Abstract
DEK is a biochemically distinct, conserved nonhistone protein that is vital to global heterochromatin integrity. In addition, DEK can be secreted and function as a chemotactic, proinflammatory factor. Here we show that exogenous DEK can penetrate cells, translocate to the nucleus, and there carry out its endogenous nuclear functions. Strikingly, adjacent cells can take up DEK secreted from synovial macrophages. DEK internalization is a heparan sulfate-dependent process, and cellular uptake of DEK into DEK knockdown cells corrects global heterochromatin depletion and DNA repair deficits, the phenotypic aberrations characteristic of these cells. These findings thus unify the extracellular and intracellular activities of DEK, and suggest that this paracrine loop involving DEK plays a role in chromatin biology.
Keywords: cellular biology, cancer, autoimmunity, juvenile arthritis
Since its initial cloning as part of the t(6;9) translocation in a subset of patients with acute myelogenous leukemia (1, 2), DEK has been shown to affect global heterochromatin integrity (3), mRNA splicing (4, 5), transcriptional control (both negative and positive) (6–8), DNA damage repair and susceptibility (9, 10), DNA replication (11), cellular differentiation (12, 13), cell viability (8), apoptosis (14), and senescence (15). DEK also plays a key role in the biology of hematopoietic and muscle stem cells (12, 16).
DEK overexpression occurs in various prevalent and difficult-to-treat neoplasms, including hepatocellular carcinoma (17), glioblastoma (18), melanoma (8, 19), bladder cancer (20), retinoblastoma (21, 22), breast cancer (23), and T-cell large granular lymphocytic leukemia (24). DEK is degraded by the F-box/tryptophan-aspartic acid (WD) repeat-containing protein 7 (Fbxw7) tumor suppressor, which affects cell division and splicing of messenger RNA (25). Elevated levels of DEK can interfere with cellular differentiation, apoptosis, senescence, and the response to chemotherapy, justifying the classification of DEK as a bona fide oncogene that plays a role in central pathways promoting tumor growth and survival (8, 19, 23).
In addition to its roles in tumor biology, which appear to be related mainly to its intracellular functions, DEK also has been implicated in the pathogenesis of autoimmune disorders, functions more attributable to its extracellular activities (discussed below). In fact, circulating autoantibodies to DEK have been identified in the serum of patients with various autoimmune diseases, including juvenile idiopathic arthritis (JIA), sarcoidosis, and systemic lupus erythematosus (SLE) (26). Furthermore, autoantibodies to DEK, as well as DEK protein itself, have been detected in synovial fluids of children with JIA (27). The presence of DEK protein and DEK autoantibodies in the extracellular space suggests a proinflammatory role for DEK. In fact, on activation, macrophages secrete DEK, which in turn can act as a chemotactic factor for neutrophils, natural killer cells, and cytotoxic T lymphocytes in the extracellular milieu (28). The extracellular signaling activities of DEK are characteristic of other nuclear factors, like high mobility group protein B1 (HMGB1), that serve as danger-associated molecular patterns, or alarmins (29), suggesting that DEK might belong to this family of proteins.
Water-soluble proteins like DEK are considered too bulky, charged, and hydrophilic to spontaneously cross a membrane by diffusion or via known membrane transporters. There is, however, a class of proteins, termed translocatory proteins, that can enter the cell via endocytosis, including human programmed cell death 5 (PDCD5), Human Immunodeficiency Virus type 1 transactivator of transcription (HIV Tat), and Drosophila Antennaepedia (30, 31). These proteins bind to negatively charged heparan sulfate-type proteoglycans (HSPGs) that are actively endocytosed via small invaginations in the plasma membrane known as caveolae. HSPGs thus serve as shuttles for translocatory proteins to enter the cell. An N-terminal fragment of DEK, spanning amino acids 78–222, exhibits a surprising ability to traverse lipid bilayers (32). This particular truncation of DEK has a high concentration of positive charges (31 arginines and lysines in the 130 amino acids of this peptide) and has been studied primarily as a tool for the cellular delivery of macromolecules. The relevance of this observation to the biology of naturally occurring, full-length DEK remained unclear, however. Here we report that full-length DEK secreted by one cell can be taken up by another cell, move to the nucleus, and function in heterochromatin biology and DNA repair, thereby potentially uniting the intracellular and extracellular activities of DEK.
