Abstract
The complexity and depth of the relationships between the three domains of life challenge the reliability of phylogenetic methods, encouraging the use of alternative analytical tools. We reconstructed a gene similarity network comprising the proteomes of 14 eukaryotes, 104 prokaryotes, 2,389 viruses and 1,044 plasmids. This network contains multiple signatures of the chimerical origin of Eukaryotes as a fusion of an archaebacterium and a eubacterium that could not have been observed using phylogenetic trees. A number of connected components (gene sets with stronger similarities than expected by chance) contain pairs of eukaryotic sequences exhibiting no direct detectable similarity. Instead, many eukaryotic sequences were indirectly connected through a “eukaryote–archaebacterium–eubacterium–eukaryote” similarity path. Furthermore, eukaryotic genes highly connected to prokaryotic genes from one domain tend not to be connected to genes from the other prokaryotic domain. Genes of archaebacterial and eubacterial ancestry tend to perform different functions and to act at different subcellular compartments, but in such an intertwined way that suggests an early rather than late integration of both gene repertoires. The archaebacterial repertoire has a similar size in all eukaryotic genomes whereas the number of eubacterium-derived genes is much more variable, suggesting a higher plasticity of this gene repertoire. Consequently, highly reduced eukaryotic genomes contain more genes of archaebacterial than eubacterial affinity. Connected components with prokaryotic and eukaryotic genes tend to include viral and plasmid genes, compatible with a role of gene mobility in the origin of Eukaryotes. Our analyses highlight the power of network approaches to study deep evolutionary events.
Keywords: mobile genetic elements, cellular evolution, network analysis, organelles, recombination
The relationships between the three domains (sensu Woese) of cellular life (Eubacteria, Archaebacteria, and Eukaryotes) have been the subject of debate ever since their definition (1, 2). In particular, the events that led to the emergence of Eukaryotes, and their relatedness to the other two domains, remain highly controversial (3–8). Progress in this area requires both methodological development and the integration of new kinds of information that have previously not been used. Early attempts to resolve these relationships used phylogenetic trees based on ribosomal RNA genes, placing Eukaryotes as the sister group of Archaebacteria (in the rRNA tree rooted on the eubacterial branch), or within Archaebacteria (1, 9–11). Subsequent more comprehensive analyses using whole genome data suggested that eukaryotic genomes also contain several genes with a sister-group relationship to eubacterial genes; indeed, analysis of the yeast and human genomes showed that eubacterium-like genes outnumber archaebacterium-like genes (12–17).
This chimerical nature of eukaryotic genomes is consistent with models of eukaryogenesis involving a fusion of an archaebacterium and a eubacterium (14, 18–23). However, other models have been formulated that might also account for the existence of two gene repertoires with affinities to Archaebacteria and Eubacteria. For instance, it has been proposed that Eukaryotes, Archaebacteria, and Eubacteria might have arisen from a eukaryote-like ancestor, with prokaryotes having undergone severe independent genome reductions owing to their ecology (the so-called Eukaryotes-early hypothesis; refs. 24–26) (but see ref. 5). Eukaryotes have also been proposed to have arisen autogenously from different eubacterial lineages, including actinobacteria (27) and the planctomycete-verrumicrobia-Chlamydia group (28–30) (but see ref. 7). Other hypotheses have proposed a central role for mobile genetic elements (MGEs) in the origin of Eukaryotes (31), with some models proposing a virus as the ancestor of the nucleus (32, 33).
In addition to phylogenetic evidence, symbiogenic hypotheses are supported by the observation that eukaryotic genes that were likely contributed by the archaebacterial partner differ significantly from those contributed by the eubacterial partner. Eukaryotic genes with archaebacterial affinities are more likely to be involved in informational processes (transcription, translation, and replication), more highly and broadly expressed, more essential (i.e., lethal upon deletion), and encode more central proteins in the protein–protein interaction network than eubacterium-derived genes (13, 15–17). These observations, however, have been based exclusively on analyses of the yeast and/or human genomes; therefore, it remains unclear whether these patterns are general to all Eukaryotes. Recently, thanks to the development of new whole-genome sequencing technologies, a broad range of eukaryote genomes have become available, providing us with an opportunity to explore both the origins and early evolution of Eukaryotes. However, along with new data, new methodological approaches are needed to explore such ancient events.
The chimerical nature of eukaryotic genomes means that some genes cluster with eubacterial genes in phylogenetic trees, whereas others cluster with archaebacterial genes, implying that a single tree cannot represent the relationships among the three domains of life. Also, extensive horizontal gene transfer (HGT), even if it affected only prokaryotic lineages, can result in many gene families exhibiting conflicting evolutionary signals. Cell-centered approaches do not take into account possible acellular partners that may have contributed to eukaryogenesis (HGT mediated by vectors like viruses or plasmids). Finally, being centered on genealogical issues, organism-centered trees do not take into account other processes (i.e., functional information indicating possible metabolic complementations between partners). Therefore, a more comprehensive analysis of the genetic material is required to study the origin of Eukaryotes.
Problematically, phylogenetic tree reconstruction is particularly challenging in the presence of highly divergent sequences (26, 34). First, it relies on gene families delimited using clustering methods such as the Markov cluster algorithm (35), which detects communities of closely related sequences from BLAST results. This approach may exclude the most divergent homologs in a family, which might be the most informative for an event as ancient as the origin of Eukaryotes. Second, multiple sequence alignments cannot be accurately constructed in the presence of a high number of substitutions. Third, long divergence times may have eroded at least part of the phylogenetic signal, or even deleted any detectable similarity between homologous sequences (34). Finally, generating phylogenetic hypotheses using very divergent sequences is very dependent on the model of sequence evolution used (e.g., ref. 36), and in practice highly divergent sequences almost always produce highly questionable placements in phylogenetic trees. Therefore, it is desirable to explore whether new data types exist that might provide new insight into deep evolutionary events.
As an alternative to phylogenetic trees, the relationships among genes can be represented more generally in the form of gene similarity networks, in which nodes and edges represent genes and similarity statements (e.g., BLAST hits), respectively (37–43). Such networks are typically composed of multiple connected components (CCs), each of which comprises a number of nodes that share similarity relationships with genes within the same CC, but not with genes outside the CC. These CCs represent groups of directly or indirectly related sequences, without the requirement that all sequences exhibit a detectable similarity to each other, thus being an extension of classical gene families. For example, within a network framework, we can think of a three-gene CC with the topology “A-B-C.” In such a CC, A exhibits detectable similarity to B, and B exhibits detectable similarity to C, but no significant similarity can be detected between A and C, e.g., as a result of a high degree of divergence and/or a fast rate of evolution. If we explicitly consider only those cases where pairwise similarity extends across the vast majority of the sequence pair—say, ≥70% of the total length— the members of the CC can be considered to be homologous. Therefore, despite the fact that all genes are homologous, and that the structure of the CC is informative about the evolution of the genes, such “workable gene families” would not be amenable to phylogenetic analysis in a single tree. By considering indirect relationships, gene similarity networks have the potential to explore deeper relationships than phylogenetic trees, thus being particularly appropriate for exploring deep evolutionary events such as the origin of Eukaryotes.
In the current study, we have used gene similarity networks to study the origin and ancient evolution of Eukaryotes. We constructed a network comprising the proteomes of 14 eukaryotes that are representative of most of the main eukaryotic lineages, 52 archaebacteria, 52 eubacteria, 2,389 viruses, and 1,044 plasmids. Among other interesting features, analysis of the structure of this complex network reveals multiple signatures of the chimerical origin of Eukaryotes as a result of an ancient event in which an archaebacterium and a eubacterium contributed genetic material, with genes descending from both ancestors preferentially exhibiting different functions and acting at different cell locations.
Results and Discussion
Construction of the Gene Similarity Network.
We constructed a database containing the nucleus-encoded proteomes of 14 eukaryotes, representative of most of the major eukaryotic supergroups (Table 1): Saccharomyces cerevisiae, Encephalitozoon intestinalis, Homo sapiens, Chlorella variabilis, Arabidopsis thaliana, Entamoeba histolytica, Plasmodium knowlesi, Tetrahymena thermophila, Phytophtora infestans, Trypanosoma cruzi, Naegleria gruberi, Giardia lamblia, and the nucleomorphs of Bigelowiella natans and Hemiselmis andersenii. Also included were the proteomes of 52 archaebacteria, 52 eubacteria, 2,389 viruses, and 1,044 plasmids (i.e., all viral genomes and all plasmid genomes corresponding to complete prokaryotic genomes available at the National Center for Biotechnology Information as of May 2011). In total, the database comprised 660,702 sequences (Dataset S1). Each sequence was used as query in a homology search against the whole database, and the results were used to construct an undirected graph. Only hits with an E-value lower than 10−5, at least 30% sequence identity, and covering at least 70% of the length of both the query and subject sequences were retained. This coverage makes it unlikely that sequence similarity is due to mere sharing of certain small protein domains. After removing sequences with no similar sequences in the dataset at these thresholds, the network consisted of 445,733 nodes connected by 7,943,719 edges (the entire dataset, including gene annotations, is available from the Dryad repository; http://datadryad.org). In total, 57.6% of these edges involve genes from the same class (archaebacterial-archaebacterial, eubacterial-eubacterial, eukaryotic-eukaryotic, plasmid-plasmid, or viral-viral). A count of the edges of each type is provided in SI Appendix, Table S1. We then used this network in a variety of ways to investigate the origin of Eukaryotes.
Table 1.
