitates from HEK-293 cells transfected with V5–Vps34, HA–KAP1, and Flag–hsp70, Flag–hsp70–K88Q or Flag–hsp70–K88R as indicated.
Diverse Autophagy-Inducing Stimuli Trigger Acetylated hsp70-Dependent, KAP1-Mediated Vps34 SUMOylation and Beclin-1 Binding During Autophagosome Formation.
Next, we determined whether Vps34 SUMOylation occurs not just in PS-induced AV formation, but is also an early event during amino acid deprivation-initiated autophagy. Stress of amino acid withdrawal also promoted hsp70 acetylation and induced binding of KAP1 to the endogenous Vps34, and induced SUMO1 conjugation to Vps34 and Vps34 SUMOylation in MCF7 cells (Fig. 4 A and B). Notably, following amino acid starvation, KAP1 was also detected in the complex along with Vps34 and acetylated hsp70 (Fig. 4B). Importantly, we also determined whether other forms of stress caused by exposure to anticancer agents such as etoposide or doxorubicin, or exposure to heat shock, UV light, or reactive oxygen species (ROS), which are well known to induce autophagy (2, 19, 32), also trigger acetylation of hsp70 and KAP1-dependent SUMOylation of Vps34 in a complex with Beclin 1. Indeed, as shown in Fig. 4C, exposure to each of these agents and stresses induced acetylation of hsp70, increasing the levels and binding of KAP1 with Vps34, thereby resulting in increased SUMO1 conjugation and SUMOylation of Vps34 in MCF7 cells. This SUMOylation is also accompanied by increased AV formation, as demonstrated by depletion of p62 expression and accumulation of LC3-II (Fig. 4C). Similar effects were also noted following treatment with rapamycin (Fig. S5). Following exposure to ROS, abrogation of LC3-II accumulation may be secondary to oxidation and inactivation of the cysteine protease ATG4 essential for LC3-II maturation in autophagosome formation (32).
Fig. 4.
Role of hsp70 in autophagosome formation in breast cancer cells. (A and B) Amino acid-starvation induces hsp70 acetylation, Vps34 SUMOylation and enhances KAP1 binding to Vps34 and acetylated hsp70. Immunoblot analyses of Vps34 (A) and KAP1 (B) immunoprecipitates from MCF7 cells cultured in Earle’s Balanced Salt Solution supplemented with a 10× amino acid mixture (+) or without amino acids (−) for 4 h. (C) Oxidative stress or chemotherapeutic agents induce Vps34 SUMOylation, hsp70 acetylation, and increased binding of KAP1 to Vps34. Immunoblot analyses of SUMO1 or Vps34 in Vps34 or KAP1 immunoprecipitates from MCF7 cells treated for 24 h, as indicated. (D) Proposed working model for autophagosome formation in MCF7 cells. Stress-induced acetylation of hsp70 increases binding to KAP1, which increases SUMOylation of Vps34. This complex binds to Beclin 1 (complexed to ATG14L, Ambra1, Bif1, or UVRAG) and promotes phagophore formation. Other complexes, such as the ULK kinase complex (ULK1/2, ATG13, FIP200), are recruited to the phagophore. Following membrane expansion, the membrane encloses the cytosolic cargos in the autophagosome.
Discussion
Our findings demonstrate that autophagy-inducing cellular stress increases intracellular levels of acetylated hsp70, which binds to Beclin-1–Vps34 complex and promotes KAP1-dependent SUMOylation and activity of Vps34 (Fig. 4D). The latter is known to be essential for AV nucleation and expansion during autophagy. Previous studies have documented that a variety of stressful stimuli can lead to increase in the levels of misfolded and polyubiquitylated proteins, which can disrupt the binding of HDAC6 with molecular chaperones, resulting in hyperacetylation of molecular chaperones such as hsp90, hsp70, and GRP78 (21, 24, 33). This is especially the case when autophagy is induced by exposure to a pan-HDAC inhibitor such as panobinostat, which confers increased sensitivity on cancer cells to autophagy inhibitors (34). Our results support a potentially dynamic, stress-induced, acetylated hsp70-dependent control of Vps34 SUMOylation (Fig. 4D). This SUMOylation was associated with an increase in the levels and lipid kinase activity of Vps34 as well as its binding to Beclin 1. These findings add to the accumulating evidence that support the role of hsp90 and hsp70 in stabilizing and promoting the activity of the molecular effectors upstream in the AV formation, such as ULK1 (15) and downstream in the AV expansion and fusion with lysosomes (27). It is clear that hsp70.1/3 DKO mice do not phenocopy the knockouts of the other autophagy genes. Based on our findings, hsp70 is required for autophagy due to many forms of stress, especially therapy-associated stress that is significantly attenuated in the hsp70.1/3 DKO. Although a previous report elucidated the role of PKD in phosphorylating and increasing the activity of Vps34 under oxidative stress (17), our findings highlight the role of SUMO1- and KAP1-dependent SUMOylation of Vps34 in augmenting the lipid kinase activity of Vps34 in response to diverse forms of autophagy-inducing cellular stress. SUMOylation-mediated increase in Vps34 activity does not contradict and is compatible with the activity of RalB and its effector Exo84 in enhancing the assembly of catalytically active ULK1 and Beclin-1–Vps34 complexes on the exocyst, which is essential during nutrient starvation-induced phagophore formation (14). Indeed, SUMOylation-mediated increase in the lipid kinase activity of Vps34 and the build-up of PI(3)P would likely collaborate with the RalB–Exo84-mediated recruitment of the activated ULK1/2-ATG13-FIP200 complex to the PAS for AV nucleation and expansion.