Results and Discussion
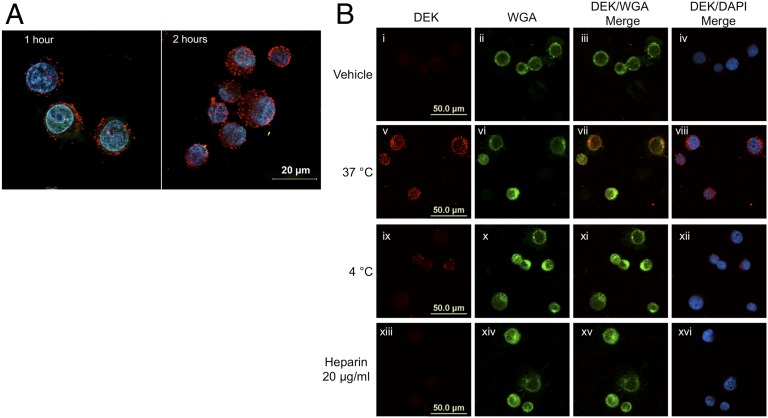
Considering that extracellular and intracellular DEK both play important biological roles, we asked whether these two functions could be connected. We began by assessing whether full-length DEK can enter a cell. We added recombinant full-length histidine (His)-tagged DEK directly to the cell culture medium of HeLa DEK knockdown (DEK-KD) cells, followed by subsequent fixation of the cells and staining for DEK. Analysis by confocal microscopy revealed DEK-positive staining, predominantly in the cytoplasm, within 1 h of incubation (Fig. 1A, Left). Importantly, nuclear DEK staining was clearly visible after 2 h of incubation with His-DEK and increased over time (Fig. 1A, Right, Fig. S1, and Movie S1).
Fig. 1.
DEK is taken up by cells in an energy-dependent process that requires surface HSPGs. (A) DEK-KD HeLa S3 cells were incubated with His-tagged recombinant DEK (20 µg/ 1 × 107 cells) for 1 or 2 h and stained with DAPI (blue) and a monoclonal DEK antibody (red). (B) HeLa DEK-KD cells were incubated with recombinant His-DEK or vehicle for 2 h under varying conditions and stained with WGA (green), a membrane-specific stain, a monoclonal DEK antibody (red), and DAPI (blue) to visualize the nuclei. (i–iv) Cells incubated with vehicle (buffer) alone. (v–viii) Cells incubated with recombinant His-DEK at 37 °C. (ix–xii) Cells incubated with recombinant His-DEK at 4 °C. (xiii–xvi) Cells pretreated with 20 μg/mL soluble heparin and then incubated with recombinant His-DEK and 20 μg/mL soluble heparin at 37 °C. Images were obtained by confocal microscopy.
We then investigated whether DEK uptake is an active process, by incubating cells at 37 °C or 4 °C and studying DEK uptake using immunofluorescence, with FITC-conjugated wheat germ agglutinin (WGA) as a membrane marker. We found substantially reduced DEK staining at 4 °C compared with 37 °C (Fig. 1B), suggesting that DEK internalization indeed appears to be an active process.
The DEK truncation 78–222 yields a polypeptide that conforms to the class of molecules known as naturally supercharged proteins, capable of cellular entry through electrostatic interactions with negatively charged moieties on the cell surface (32). To test whether this also holds true for WT DEK (82 arginines and lysines in 375 amino acids, and thus a substantially reduced overall presence of positively charged amino acids compared with the truncation 87–222), we incubated HeLa DEK-KD cells with heparin, a soluble competitor of surface HSPGs, and recombinant human WT DEK. A soluble heparin concentration of 20 µg/mL was sufficient to suppress binding and internalization of recombinant DEK (Fig. 1B and Fig. S2), suggesting that HSPGs are important for initial binding of DEK to cellular membranes.