Genes of archaebacterial and eubacterial ancestry in eukaryotic genomes
| Supergroup | Genome | Total genes | Archaebacterial | Eubacterial | Ambiguous | ESPs |
| Opisthokonts | S. cerevisiae (fungus) | 5,861 (4,641) | 251 (149) | 463 (286) | 212 (130) | 4,935 (4188) |
| E. intestinalis (fungus) | 1,833 (1,672) | 171 (120) | 78 (66) | 33 (29) | 1,551 (1481) | |
| H. sapiens (animal) | 21,973 (10,986) | 408 (163) | 1,074 (445) | 419 (152) | 20,072 (10,452) | |
| Plants | C. variabilis (green alga) | 9,780 (7,979) | 251 (179) | 1,103 (707) | 374 (233) | 8,052 (7,097) |
| A. thaliana (land plant) | 27,225 (12,753) | 483 (200) | 1,855 (719) | 685 (225) | 24,202 (11,958) | |
| Amoebozoans | E. histolytica (amoeba) | 8,150 (5,518) | 348 (159) | 283 (136) | 171 (91) | 7,348 (5,216) |
| Cercozoa | B. natans (nucleomorph) | 283 (271) | 37 (37) | 9 (7) | 5 (4) | 232 (225) |
| Chromalveolates | P. knowlesi (apicomplexa) | 5,102 (4,560) | 156 (111) | 168 (134) | 76 (57) | 4,702 (4,309) |
| T. thermophila (ciliate) | 24,725 (18,552) | 203 (129) | 515 (274) | 239 (106) | 23,768 (18,191) | |
| H. andersenii (nucleomorph) | 471 (408) | 86 (64) | 19 (17) | 7 (6) | 359 (327) | |
| P. infestans (oomycete) | 17,797 (12,600) | 262 (150) | 898 (452) | 358 (178) | 16,279 (12,041) | |
| JEH | T. cruzi (euglenozoan) | 19,607 (9,376) | 390 (145) | 579 (255) | 279 (107) | 18,359 (9,005) |
| N. gruberi (heterolobosean) | 15,711 (11,755) | 272 (172) | 818 (443) | 338 (165) | 14,283 (11,172) | |
| POD | G. lamblia (diplomonad) | 7,364 (6,327) | 170 (119) | 115 (89) | 66 (56) | 7,013 (6102) |
| Total | 165,882 (95,317) | 3,488 (420) | 7,977 (1395) | 3,262 (451) | 151,155 (94,182) |
ESPs, eukaryotic-specific proteins; JEH, Jakobids-Euglenozoa-Heterolobosea; POD, Parabasalids-Oxymonads-Diplomonads. Values outside parentheses represent numbers of genes, and numbers within parentheses represent the number of different connected components to which these genes belong. For each pair of eubacterial-archaebacterial values, the highest value is represented in boldface.
The network is composed of 57,123 CCs, of which the two biggest contain 5,899 and 2,412 nodes. The biggest one mostly contains members of the ABC transporter gene family, and the second one mostly contains dehydrogenases and reductases. We classified the CCs according to their content in sequences from the three domains of cellular life. Among CCs containing genes derived from the three domains, 7,595, 11,480, and 16,326 contain only archaebacterial, eubacterial, and eukaryotic genes, respectively, 115 contain both eukaryotic and archaebacterial sequences (to the exclusion of eubacterial sequences), 781 contain eukaryotic and eubacterial genes (to the exclusion of archaebacterial genes), 2,005 contain archaebacterial and eubacterial sequences (to the exclusion of eukaryotic sequences), and 895 contain genes belonging to the three domains of cellular life (SI Appendix, Fig. S1A). The remaining 17,926 CCs contain exclusively sequences derived from MGEs (viruses and/or plasmids), corresponding to the idea of genetic worlds as advanced by Halary et al. (39). The observation that the number of CCs that contain eukaryotic plus eubacterial genes is 6.79-fold higher than the number of CCs including eukaryotic plus archaebacterial genes is consistent with previous observations that eukaryotic genomes contain a higher fraction of eubacterial homologs than of archaebacterial homologs (13, 15–17). It should be noted, however, that previous analyses have been based on relatively big eukaryotic genomes that are rich in genes of eubacterial ancestry (human and yeast; see Eukaryotic Genomes Exhibit Different Proportions of Genes of Archaebacterial and Eubacterial Ancestry).
Analysis of the Topology of the Network.
The 895 CCs that contain representatives of the three domains of cellular life may contain information on the relationships among these taxa. These CCs significantly outnumber the gene families used in previous analyses of the relationships among the three domains of life (e.g., refs. 44 and 45). A total of 15,324 eukaryotic sequences belong to such CCs, compared with 1,849 eukaryotic genes belonging to Eukaryotes+Archaebacteria CCs, 4,790 in Eukaryotes+Eubacteria CCs, and 66,719 in Eukaryotes-only CCs. To distinguish among competing hypotheses on the origin of Eukaryotes, we examined the topology of these CCs.
Of these CCs, a total of 208 contain at least one pair of eukaryotic sequences for which the shortest path connecting them involves an archaebacterial and a eubacterial sequence (“eukaryoteA-archaebacterium-eubacterium-eukaryoteB”, “EA-A-B-EB”; Fig. 1; see SI Appendix, Fig. S2 for all such CCs) (a reanalysis of which pairs of eukaryotic sequences were required to belong to the same genome resulted in 105 EA-A-B-EB CCs). In such paths, neither EA and EB, or EA and B, or EB and A, exhibit significant similarity according to the criteria used (E-value < 10−5, ≥ 30% identity, ≥ 70% coverage), implying that a phylogenetic tree involving all these sequences cannot be constructed, despite the facts that these sequences may be homologous and that the structure of the CC contains relevant evolutionary information about the origin of Eukaryotes. To confirm the notion that eukaryotic genes at the extremes of these E-A-B-E paths are homologous, we examined their Pfam domain composition. We found that 190 out of these 208 CCs contain E-A-B-E paths in which eukaryotic genes, despite not being linked in the network, encode the same protein domains, or belong to the same Pfam family, thereby confirming that they are distant homologs.
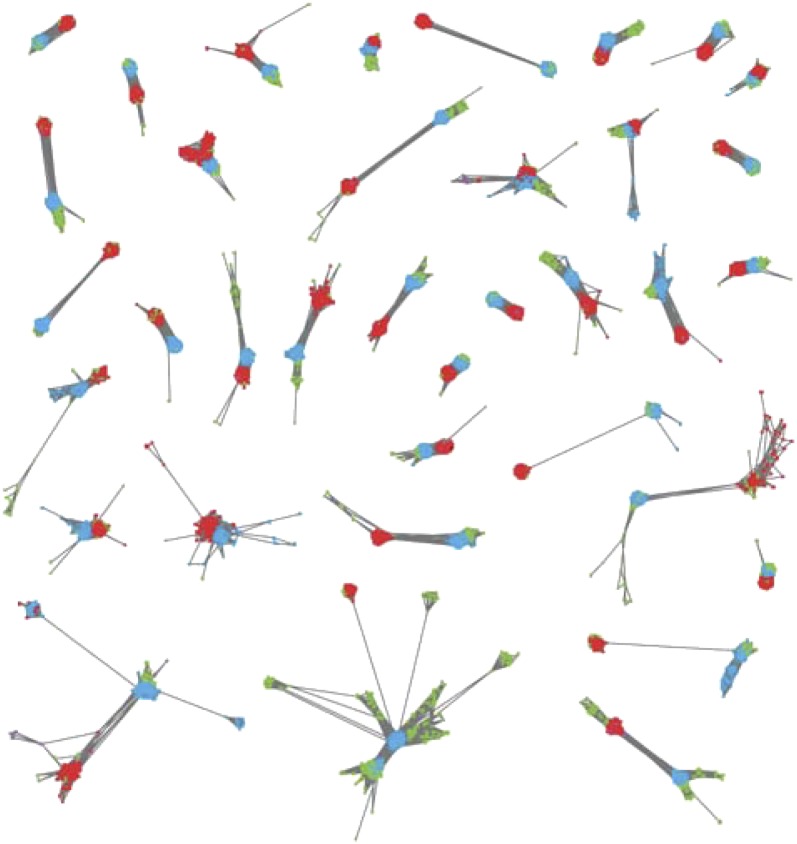
Fig. 1.
Selection of connected components containing a eukaryote–archaebacterium–eubacterium–eukaryote path. Eubacterial genes are represented in red, archaebacterial genes in blue, eukaryotic genes in green, plasmid genes in purple, and virus genes in black. Nodes were automatically distributed within each connected component using the edge-weighted spring-embedded visualization algorithm. This algorithm tends to place highly connected nodes and their neighbors close together. For a visualization of all CCs with an E-A-B-E topology, see SI Appendix, Fig. S1.