In Vps34, the C-terminal helix (kα12 residues 878–886) was shown to fold over the catalytic loop in a closed auto-inhibitory, off the membrane (kα12-in) conformation (18). The loop between the last two helices kα11 and kα12 may act as a hinge that enables a closed to open conformation transition of the kα12, i.e., activating, on-membrane, kα12-out conformation (18). Our model (Fig. 4D) also implicates a potentially central role for the SUMOylation of Lys840 in the loop region between kα11 and kα12 of Vps34, because this may support the open, activating and on-membrane (kα12-out) conformation of Vps34, which is known to be essential for promoting Vps34 activity (18). However, it is difficult to exclude the possibility that SUMOylation of Vps34 also regulates the interaction between Vps34 and Vps15, which is required for Vps34 activity, especially because the C-terminal domain (837–864) in Vps34 is critical for Vps34 to bind with the kinase and HEAT domain of Vps15 (35). Regardless of the precise mechanism involved, it is clear that the levels of acetylated hsp70 and levels and activity of KAP1, as well as the levels of SUMO1 and SENP5, are likely to regulate the SUMOylation of Lys840 in Vps34. Therefore, our findings highlight the SUMOylation status of Lys840 as the potential determinant of the hinge function of the catalytic loop, thereby controlling this conformational switch and the catalytic lipid kinase activity of Vps34 (18).
Methods
Reagents.
Panobinostat (PS) was kindly provided by Novartis Pharmaceuticals. All chemicals were obtained from Sigma-Aldrich. Antibodies for Flag-tag, HA-tag, Myc-tag, acetylated lysine, hsp70, HDAC6, and β-actin were obtained as described (36, 37). Polyclonal antibodies specifically recognizing hsp70 acetylated at Lys-126 or Lys-159 were generated by Alpha Diagnostic. The following antibodies were purchased from commercial sources: anti-SUMO1 (Invitrogen), anti-Vps34 (Echelon Biosciences), anti-KAP1 (Novus), anti-V5 epitope (MBL), anti-LC3B (Cell Signaling), anti-Beclin 1 (BD Biosciences).
Plasmid Expression Vectors.
His-tagged and HA-tagged SUMO1, HA–p300, pCDNA3.1-HDAC6, HDAC6 siRNA, HA-tagged KAP1, pCDNA3.1-hsp70AS (anti-sense), and pRNATin-H1.2/Neo vector containing hsp70 siRNA were developed as described (24, 36–39). The cDNA encoding mRFP-GFP-LC3 (tfLC3) was excised from ptfLC3 (Addgene, 21074) and subcloned into the lentiviral vector pCDH1-MCS1-EF1-puro (System Bioscience, CD510A-1). All other cDNAs for cloning and KAP1 siRNA were obtained from Origene. Vps34 and hsp70 mutants were generated by site-directed mutagenesis using the Quick Change XL kit from Stratagene. Plasmid constructs generated during this study are listed in Table S1.
Cell Culture.
HEK-293, MCF7, wild-type, and hsp70.1/3−/− MEFs (40) were maintained in DMEM with 10% (vol/vol) FBS. Cells were passaged two to three times per week. Exponentially growing cells were used for all described experiments.
Immunoprecipitation and Immunoblot Analyses.