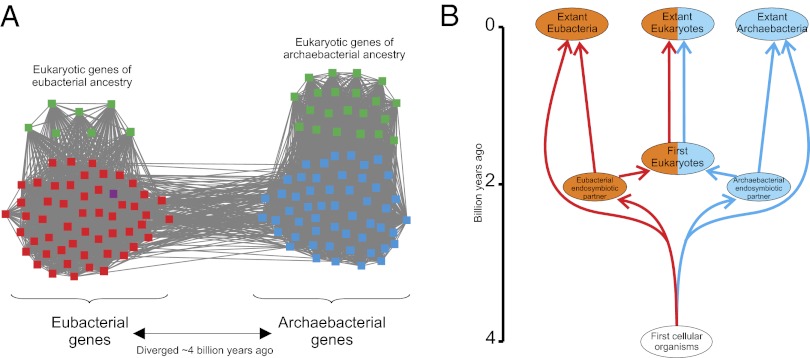
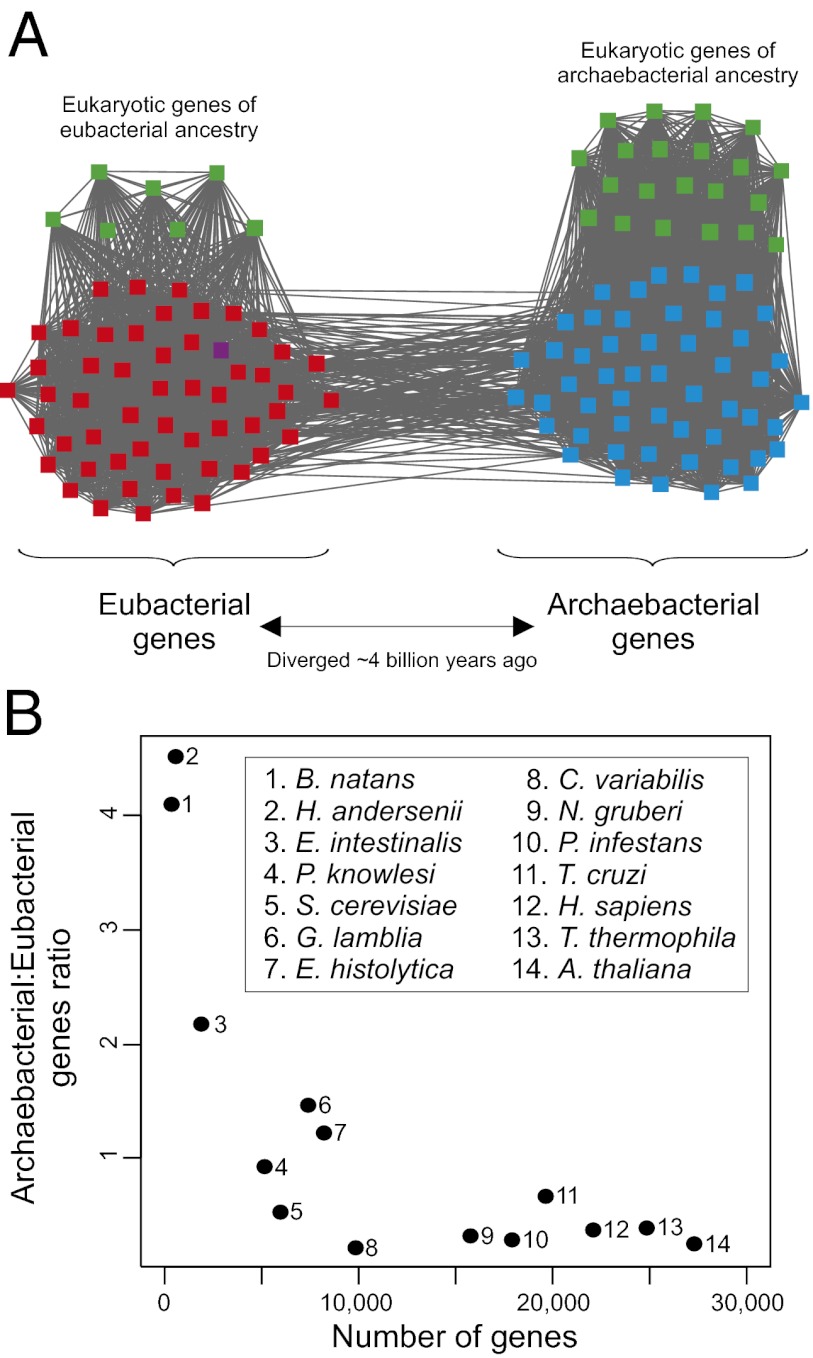
We interpret such CCs as a compelling signature of the chimerical origin of (at least extant) Eukaryotes as a result of a process in which a eubacterium and an archaebacterium contributed genetic material (14, 18–21). In such CCs, the eukaryotic sequences that are directly linked to archaebacterial and eubacterial sequences may represent, respectively, genes contributed by the archaebacterial and eubacterial ancestors during endosymbiosis (which is thought to have taken place ∼2 billion y ago (Gya); refs. 46–48), and the archaebacterial-eubacterial link may trace back to the most recent common ancestors (MRCAs) of Eubacteria and Archaebacteria (which are thought to have existed ∼4 Gya; ref. 49) (Fig. 2). Therefore, eukaryotic genes contributed by the archaebacterial (eubacterial) endosymbiotic partner may have diverged from their orthologs in extant archaebacterial (eubacterial) genomes ∼2 Gya and may have diverged from their orthologs in extant Eubacteria (Archaebacteria) ∼4 Gya (Fig. 2). Likewise, the MRCA of a pair of eukaryotic genes contributed by the eubacterial and archaebacterial ancestors may have existed ∼4 Gya (Fig. 2). Eukaryotic genes exhibit a faster rate of evolution than prokaryotic genes (50), which may account for the fact that eukaryotic sequences exhibit detectable homology to sequences from which they diverged ∼2 Gya, but not to those from which they diverged ∼4 Gya, whereas prokaryotic sequences can retain some similarity to homologs from which they diverged ∼4 billion y ago.
Fig. 2.
Connected component with eukaryotic genes likely contributed by the archaebacterial and eubacterial ancestors (A), and the likely evolutionary history of the gene family (B). In this connected component, eukaryotic genes contributed by one domain do not exhibit detectable similarity to eukaryotic genes contributed by the other domain. The shortest paths linking eukaryotic genes of eubacterial and archaebacterial affinity involve an archaebacterial and a eubacterial sequence (resulting in a eukaryote–archaebacteria–eubacteria–eukaryote path). Eubacterial genes are represented in red, archaebacterial genes in blue, eukaryotic genes in green, and plasmid genes in purple.
A fraction of eukaryotic genes in EA-A-B-EB CCs may have been contributed by prokaryotes other than the two endosymbiotic partners through posteukaryogenesis HGT. In particular, plant (C. variabilis and A. thaliana) genes of eubacterial affinity may also have been incorporated through endosymbiotic gene transfer (EGT) from the proto-chloroplast, which is thought to have descended from a eubacterial endosymbiont ∼1.5 Gya (51). However, a reanalysis excluding plant genes still resulted in 142 CCs with at least one EA-A-B-EB shortest path, indicating that the presence of these CCs is, for the most part, not the result of the acquisition of chloroplasts by plants.
Visual inspection of the 208 E-A-B-E CCs revealed the presence of 36 CCs with a topology that is clearly consistent with endosymbiotic theory (Figs. 1 and 2A). In each of these CCs, the eubacterial and archaebacterial domains are represented by two distinguishable clusters; i.e., proteins from each prokaryotic domain are preferentially connected to those of the same domain (average conductance for archaebacterial and eubacterial sequences, respectively, 0.257 and 0.163). Each of these modules is connected to both eukaryotic sequences and to the other module. As a result, these CCs contain two groups of eukaryotic genes: one that is connected to archaebacterial genes, and another that is connected to eubacterial genes, most likely corresponding to genes contributed by the archaebacterial and eubacterial ancestors, respectively. Despite being most likely homologous, both groups do not exhibit detectable similarity as a result of a long divergence time and/or a fast rate of evolution. In agreement with this hypothesis, 34 of these 36 CCs exhibit pairs of eukaryotic proteins that, despite being not linked in the network, exhibit an equivalent domain composition or belong to the same Pfam family.
To discard the possibility that these CCs might be in part the result of posteukaryogenesis HGT events from prokaryotes to Eukaryotes, we examined the eukaryotic species represented at both sides of these E-A-B-E CCs (i.e., the species represented in the group of eukaryotic genes connected to archaebacterial genes, and those represented among eukaryotic genes connected to eubacterial genes). Out of the 36 CCs represented in Fig. 1, 26 contain representatives of at least two eukaryotic supergroups at both sides. Given the fast radiation of the major eukaryotic lineages, which resulted in a star-like eukaryote tree, this observation suggests that these E-A-B-E patterns are the result of the primary endosymbiosis, rather than of posteukaryogenesis HGT.
These 36 CCs contain a total of 72 S. cerevisiae genes. Out of these genes, a total of 51 are involved in translation (including 41 ribosomal proteins, two translation initiation factors, and three tRNA synthetases). Among genes not involved in translation, 15 are part of the proteasome. Out of the 72 S. cerevisiae genes, 13 are linked only to eubacterial genes (to the exclusion of archaebacterial genes), 42 are linked only to archaebacterial genes (to the exclusion of eubacterial genes), 9 are linked to genes from both prokaryotic domains (although all of them are preferentially linked to genes of one of the domains), and 8 are linked only to other eukaryotic genes. Consistent with a eubacterial ancestry of extant mitochondria (18), among the 13 yeast genes that are linked only to eubacterial sequences, 10 are annotated as proteins targeted to the mitochondrion. Conversely, among the 42 yeast genes that are linked only to archaebacterial sequences, only 2 are targeted to the mitochondrion. For a full description of the genes in these CCs, see Dataset S2.
Such a clear topology is expected for gene families whose members were rarely (or not at all) involved in HGT between Eubacteria and Archaebacteria. Factors such as HGT may have resulted in CCs with more complex topologies (SI Appendix, Fig. S2). For instance, interdomain HGT between Archaebacteria and Eubacteria may have resulted in at least one of the modules containing sequences of both domains.
In addition to HGT, other factors might render difficult the observation of this kind of clear E-A-B-E CCs. Genes widely vary in their rates of evolution (17, 52, 53), and this variability most likely has an effect on the topology of the CCs. In gene families with low evolutionary rates, eukaryotic genes contributed by one prokaryotic ancestor may retain detectable similarity to their orthologs in prokaryotes from the same domain (∼2 billion years of divergence), to orthologs in prokaryotes from the other domain (∼4 billion years), and to genes contributed by the other prokaryotic ancestor (∼4 billion years), resulting in a clique-like topology (i.e., with each node being connected to all, or most, other nodes in the CC). However, for faster-evolving gene families, sequence similarity between eukaryotic sequences and their homologs may be detectable only up to a certain divergence time threshold, which will depend on the rate of evolution. In gene families of intermediate evolutionary rate, sequence similarity may be detectable after 2 billion years, but not after 4 billion years of divergence. CCs involving eukaryotic sequences plus representatives of only one prokaryotic domain might therefore correspond to gene families with such intermediate rates of evolution. These CCs might have initially belonged to EA-A-B-EB CCs that, owing to fast evolution, split into EA-A and B-EB CCs. In agreement with this hypothesis, 28 out of the 115 E+A CCs contain eukaryotic genes that share their domain composition or Pfam family membership with genes in E+B CCs, pointing to a potential distant homology. In even faster-evolving gene families, sequence similarity may not be detectable even after only 2 billion years of divergence, which may account for the numerous eukaryote-specific CCs. Indeed, it has been shown that eukaryotic-specific proteins (ESPs) tend to evolve faster than those with detectable prokaryotic homologs (17). Alternatively, ESPs might represent eukaryotic innovations, or might have been contributed by a third, noneubacterial and nonarchaebacterial prokaryote without living descendants (15). CCs including sequences from Eukaryotes and only one prokaryotic domain might also represent (i) gene families that were not shared by the two endosymbiotic partners, owing to family gain/loss after the divergence of Archaebacteria and Eubacteria; (ii) gene families that were shared between these two ancestors, but are no longer shared between extant archaebacterial and eubacterial genomes, owing to family loss in the past 2 billion years; or (iii) posteukaryogenesis HGT events between prokaryotes and Eukaryotes.
Eukaryotic Genes Tend to Have Either Archaebacterial or Eubacterial Neighbors in the Network.
To determine how eubacterial-like or how archaebacterial-like each eukaryote gene is, for each of the 14,727 eukaryotic genes with detectable prokaryotic homologs, we computed the number of archaebacterial and eubacterial nodes to which it was directly connected in the network (degreeA and degreeB, respectively). We also computed the proportion of prokaryotic hits that are eubacterial [pB = degreeB/(degreeA + degreeB)]. Remarkably, pB exhibits a markedly bimodal distribution (SI Appendix, Fig. S3), with 5,565 eukaryotic genes being exclusively connected to eubacterial genes (pB = 1) and 2,774 having only archaebacterial homologs (pB = 0). Conversely, genes with a similar number of archaebacterial and eubacterial homologs are less frequent (SI Appendix, Fig. S3). Separate analysis of the proteomes of each of the 14 eukaryotic species resulted in similar results (SI Appendix, Fig. S3).
To discard certain network features as the underlying factors of this bimodality, we repeated our analyses on different subsets of our dataset. First, out of the 14,727 eukaryotic genes with prokaryotic homologs, 2,297 exhibit detectable similarity to a single prokaryotic sequence. These genes are bound to exhibit a pB value of either 0 or 1, which may contribute to the bimodality of the distribution of pB. To discard this possibility, analyses were repeated on the 7,919 eukaryotic genes with at least 10 prokaryotic detectable homologs (degreeA + degreeB ≥ 10), with similar results (SI Appendix, Fig. S3). Among these genes, the most frequent value for pB is 0 (1,510 eukaryotic genes exhibit archaebacterial hits exclusively), followed by pB = 1 (1,293 eukaryotic genes have only eubacterial homologs). Second, a total of 896 CCs involve eukaryotic genes and prokaryotic genes belonging to one domain only (i.e., either eubacterial or archaebacterial genes; SI Appendix, Fig. S1A). Because eukaryotic genes in these CCs can be linked only to prokaryotic genes of one domain, this feature could also potentially account for the observed bimodality of pB. However, similar results were obtained when analyses were restricted to genes belonging to CCs containing representatives of the three domains of cellular life (SI Appendix, Fig. S3). Therefore, the bimodal character of pB is independent of these network features.
This marked bimodality of the proportion of prokaryotic genes that are eubacterial (or archaebacterial) indicates that eukaryotic genes highly linked to genes in one prokaryotic domain (i.e., likely contributed by a prokaryote of this domain) tend not to be linked to genes of the other prokaryotic domain. This trend reveals the presence of two markedly different groups of genes within eukaryotic genomes: one that is strongly linked to archaebacterial genes and another with strong affinities to eubacterial sequences. We interpret this observation as another reflection of the chimerical nature of eukaryotic cells.
Taken together, our observations support a chimerical origin of Eukaryotes and seem difficult to reconcile with both the Eukaryotes-early hypothesis (24–26) or with an autogenous origin of Eukaryotes from a single prokaryotic lineage (27–30). Under the former scenario, eukaryotic genes are expected to be similarly connected to prokaryotic genes of both domains, thus making unlikely the presence of CCs with the EA-A-B-EB topology. Under the latter, eukaryotic genes would be preferentially linked to a single prokaryotic domain.
Chimerical Nature of Ancient Eukaryotic Genomes.
Although the patterns described so far are compatible with an endosymbiotic origin of Eukaryotes, they would also be compatible with a series of smaller HGT events from prokaryotes to Eukaryotes after eukaryogenesis (54, 55), or a progressive integration of a prokaryotic consortium of archaebacteria and eubacteria into a superorganism. To determine whether ancient eukaryotic genomes were chimerical, we repeated our analyses on the subset of eukaryotic genes that were most likely present in these genomes. The deep relationships among the major eukaryotic lineages are currently unresolved, possibly as a result of a fast radiation of Eukaryotes after eukaryogenesis, resulting in a star-like eukaryotic tree. Therefore, we assumed that CCs containing representatives of at least three out of the seven major eukaryotic supergroups included in the analysis (Table 1) were likely present in the MRCAs of extant Eukaryotes.
When we restricted our analyses to this subset of relatively ancient eukaryotic gene families, we obtained similar results to those obtained from the whole network: pB exhibits a markedly bimodal distribution, regardless of the studied genome (SI Appendix, Fig. S3), and 192 CCs exhibit an EA-A-B-EB shortest path. These results indicate that ancient eukaryotic genomes (and probably the first eukaryotic organisms) had a chimerical nature, and therefore that the patterns observed in extant eukaryotic genomes are not the result of posteukaryogenesis HGT events.
Among CCs containing representatives of three or more eukaryotic supergroups, 187 contain eukaryotic and eubacterial genes (to the exclusion of archaebacterial sequences), and 83 contain eukaryotic and archaebacterial genes (to the exclusion of eubacterial sequences) (SI Appendix, Fig. S1B). Although the former outnumber the latter, the difference is not as marked as observed in the entire network: a 2.25-fold difference among ancient genes (SI Appendix, Fig. S1B), versus a 6.79-fold difference in the entire network (SI Appendix, Fig. S1A). This smaller difference suggests that the eubacterial:archaebacterial ratio might not have been as high in ancient eukaryotic genomes as in extant Eukaryotes. Remarkably, the number of CCs that contain eukaryotic and archaebacterial sequences in the entire network (a total of 115 CCs; SI Appendix, Fig. S1A) is comparable with the number of such CCs among “ancient” CCs (83 CCs; SI Appendix, Fig. S1B) whereas the number of CCs that contain eukaryotic and eubacterial sequences exhibits a higher variation (781 in the entire network versus only 187 in ancient CCs). Taken together, these observations suggest a scenario in which the number of gene families of archaebacterial ancestry remained relatively constant during the evolution of Eukaryotes, being retained in most of the eukaryotic lineages, whereas the number of eubacterium-derived families underwent extensive modification, with an important number of gene families being absent in some eukaryotic lineages (see next section for further results supporting this scenario). The increase over time of the overall proportion of eubacterial genes in the nuclear genomes of the studied eukaryotes might be the result of posteukaryogenesis HGT from Eubacteria or independent EGT from the mitochondrial and chloroplastic genomes in the different eukaryotic lineages. This dynamism of the number of eukaryotic genes of eubacterial ancestry is consistent with previous observations pointing out the essentiality of eukaryotic genes of archaebacterial ancestry versus the greater evolvability of eubacterium-derived genes (13, 15–17).
Eukaryotic Genomes Exhibit Different Proportions of Genes of Archaebacterial and Eubacterial Ancestry.
We classified each eukaryotic gene according to its prokaryotic affinity. Genes with pB < 0.3 were conservatively considered to be of likely archaebacterial ancestry, and those with pB > 0.7 were deemed eubacterial. The remaining genes were considered of ambiguous ancestry, and thus not considered in this section. Out of the 14,727 eukaryotic genes with detectable prokaryotic homologs, 3,488 were classified as archaebacterial and 7,977 as eubacterial. When this analysis was conducted separately for genes belonging to each of the 14 eukaryotic species studied, the higher content in eubacterial genes was confirmed for 9 species (S. cerevisiae, H. sapiens, C. variabilis, A. thaliana, P. knowlesi, T. thermophila, P. infestans, T. cruzi, and N. gruberi). Surprisingly, E. intestinalis, E. histolytica, G. lamblia, and the nucleomorphs of B. natans, and H. andersenii exhibit more genes of archaebacterial affinity than genes of eubacterial affinity (Table 1). The same trends were recovered when only ancient eukaryotic genes (i.e., those belonging to CCs with representatives of three or more eukaryotic supergroups) were considered (SI Appendix, Table S2). The finding of Eukaryotic genomes with more genes of archaebacterial than eubacterial ancestry has not been described previously.