Immunoprecipitation of Flag, Beclin 1, hsp70, Vps34, Myc-tag, or V5-tag from cell lysates were performed as described (24).
Ni-Beads Pull-Down Assays.
To detect the effects of lysine to arginine mutation on Flag–Vps34 conjugation with His–SUMO1, Flag–Vps34 wt and mutant cDNAs were transfected into HEK-293 cells for 48 h. Purification of His-tagged proteins was performed as described (36). The eluted proteins were immunoblotted with anti-Flag antibodies.
Immunofluorescence Confocal Microscopy.
MCF7 cells were grown on coverslips and treated with 50 nM PS for 24 h. Immunofluorescence confocal microscopy was performed as described (24, 34). Briefly, permeabilized cells were incubated with mouse anti-SUMO1 and rabbit anti-Vps34 antibodies for 2 h at room temperature. Cells were washed with 1× PBS and incubated with Alexa Fluor 594-conjugated anti-rabbit and Alexa Fluor 488-conjugated anti-mouse secondary antibodies (Invitrogen) in the dark for 1 h at room temperature. The slides were washed with 1× PBS and mounted with Vectashield containing DAPI. For LC3 staining, cells were incubated with rabbit anti-LC3 II antibody followed by Alexa Fluor 488-conjugated anti-rabbit secondary antibody. For the detection of acidic vesicular organelles (AVOs), MCF7 cells were stained with 1 µg/mL of acridine orange as described (41). Images were acquired with an LSM510 confocal microscope with a 63× objective (Carl Zeiss). To monitor autophagic flux, recombinant lentivirus encoding tfLC3 was produced and transduced into MCF-7 cells as described (42). After treatment with PS or control DMSO, the cells were permeabilized with 100 µg/mL digitonin for 2 min to clearly visualize membrane-bound form of LC3 (43) and fixed in 4% paraformaldehyde in PBS for 10 min. Images were obtained using an OLYMPUS IX81 deconvolution microscope and analyzed using SlideBook 5.0 software (Intelligent Imaging Innovations).
SUMOylation, Acetylation, Deacetylation, and PI(3)KC3 Lipid Kinase Activity Assays.
To measure Vps34 SUMOylation in vitro, Flag–Vps34 was produced using a TNT coupled reticulocyte lysate system (Promega) and purified using M2 affinity beads. In vitro SUMOylation reactions were performed with a kit from LAE Biotech International, as described (36).
To detect acetylation of hsp70, an in vitro acetylation assay was performed as described (24). To detect the deacetylation of hsp70 by HDAC6, we used a modification of the method described in ref. 37. Acetylated hsp70 was purified from PS-treated HEK-293/Flag hsp70 cells using an anti-M2 affinity column. Acetylated hsp70 was eluted in equilibration buffer containing 100 μg/mL Flag peptide and dialyzed with deacetylase assay buffer (36). Next, acetylated Flag–hsp70 was incubated in deacetylase assay buffer with or without in vitro translated HDAC6 in the presence of 400 nM TSA or 100 nM PS at 37 °C for 30 min. SDS sample buffer was added, and the reactions were boiled before SDS/PAGE.
To measure in vitro PI(3)KC3 lipid kinase, in vitro translated Flag–Vps34 was in vitro SUMOylated as described above, then M2 agarose beads were added and the reactions were incubated at 37 °C for 2 h. The beads were washed and preincubated, and PI(3)KC3 kinase activity assays were performed as described (13).
Mass Spectrometry Analysis.
To determine the lysine residues on hsp70 that are acetylated by PS treatment, we used the method described in ref. 24. To determine the components of a PS-induced Vps34 complex, MCF7 cells were treated with PS for 24 h and Vps34 was immunoprecipitated. Immunoprecipitates were washed with lysis buffer and resolved on a 7% SDS/PAGE gel. Proteins were visualized by silver staining. Distinct bands were excised from the gel and subjected to mass spectrometric analysis.
DNA Fragmentation and Cell Death Assays.
To determine the effects of 3-MA or chloroquine on PS-induced apoptosis, MCF7 cells were treated with PS and 3-MA or chloroquine for 48 h then analyzed by Cell Death Detection ELISAPLUS (Roche) according to the manufacturer’s protocol. To determine the effects of 3-MA on PS-induced cell death, MCF7 cells were treated with PS and 3-MA for 48 h. Cell death was analyzed by staining cells with propidium iodide, followed by flow cytometry on an Accuri CFlow6 flow cytometer.