Genes differ in their propensity to duplicate, which might potentially be affecting these results. For example, eukaryotic genes of eubacterial origin are more likely to present duplicates than those of archaebacterial ancestry (16, 17). To discard this possibility, we considered, in addition to the number of genes in each category, the number of different CCs to which these genes belong, given that genes resulting from a duplication event likely fall within the same CC. Again, the same trends were recovered (Table 1), indicating that our observations are not affected by the different duplicabilities of eukaryotic genes of eubacterial and archaebacterial ancestry.
Importantly, genomes with a high content of genes of archaebacterial affinity do not cluster together in the currently accepted eukaryotic phylogeny. For instance, our dataset includes two fungi, of which one presents a higher number of eubacterium-derived genes (S. cerevisiae), and the other contains a higher number of genes of archaebacterial affinity (E. intestinalis). Similarly, unikonts include two organisms with a predominance of eubacterial genes (S. cerevisiae and H. sapiens), and another two with a high content in archaebacterial genes (E. intestinalis and E. histolytica). Finally, the highest eubacterial-to-archaebacterial gene ratios correspond to the alga C. variabilis and to the land plant A. thaliana whereas the lowest ratios correspond to the nucleomorphs of B. natans and H. andersenii, which are thought to have derived from algae. Therefore, the heterogeneity observed in the archaebacterial-to-eubacterial content ratio of the studied eukaryotes may respond to the particular ecological conditions in which each organism lives, rather than to their shared genealogy.
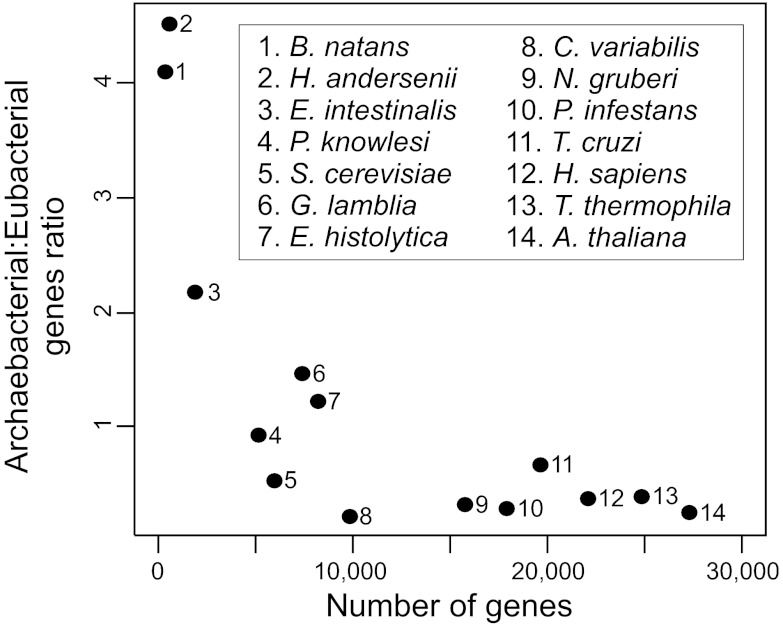
Remarkably, eukaryotic genomes with more genes of archaebacterial affinity than genes of eubacterial affinity rank among the smallest included in the analyses. Indeed, the archaebacterial:eubacterial ratio negatively correlates with the total number of genes in a genome (Spearman’s rank correlation coefficient, ρ = −0.771, P = 0.002; Fig. 3). This observation might be the result of eukaryotic genes of eubacterial ancestry being preferentially lost during genome reductions and/or gained during genome expansions, consistent with the higher evolvability of this set of genes (13, 15–17). In agreement with this hypothesis, microsporidia (including E. intestinalis), and in particular nucleomorphs, are the result of extensive genome reductions, and E. histolytica has experienced genome reduction involving most mitochondrial pathways (56–58). Nucleomorphs are highly reduced eukaryotic nuclei present in the plastids of certain secondarily photosynthetic eukaryotes (for a review, see ref. 57). They once were the nuclei of unicellular eukaryotic algae (a green alga in the case of B. natans, and a red alga in the case of H. andersenii), which were engulfed by nonphotosynthetic eukaryotes. These independent endosymbiotic events were followed by extensive gene losses and endosymbiotic EGTs to the hosts’ nuclear genomes, resulting in numbers of genes as small as 283 (B. natans) and 471 (H. andersenii; Table 1). Despite this dramatic reduction, nucleomorph genomes have retained a representation of the eubacterial and archaebacterial gene repertoires (Table 1; SI Appendix, Fig. S3). Remarkably, the B. natans and H. andersenii genomes contain as little as 9 and 19 genes of likely eubacterial ancestry, again consistent with the high degree of dispensability of eukaryotic genes of eubacterial ancestry. Interestingly, the archaebacterial:eubacterial content ratio is very similar for both genomes (4.11 for B. natans and 4.53 for H. andersenii), suggesting a predictability of this ratio during strong genome reduction.
Fig. 3.
Correlation between the number of genes of each eukaryotic genome and the archaebacterial-to-eubacterial gene ratio.
On the contrary, S. cerevisiae, H. sapiens, and A. thaliana have experienced whole genome duplication events, and the T. thermophila, P. infestans, and T. cruzi genomes have experienced important genome expansions (59–61). It should be noted, in addition, that the high content of genes of eubacterial affinity in the C. variabilis and A. thaliana genomes may be in part the result of EGT from the proto-chloroplast (of cyanobacterial ancestry) to plant genomes, and the low number of eubacterium-derived genes in the G. lamblia and E. histolytica genomes might be explained by the loss of mitochondria in these organisms (62–64).
Genes of Archaebacterial and Eubacterial Ancestry Perform Different Tasks in Eukaryotic Cells.
We considered whether genes of archaebacterial and eubacterial affinity perform different tasks in eukaryotic cells. For that purpose, each eukaryotic gene in the network was assigned to one (or a few) functional categories based on its similarities to the eukaryotic clusters of orthologous genes (KOGs). Among the 3,488 genes of likely archaebacterial ancestry, 1,832 are involved in “informational” processes (i.e., those involved in the “information storage and processing” supercategory), and 1,289 are involved in “operational” processes (“cellular processes” and “metabolism” supercategories). The remaining genes are of unknown, or poorly characterized, function. Among the 7,977 genes deemed as eubacterial, 870 are involved in informational processes, and 4,955 are involved in operational processes. Therefore, eukaryotic genes of archaebacterial and eubacterial affinities are clearly enriched in informational and operational functions, respectively (Fisher’s exact test, P < 10−6; SI Appendix, Fig. S3). This result mirrors previous observations in the yeast and human genomes (13, 15–17).
We evaluated the consistency of this enrichment across the different functional categories. The proportion of genes involved in each of the informational categories (“translation, ribosomal structure, and biogenesis;” “RNA processing and modification;” “transcription;” “replication, recombination, and repair;” and “chromatin structure and dynamics”) is at least twice as high among eukaryotic genes of archaebacterial affinity than among those of eubacterial affinity (SI Appendix, Table S3). Conversely, for all categories belonging to the supercategory “metabolism” (“energy production and conversion;” “carbohydrate transport and metabolism;” “amino acid transport and metabolism;” “nucleotide transport and metabolism;” “coenzyme transport and metabolism;” “lipid transport and metabolism;” “inorganic ion transport and metabolism;” and “secondary metabolites biosynthesis, transport, and catabolism”), the proportion is higher among eubacterial genes than among archaebacterial genes (SI Appendix, Table S3). As for categories belonging to the cellular processes supercategory, the proportion is higher among eubacterial genes for “nuclear structure;” “defense mechanisms;” “cell wall/membrane/envelope biogenesis;” “cytoskeleton;” and “intracellular trafficking, secretion, and vesicular transport;” and higher among archaebacterial genes for “cell cycle control, cell division, chromosome partitioning;” “signal transduction mechanisms;” and “posttranslational modification, protein turnover, chaperones” (SI Appendix, Table S3).
We finally evaluated the enrichment of genes of archaebacterial and eubacterial affinities in informational and operational functions separately for genes belonging to each of the 14 eukaryotic genomes included in the analysis. Similar results were obtained in all species: the proportion of informational genes was always higher among archaebacterial genes whereas the proportion of operational genes was always higher among eubacterial genes (SI Appendix, Table S3 and Fig. S3). These results allow generalizing previous observations in Opisthokonts (13, 15–17) to all of the major eukaryotic groups studied, thereby indicating that the archaebacterial and eubacterial eukaryogenesis partners had a more important contribution to the informational and operational apparatuses of the first eukaryotic cells, respectively.
Proteins Encoded by Eukaryotic Genes of Archaebacterial and Eubacterial Ancestry Are Enriched in Different Cell Compartments, yet Intertwined.
We considered whether eukaryotic genes of archaebacterial and eubacterial ancestry preferentially act in different subcellular compartments. The yeast and human proteomes were used as reference as they are the most comprehensively annotated in the analysis. Comparison of the subcellular compartments of the proteins encoded by the 251 yeast genes of likely archaebacterial ancestry and those encoded by the 463 yeast genes of likely eubacterial ancestry revealed that the archaebacterial gene set is enriched in genes acting at the nucleus and the cytosol. The enrichment of the archaebacterial repertoire in genes encoding nucleus-localized proteins is consistent with this repertoire being enriched in genes participating in transcription and replication (12, 13, 15–17). Conversely, the eubacterial gene set is enriched in genes acting at the mitochondrion and the peroxisome (Table 2). The enrichment of the eubacterial gene set in genes encoding mitochondrial proteins is consistent with a eubacterial origin of mitochondria (18). The enrichment of the eubacterial gene set in genes encoding proteins targeted to the peroxisome would be consistent with either a eubacterial endosymbiont being the ancestor of peroxisomes, or with peroxisomes having borrowed proteins originally targeted to the mitochondrion (for a review, see ref. 65). The proportion of genes that act at the cell membrane and that of genes that act at the endoplasmic reticulum is not significantly different among yeast genes of archaebacterial and eubacterial affinity (Table 2). Nevertheless, the proportion is higher among eubacterial genes in both cases: proteins targeted to the cell membrane include 4 proteins classified as archaebacterial, and 18 classified as eubacterial, and those targeted to the endoplasmic reticulum include 5 proteins of archaebacterial affinity and 18 deemed as eubacterial. These observations are in line with eukaryotic membrane lipids being eubacterial-like and presenting an opposite chirality to those of Archaebacteria. Consistent results were obtained for the human proteome: archaebacterium-like human proteins are enriched for proteins locating to the nucleus and the cytosol, and eubacterium-like proteins tend to locate to the mitochondrion, endoplasmic reticulum, vacuole, and peroxisome (SI Appendix, Table S4). However, it is also necessary to point out that no organelle was found associated with genes of only one affinity and that the intertwining of genes of different ancestry is a feature of eukaryote cells.
Table 2.
Subcellular location of yeast proteins encoded by genes of archaebacterial and eubacterial ancestry
| Location | % among archaebacterial genes | % among eubacterial genes | P |
| Cell wall | 0.00 | 1.30 | 0.096 |
| Plasma membrane | 1.59 | 3.89 | 0.113 |
| Cytosol | 12.75 | 6.05 | 0.003** |
| Endoplasmic reticulum | 1.99 | 3.89 | 0.191 |
| Mitochondrion | 9.16 | 37.58 | 1.65 × 10−17*** |
| Peroxisome | 0.00 | 2.16 | 0.017* |
| Vacuole | 1.20 | 3.67 | 0.060 |
| Nucleus | 41.83 | 19.01 | 1.31 × 10−10*** |
P values correspond to the Fisher’s exact test. *P < 0.05; **P < 0.01; ***P < 0.001.
Evaluating the Potential Role of Gene Mobility and Mobile Genetic Elements in the Evolution of Eukaryotes.
We next classified CCs not only on the basis of their content in archaebacterial, eubacterial and eukaryotic genes, but also according to whether or not they contain sequences derived from MGEs (viruses or plasmids). CCs that include MGEs probably represent gene families capable of undergoing mobilization. Among the 1,791 CCs that include both eukaryotic and prokaryotic sequences, 1,189 (i.e., 66.4%) include MGE sequences as well (SI Appendix, Fig. S1C). The proportion is higher for the 895 CCs that contain representatives of the three domains of life (87.4%), but it is lower among the 2,005 CCs that contain both archaebacterial and eubacterial sequences (60.1%), and even lower for the 19,075 CCs containing only archaebacterial or only eubacterial sequences (32.3%). Furthermore, among the 1,297 CCs containing both eukaryotic and MGE sequences, the proportion of both types of sequences is positively correlated (ρ = 0.264, P < 10−15); i.e., CCs with a high proportion of eukaryotic genes tend to contain also a high fraction of MGE genes. A possible explanation for these observations would be that eukaryotic genes might have been contributed by a flow of HGT from prokaryotes mediated by MGEs. Alternatively, such genes may have been directly contributed by prokaryotic genomes (e.g., by a fusion event). Arguably, gene families of archaebacterial or eubacterial ancestry that were capable of establishing in eukaryotic genomes after eukaryogenesis (presumably, those that were capable of successfully adjusting to the new eukaryotic genomic context) were also susceptible to engage in mobilization, and therefore likely to present representatives in the genomes of MGEs.
It has been proposed that viruses contributed a number of aspects of eukaryotic cell biology, including the nucleus (for review, see refs. 31 and 66; but see ref. 4). For example, a poxvirus has been proposed as the ancestor of the nucleus, based on the structural and physiological similarities between virion factories and the nucleus (32, 33). If the nucleus was the descendant of an ancient virus, one would expect that eukaryotic proteins encoded by genes with detectable viral homologs (i.e., those directly linked to viral sequences) would preferentially locate to the nucleus. A total of 61 yeast genes are directly linked to viral genes in our network, of which 13 (i.e., 21%) encode proteins that locate to the nucleus. This proportion is, however, equivalent to that for the rest of the yeast genome (among yeast genes without viral homologs, 21% encode proteins that are targeted to the nucleus). Similar results were obtained when only the 50 yeast genes with homologs in nucleocytoplasmic large DNA viruses were considered: among these genes, 11 (i.e., 22%) encode proteins that locate to the nucleus, a proportion that is indistinguishable from that for the rest of the yeast genome (21%; Fisher’s exact test, P = 0.860). Therefore, our observations do not support a viral ancestry of the nucleus. Notwithstanding these observations, a more modest yet general contribution of viruses to the biology of the nucleus, for instance via a series of small HGT events, cannot be discarded.
Of particular interest are eukaryotic genes that present homologs in viral, but not in prokaryotic, genomes, as these are the most likely to have been contributed by viruses rather than by prokaryotes (alternatively, they can be eukaryotic-specific proteins that were acquired by viruses). Our network includes 21 yeast genes with these characteristics, out of which 20 are ancient (i.e., present in CCs with representatives of three or more eukaryotic supergroups), and therefore are probably not the result of recent acquisitions from viruses. These 20 genes include 8 members of the ubiquitin pathway, 5 proteins involved in translation (2 ribosomal proteins, 2 elongation factors, and an initiation factor), 2 involved in transcription (including the largest subunit of RNA polymerase II), and 2 involved in replication (type II topoisomerase, and PCNA, which interacts with DNA polymerase δ). For a full list of these genes, see SI Appendix, Table S5.
Conclusion
Here, we have used an analytic tool (gene similarity networks) to study the origin and early evolution of Eukaryotes. Usage of this device allowed us to conduct a more comprehensive analysis than traditional phylogenetic methods, by incorporating a unique kind of datum—extended similarity information—which is systematically removed when constructing traditional phylogenetic trees. Not only have we been able to use this kind of information to trace eukaryote origins, but we have also been able to use homology information to track the subsequent evolution of eukaryote genomes, to provide information on genome evolutionary dynamics, and to raise the possibility that the first recognizable eukaryote had a more balanced collection of eubacterial and archaebacterial genes. We have also been able to show that there is little or no support for certain proposals for eukaryote origins because they do not parsimoniously fit the observed data.
Results presented here provide multiple lines of evidence supporting endosymbiotic theories (14, 18–21). In particular, our network approach uncovers a number of signatures of the chimerical nature of eukaryotic genomes that could not have been disentangled using tree approaches. Remarkably, our gene similarity network contains a considerable number of CCs with a eukaryote–archaebacterium–eubacterium–eukaryote topology (Figs. 1 and 2). This topology is in good agreement with endosymbiotic theories. Eukaryotic sequences linked to archaebacterial sequences likely represent genes contributed by the archaebacterial endosymbiotic partner whereas those linked to eubacterial sequences may represent eukaryotic genes of eubacterial ancestry. Approximately 4 billion y of divergence may have erased sequence similarity between eukaryotic sequences contributed by one domain and eukaryotic sequences contributed by the other domain, or prokaryotic genes from the other domain. As a result, such CCs are not amenable for phylogenetic analysis in a single tree, despite the facts that the sequences are most likely homologous and that the topology of these CCs contain valuable evolutionary information that can be explored using network methods.
The presence of CCs with a eukaryote–archaebacterium–eubacterium–eukaryote topology seems difficult to reconcile with alternative scenarios for the relationships among the three domains of cellular life, such as the Eukaryotes-early hypothesis. According to this model, the first life forms would have been eukaryote-like, and Archaebacteria and Eubacteria would have arisen from these organisms by independent severe genome reductions, as a result of their particular ecology (24–26). Under this scenario, eukaryotic genes would be expected to be equally linked to their homologs in Archaebacteria and Eubacteria. Our observations also seem incompatible with autogenous hypotheses placing a single prokaryotic lineage as the ancestor of Eukaryotes (27–30). Under such scenarios, eukaryotic genes would be expected to be mostly linked to prokaryotic genes of the involved domain.
In addition to the particularly appealing eukaryote–archaebacterium–eubacterium–eukaryote CCs, the network contains additional signatures of the chimerical nature of Eukaryotes in other kinds of CCs. Although these signatures are not as easy to visualize, they can be recovered from statistical analysis of the network edges. Remarkably, the proportion of prokaryotic homologs of a given eukaryotic gene that are eubacterial (pB) is strongly bimodal (SI Appendix, Fig. S3), implying that eukaryotic genes that are highly linked to genes of a given prokaryotic domain tend not to be linked to genes of the other prokaryotic domain. As a result, eukaryotic genes with a similar number of archaebacterial and eubacterial homologs are underrepresented. This observation is also in agreement with Eukaryotes being the result of a fusion of an archaebacterium and a eubacterium.
Although our network analyses strongly support a chimerical nature of extant and ancient eukaryotic genomes, these observations alone cannot rule out a scenario in which Eukaryotes would have arisen before endosymbiosis. Under such a scenario (the so-called proto-eukaryote hypothesis; e.g., ref. 67), a lineage of amitochondriate, nucleated proto-eukaryotes would have existed, before the acquisition of the mitochondrion. Multiple lines of evidence, however, have been used to criticize this particular fusion model. First, all extant Eukaryotes display mitochondria, or the relics of mitochondria (68). Second, it has been argued that the energy generated by mitochondria may have been essential to allow the dramatic increase in cell size at the origin of Eukaryotes, suggesting that the fusion event was a key requirement for the origin of Eukaryotes (21). Finally, our analysis of yeast and human cell compartments shows that most compartments (with the only exception of the yeast cell wall and the peroxisome, which seem to contain mostly proteins of eubacterial ancestry) contain proteins of both archaebacterial and eubacterial ancestry. This mixed ancestry of most cell compartments is consistent with an early, rather than a late, integration of the archaebacterial and eubacterial gene set. Of particular interest is the nucleus, which includes 105 and 88 proteins with affinities to archaebacteria and eubacteria, respectively. This mixed ancestry of the nucleus is in agreement with previous analyses revealing a mixed ancestry of the nucleolus, the nuclear envelope and the nuclear pore complex, and suggests that the nucleus, a typical feature of all Eukaryotes, arose after rather than before endosymbiosis (4, 69, 70).