Supplementary Material
Acknowledgments
Y.Y. was supported by Grants STCSM11PJ1401500 and CNSF81272391. T.K.P. was supported by National Institutes of Health Grant R01CA154320.
Footnotes
Conflict of interest statement: Peter Atadja is an employee of Novartis Institute for Biomedical Research. All other authors declare no competing financial interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217692110/-/DCSupplemental.
References
- 1.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuervo AM. The plasma membrane brings autophagosomes to life. Nat Cell Biol. 2010;12(8):735–737. doi: 10.1038/ncb0810-735. [DOI] [PubMed] [Google Scholar]
- 3.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12(9):831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Klionsky DJ. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22(2):124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PLoS ONE. 2010;5(11):e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331(6016):456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu EY, Ryan KM. Autophagy and cancer—issues we need to digest. J Cell Sci. 2012;125(Pt 10):2349–2358. doi: 10.1242/jcs.093708. [DOI] [PubMed] [Google Scholar]
- 9.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol. 2009;186(6):773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 11.Sun Q, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105(49):19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex—at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20(6):355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9(10):1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodemann BO, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144(2):253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joo JH, et al. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell. 2011;43(4):572–585. doi: 10.1016/j.molcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuya T, et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell. 2010;38(4):500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenberg-Lerner A, Kimchi A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ. 2012;19(5):788–797. doi: 10.1038/cdd.2011.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller S, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327(5973):1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy JM, Thorburn A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol Ther. 2011;131(1):130–141. doi: 10.1016/j.pharmthera.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2004;101(52):18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao R, et al. Treatment with panobinostat induces glucose-regulated protein 78 acetylation and endoplasmic reticulum stress in breast cancer cells. Mol Cancer Ther. 2010;9(4):942–952. doi: 10.1158/1535-7163.MCT-09-0988. [DOI] [PubMed] [Google Scholar]
- 22.Gammoh N, et al. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc Natl Acad Sci USA. 2012;109(17):6561–6565. doi: 10.1073/pnas.1204429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreou AM, Tavernarakis N. SUMOylation and cell signalling. Biotechnol J. 2009;4(12):1740–1752. doi: 10.1002/biot.200900219. [DOI] [PubMed] [Google Scholar]
- 24.Yang Y, et al. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 2008;68(12):4833–4842. doi: 10.1158/0008-5472.CAN-08-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, et al. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008;15(9):1460–1471. doi: 10.1038/cdd.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28(18):5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36(1):15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Kwon SW, Anselmo A, Kaur K, White MA. Broad spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J Biol Chem. 2004;279(20):20999–21002. doi: 10.1074/jbc.M401541200. [DOI] [PubMed] [Google Scholar]
- 29.Kim HE, Jiang X, Du F, Wang X. PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1. Mol Cell. 2008;30(2):239–247. doi: 10.1016/j.molcel.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Friedman JR, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10(16):2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov AV, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28(5):823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scherz-Shouval R, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26(7):1749–1760. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyault C, et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21(17):2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao R, et al. Combination of pan-histone deacetylase inhibitor and autophagy inhibitor exerts superior efficacy against triple-negative human breast cancer cells. Mol Cancer Ther. 2012;11(4):973–983. doi: 10.1158/1535-7163.MCT-11-0979. [DOI] [PubMed] [Google Scholar]
- 35.Budovskaya YV, Hama H, DeWald DB, Herman PK. The C terminus of the Vps34p phosphoinositide 3-kinase is necessary and sufficient for the interaction with the Vps15p protein kinase. J Biol Chem. 2002;277(1):287–294. doi: 10.1074/jbc.M109263200. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9(11):1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bali P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: A novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280(29):26729–26734. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 38.Ziv Y, et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8(8):870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 39.Guo F, et al. Abrogation of heat shock protein 70 induction as a strategy to increase antileukemia activity of heat shock protein 90 inhibitor 17-allylamino-demethoxy geldanamycin. Cancer Res. 2005;65(22):10536–10544. doi: 10.1158/0008-5472.CAN-05-1799. [DOI] [PubMed] [Google Scholar]
- 40.Hunt CR, et al. Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol Cell Biol. 2004;24(2):899–911. doi: 10.1128/MCB.24.2.899-911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paglin S, et al. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001;61(2):439–444. [PubMed] [Google Scholar]
- 42.Young MM, et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012;287(15):12455–12468. doi: 10.1074/jbc.M111.309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.