Eukaryotic genes of archaebacterial ancestry are known to differ from those of eubacterial ancestry in several ways: in general, eukaryotic genes derived from the archaebacterial ancestor are more likely to be involved in informational processes, more highly and broadly expressed, more essential, and to encode more highly connected proteins in the protein–protein interaction network than eubacterium-derived genes (13, 15–17). These differences provide further support for a chimerical origin of Eukaryotes and argue against alternative scenarios such as the Eukaryotes-early hypothesis. For these differences to be compatible with the Eukaryotes-early hypothesis, Archaebacteria would have had to somehow retain proto-eukaryotic genes that in modern eukaryotic genomes perform informational functions, are unlikely to be lost or to undergo duplication, are highly and broadly expressed, and encode highly connected proteins. Conversely, Eubacteria would have had to retain genes that in extant Eukaryotes perform operational tasks, are expressed at lower levels and in a narrower range of tissues, and encode more peripheral proteins to the protein–protein interaction network. It seems very unlikely that such an asymmetrical repartition of proto-eukaryotic genes among Archaebacteria and Eubacteria would have resulted in viable organisms. Similarly, these differences between eukaryotic genes of archaebacterial and eubacterial ancestry would not be expected if Eukaryotes had arisen autogenously from a prokaryotic lineage.
These differences, however, had not been evaluated until now in eukaryotes other than yeast and humans, leaving open the possibility that they could represent an opisthokont-specific feature. In the present analysis, the enrichment of eukaryotic genes of archaebacterial and eubacterial affinity in informational and operational functions, respectively, is confirmed for all 14 eukaryotic genomes studied, which are representative of most of the major eukaryotic groups (SI Appendix, Table S3 and Fig. S3). The consistency of these observations across all studied eukaryotic groups strongly suggests that they existed at the origin of Eukaryotes, implying that the archaebacterial and eubacterial eukaryogenesis partners contributed different functional parts of the first eukaryotic cells. Therefore, the early history of Eukaryotes is likely better understood as the stabilization of a functional partnership rather than solely as a series of divergences. It should be noticed, however, that despite this general tendency, both endosymbiotic partners seem to have contributed genes from both functional categories.
Other features previously observed in Opisthokonts, on the contrary, are not generalizable to all Eukaryotes. The yeast and human genomes exhibit a clearly higher number of genes of eubacterial ancestry than of archaebacterium-derived genes (13, 15–17). However, our analyses reveal the existence of eukaryotes with more genes of archaebacterial affinity than eubacterium-like genes (Table 1). This observation raises questions about the relative contribution of the archaebacterial and eubacterial eukaryogenesis partners to the first eukaryotic genomes. The differences observed in the proportion of archaebacterial and eubacterial genes across the different eukaryotes studied do not seem to be related to their phylogenetic relationships, and therefore, these differences might respond to the different ecological environments in which the studied organisms live, rather than to phylogenetic constraints. Remarkably, the archaebacterial-to-eubacterial gene content ratio seems to be related to genome size, with smaller genomes containing a higher proportion of genes of archaebacterial affinity. Genomic data for eukaryotes other than Opisthokonts and plants are currently limited. The future availability of a wider range of protist genomes, together with a better resolution of the phylogeny of Eukaryotes, may enable an accurate mapping of the variation in the sizes of the archaebacterial and eubacterial gene repertoires and a better understanding of the factors underlying this variation.
The number of genes of archaebacterial affinity is fairly similar across most of the studied eukaryotic genomes (with the only exception of nucleomorphs, whose genomes are extremely reduced) whereas the number of genes of eubacterial affinity is much more variable (Table 1). These observations indicate that the eubacterial gene repertoire is more evolvable than the more static archaebacterial gene set. This finding is in line with previous observations that eukaryotic genes of eubacterial ancestry are less likely to be essential, less selectively constrained, and more likely to undergo duplication than eukaryotic genes of archaebacterial ancestry (13, 15–17).
Our analyses reveal further differences between eukaryotic genes contributed by both eukaryogenesis partners, showing that proteins encoded by genes derived from both ancestors tend to locate to different subcellular compartments. In particular, yeast genes of archaebacterial affinity are enriched in genes acting at the nucleus and the cytosol whereas those of eubacterial affinity preferentially act at the mitochondrion, the cell wall, the vacuole, and the peroxisome (Table 2). These observations shed more light on the contributions of both endosymbiotic partners to eukaryotic cells. These parts are now so intertwined in Eukaryotes that it indicates a long and complex stabilization.
The analysis of the evolutionary affinities of eukaryotic genes acting at the different cell compartments also argues against other alternative scenarios regarding the origin of Eukaryotes. In particular, hypotheses placing a virus as the ancestor of the nucleus (32, 33) are not supported by our observation that yeast genes with viral homologs do not preferentially encode proteins that are targeted to the nucleus (indeed, the proportion of genes encoding nuclear proteins is the same for those that have viral homologs and for those that do not have viral homologs in the yeast genome).
Taken together, results presented here highlight the suitability of gene similarity networks as a powerful tool for studying the origin of Eukaryotes, and evolution in general, especially when it comes to studying deep evolutionary events and introgressive descent. Networks can complement trees in evolutionary analyses by providing a wider picture of the relationships among sequences and organisms. Without a doubt, gene similarity networks are tools, whose power and potential pitfalls remain to be explored. In any case, we would like to emphasize that by no means are similarity networks expected to replace phylogenetic trees in evolutionary analyses. Both trees and networks (and, ideally, a combination of both) will continue to shed light on questions such as the origin and evolution of Eukaryotes.
Methods
Age of Connected Components.
CCs were classified as ancient” if they comprised representatives of at least three different eukaryotic supergroups. For that purpose, the eukaryotic species included in the analysis were classified into seven supergroups according to refs. 71 and 72 (Table 1). For the purpose of age classification, nucleomorphs were considered as Plants, as they are thought to have derived from algae (57).
Eukaryote–Eubacteria–Archaebacteria–Eukaryote Connected Components.
The Dijkstra algorithm, as implemented in the “Graph” module for PERL, was applied to determine the shortest path between each pair of eukaryotic genes in the same CC. CCs containing a eukaryote–archaebacterium–eubacterium–eukaryote shortest path were classified as such.
Functional Information.
Each eukaryotic gene was used as query in an RPS-BLAST search against the KOG profiles. Genes were then assigned to one (or in some cases, a few) functional category(ies) according to their best-matching KOG. Genes whose categories include translation, ribosomal structure, and biogenesis; RNA processing and modification; transcription; DNA replication, recombination, and repair; or chromatin structure and dynamics were classified as informational. Genes pertaining to the remaining categories were considered operational. Genes pertaining to no KOG, or to categories “general function prediction only” or “function unknown,” exclusively, remained unclassified.
Subcellular Locations.
The subcellular locations of each yeast and human protein were obtained from the Gene Ontology database.
Supplementary Material
Acknowledgments
We thank four anonymous referees for helpful comments. This work was supported by Science Foundation Ireland Grant 09/RFP/EOB2510 (to J.O.M.), a mobility grant from the Royal Irish Academy (to D.A.-P.), and a Ulysses mobility grant from Egide and the Irish Research Council for Science, Engineering, and Technology (to E.B. and J.O.M.). In addition, computational facilities were provided by the Irish Centre for High End Computing and National University of Ireland Maynooth High Performance Computing Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.B. is a guest editor invited by the Editorial Board.
Data deposition: The survey sequence data have been deposited in the Dryad database, http://datadryad.org (doi no. 10.5061/dryad.qr81p).
See Author Summary on page 6624 (volume 110, number 17).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211371110/-/DCSupplemental.
References
- 1.Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc Natl Acad Sci USA. 1977;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440(7084):623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 4.Martin W. Archaebacteria (Archaea) and the origin of the eukaryotic nucleus. Curr Opin Microbiol. 2005;8(6):630–637. doi: 10.1016/j.mib.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Martin W, et al. The evolution of eukaryotes. Science. 2007;316(5824):542–543, author reply 542–543. doi: 10.1126/science.316.5824.542c. [DOI] [PubMed] [Google Scholar]
- 6.Martin W, Hoffmeister M, Rotte C, Henze K. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem. 2001;382(11):1521–1539. doi: 10.1515/BC.2001.187. [DOI] [PubMed] [Google Scholar]
- 7.McInerney JO, et al. Planctomycetes and eukaryotes: A case of analogy not homology. Bioessays. 2011;33(11):810–817. doi: 10.1002/bies.201100045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gribaldo S, Poole AM, Daubin V, Forterre P, Brochier-Armanet C. The origin of eukaryotes and their relationship with the Archaea: Are we at a phylogenomic impasse? Nat Rev Microbiol. 2010;8(10):743–752. doi: 10.1038/nrmicro2426. [DOI] [PubMed] [Google Scholar]
- 9.Lake JA, Henderson E, Oakes M, Clark MW. Eocytes: A new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci USA. 1984;81(12):3786–3790. doi: 10.1073/pnas.81.12.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lake JA. Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences. Nature. 1988;331(6152):184–186. doi: 10.1038/331184a0. [DOI] [PubMed] [Google Scholar]
- 11.Gouy M, Li WH. Phylogenetic analysis based on rRNA sequences supports the archaebacterial rather than the eocyte tree. Nature. 1989;339(6220):145–147. doi: 10.1038/339145a0. [DOI] [PubMed] [Google Scholar]
- 12.Horiike T, Hamada K, Kanaya S, Shinozawa T. Origin of eukaryotic cell nuclei by symbiosis of Archaea in Bacteria is revealed by homology-hit analysis. Nat Cell Biol. 2001;3(2):210–214. doi: 10.1038/35055129. [DOI] [PubMed] [Google Scholar]
- 13.Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci USA. 1998;95(11):6239–6244. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pisani D, Cotton JA, McInerney JO. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol. 2007;24(8):1752–1760. doi: 10.1093/molbev/msm095. [DOI] [PubMed] [Google Scholar]
- 15.Esser C, et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21(9):1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- 16.Cotton JA, McInerney JO. Eukaryotic genes of archaebacterial origin are more important than the more numerous eubacterial genes, irrespective of function. Proc Natl Acad Sci USA. 2010;107(40):17252–17255. doi: 10.1073/pnas.1000265107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez-Ponce D, McInerney JO. The human genome retains relics of its prokaryotic ancestry: Human genes of archaebacterial and eubacterial origin exhibit remarkable differences. Genome Biol Evol. 2011;3:782–790. doi: 10.1093/gbe/evr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14(3):255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 19.Zillig W, Schnabel R, Stetter KO. Archaebacteria and the origin of the eukaryotic cytoplasm. Curr Top Microbiol Immunol. 1985;114:1–18. doi: 10.1007/978-3-642-70227-3_1. [DOI] [PubMed] [Google Scholar]
- 20.Rivera MC, Lake JA. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431(7005):152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- 21.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467(7318):929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 22.Forterre P. A new fusion hypothesis for the origin of Eukarya: Better than previous ones, but probably also wrong. Res Microbiol. 2011;162(1):77–91. doi: 10.1016/j.resmic.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 23.McInerney JO, Pisani D, Bapteste E, O’Connell MJ. The Public Goods Hypothesis for the evolution of life on Earth. Biol Direct. 2011;6:41. doi: 10.1186/1745-6150-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurland CG, Collins LJ, Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312(5776):1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- 25.Doolittle WF. Revolutionary concepts in evolutionary cell biology. Trends Biochem Sci. 1980;5:146–149. [Google Scholar]
- 26.Forterre P, Philippe H. Where is the root of the universal tree of life? Bioessays. 1999;21(10):871–879. doi: 10.1002/(SICI)1521-1878(199910)21:10<871::AID-BIES10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 27.Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52(Pt 2):297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 28.Devos DP, Reynaud EG. Evolution. Intermediate steps. Science. 2010;330(6008):1187–1188. doi: 10.1126/science.1196720. [DOI] [PubMed] [Google Scholar]
- 29.Reynaud EG, Devos DP. Transitional forms between the three domains of life and evolutionary implications. Proc Biol Sci. 2011;278(1723):3321–3328. doi: 10.1098/rspb.2011.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santarella-Mellwig R, et al. The compartmentalized bacteria of the planctomycetes-verrucomicrobia-chlamydiae superphylum have membrane coat-like proteins. PLoS Biol. 2010;8(1):e1000281. doi: 10.1371/journal.pbio.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forterre P, Prangishvili D. The great billion-year war between ribosome- and capsid-encoding organisms (cells and viruses) as the major source of evolutionary novelties. Ann N Y Acad Sci. 2009;1178:65–77. doi: 10.1111/j.1749-6632.2009.04993.x. [DOI] [PubMed] [Google Scholar]
- 32.Bell PJ. Viral eukaryogenesis: Was the ancestor of the nucleus a complex DNA virus? J Mol Evol. 2001;53(3):251–256. doi: 10.1007/s002390010215. [DOI] [PubMed] [Google Scholar]
- 33.Takemura M. Poxviruses and the origin of the eukaryotic nucleus. J Mol Evol. 2001;52(5):419–425. doi: 10.1007/s002390010171. [DOI] [PubMed] [Google Scholar]
- 34.Gribaldo S, Philippe H. Ancient phylogenetic relationships. Theor Popul Biol. 2002;61(4):391–408. doi: 10.1006/tpbi.2002.1593. [DOI] [PubMed] [Google Scholar]
- 35.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30(7):1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci USA. 2008;105(51):20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adai AT, Date SV, Wieland S, Marcotte EM. LGL: Creating a map of protein function with an algorithm for visualizing very large biological networks. J Mol Biol. 2004;340(1):179–190. doi: 10.1016/j.jmb.2004.04.047. [DOI] [PubMed] [Google Scholar]
- 38.Fondi M, Fani R. The horizontal flow of the plasmid resistome: Clues from inter-generic similarity networks. Environ Microbiol. 2010;12(12):3228–3242. doi: 10.1111/j.1462-2920.2010.02295.x. [DOI] [PubMed] [Google Scholar]
- 39.Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci USA. 2010;107(1):127–132. doi: 10.1073/pnas.0908978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dagan T. Phylogenomic networks. Trends Microbiol. 2011;19(10):483–491. doi: 10.1016/j.tim.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 41.Dagan T, Roettger M, Bryant D, Martin W. Genome networks root the tree of life between prokaryotic domains. Genome Biol Evol. 2010;2:379–392. doi: 10.1093/gbe/evq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 2011;21(4):599–609. doi: 10.1101/gr.115592.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamminen M, Virta M, Fani R, Fondi M. Large-scale analysis of plasmid relationships through gene-sharing networks. Mol Biol Evol. 2012;29(4):1225–1240. doi: 10.1093/molbev/msr292. [DOI] [PubMed] [Google Scholar]
- 44.Bapteste E, et al. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc Natl Acad Sci USA. 2002;99(3):1414–1419. doi: 10.1073/pnas.032662799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311(5765):1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 46.Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA. 2011;108(33):13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knoll AH, Javaux EJ, Hewitt D, Cohen P. Eukaryotic organisms in Proterozoic oceans. Philos Trans R Soc Lond B Biol Sci. 2006;361(1470):1023–1038. doi: 10.1098/rstb.2006.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen B, Fletcher IR, Brocks JJ, Kilburn MR. Reassessing the first appearance of eukaryotes and cyanobacteria. Nature. 2008;455(7216):1101–1104. doi: 10.1038/nature07381. [DOI] [PubMed] [Google Scholar]
- 49.Feng DF, Cho G, Doolittle RF. Determining divergence times with a protein clock: Update and reevaluation. Proc Natl Acad Sci USA. 1997;94(24):13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hedges SB, et al. A genomic timescale for the origin of eukaryotes. BMC Evol Biol. 2001;1:4. doi: 10.1186/1471-2148-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butterfield NJ. Bangiomorpha pubescens n. gen., n. sp.: Implications for the evolution of sex, multicellularity, and the Mesoproterozoic/Neoproterozoic radiation of eukaryotes. Paleobiology. 2000;26:386–404. [Google Scholar]
- 52.Alvarez-Ponce D. The relationship between the hierarchical position of proteins in the human signal transduction network and their rate of evolution. BMC Evol Biol. 2012;12:192. doi: 10.1186/1471-2148-12-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li WH, Wu CI, Luo CC. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2(2):150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 54.Doolittle WF. You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998;14(8):307–311. doi: 10.1016/s0168-9525(98)01494-2. [DOI] [PubMed] [Google Scholar]
- 55.Bapteste E, Walsh DA. Does the “Ring of Life” ring true? Trends Microbiol. 2005;13(6):256–261. doi: 10.1016/j.tim.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 56.Loftus B, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433(7028):865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 57.Moore CE, Archibald JM. Nucleomorph genomes. Annu Rev Genet. 2009;43:251–264. doi: 10.1146/annurev-genet-102108-134809. [DOI] [PubMed] [Google Scholar]
- 58.Texier C, Vidau C, Viguès B, El Alaoui H, Delbac F. Microsporidia: A model for minimal parasite-host interactions. Curr Opin Microbiol. 2010;13(4):443–449. doi: 10.1016/j.mib.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 59.Eisen JA, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4(9):e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haas BJ, et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461(7262):393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 61.El-Sayed NM, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309(5733):409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 62.Tovar J, et al. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426(6963):172–176. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- 63.Mai Z, et al. Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol Cell Biol. 1999;19(3):2198–2205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tovar J, Fischer A, Clark CG. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999;32(5):1013–1021. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- 65.Gabaldón T. Peroxisome diversity and evolution. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):765–773. doi: 10.1098/rstb.2009.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006;117(1):5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 67.Glansdorff N, Xu Y, Labedan B. The last universal common ancestor: Emergence, constitution and genetic legacy of an elusive forerunner. Biol Direct. 2008;3:29. doi: 10.1186/1745-6150-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Giezen M. Hydrogenosomes and mitosomes: Conservation and evolution of functions. J Eukaryot Microbiol. 2009;56(3):221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- 69.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3(12):1612–1637. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 70.Staub E, Fiziev P, Rosenthal A, Hinzmann B. Insights into the evolution of the nucleolus by an analysis of its protein domain repertoire. Bioessays. 2004;26(5):567–581. doi: 10.1002/bies.20032. [DOI] [PubMed] [Google Scholar]
- 71.Fritz-Laylin LK, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140(5):631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 72.Rodríguez-Ezpeleta N, et al. Toward resolving the eukaryotic tree: The phylogenetic positions of jakobids and cercozoans. Curr Biol. 2007;17(16):1420–1425. doi: 10.1016/j.cub.2007.07.036. [DOI] [PubMed] [Google Scholar]