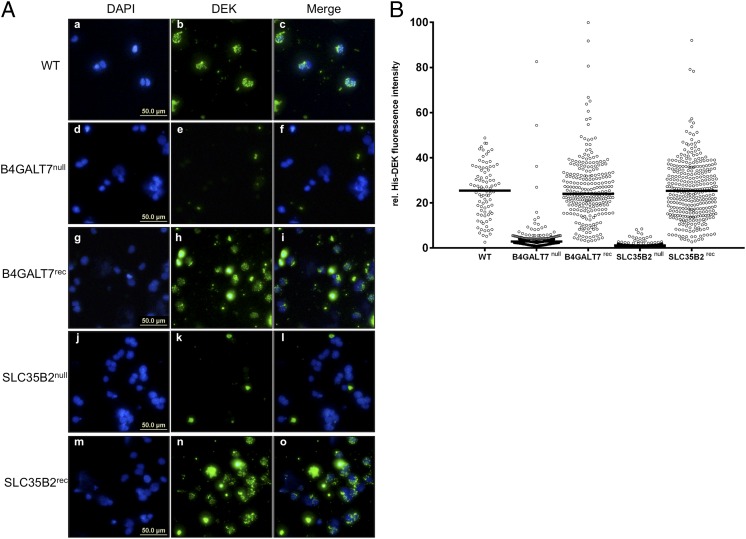
To further confirm that HSPGs are necessary for cellular DEK uptake, we turned to a previously described elegant genetic system (33). We used HAP1 cells and HAP1 cells deficient in either B4GALT7 (B4GALT7null) or SLC35B2 (SLC35B2null) (33), genes that specify part of the biochemical pathway required for the synthesis and translocation of HSPGs. The HAP1 cell line is derived from the haploid myeloid KBM7 cell line that was originally isolated from a patient with leukemia (34). Incubation of His-DEK with these mutant cell lines revealed that cells with defects in HSPG synthesis and localization cannot internalize DEK (Fig. 2 A, d–f, and j–l and B). To further validate this observation, we used genetic reconstitution with functional B4GALT7 (B4GALT7rec) or SLC35B2 (SLC35B2rec), which resulted in restoration of DEK uptake comparable to the WT phenotype (Fig. 2 A, g–i and m–o and B). Thus, genetic studies demonstrate that HSPGs are required for cellular entry of DEK.
Fig. 2.
Cells defective in essential enzymes required for HSPG synthesis or positioning exhibit significantly reduced DEK uptake. (A) WT or mutant HAP1 cells incubated with recombinant His-DEK at 37 °C for 2 h and stained with DAPI (blue) or a monoclonal DEK antibody (green). (a–c) WT HAP1 cells. (d–f) HAP1 cells deficient in B4GALT7. (g–i) HAP1 B4GALT7null cells reconstituted with functional B4GALT7. (j–l) HAP1 cells deficient in SLC35B2. (m–o) HAP1 SLC35B2null cells reconstituted with functional SLC35B2. (B) Graphical representation of His-DEK–specific intensities in the nucleus. Multiple micrographs, as shown in A, were subjected to an automated intensity measurement workflow using the KNIME image processing software. More details are shown in Fig. 4 and Figs. S3 and S4. Segmentation of nuclei was carried out in the DAPI channel, followed by measurement of the His-DEK–specific intensities within the assigned segments using the FITC channel. Signals originating from cell clumps, debris, dead cells, or background signals owing to nonspecific extracellular aggregation of His-DEK were excluded from the measurements. Median values for the relative (rel.) His-DEK fluorescence intensity of all samples in a given group are indicated by the horizontal lines in the scatter dot plot.
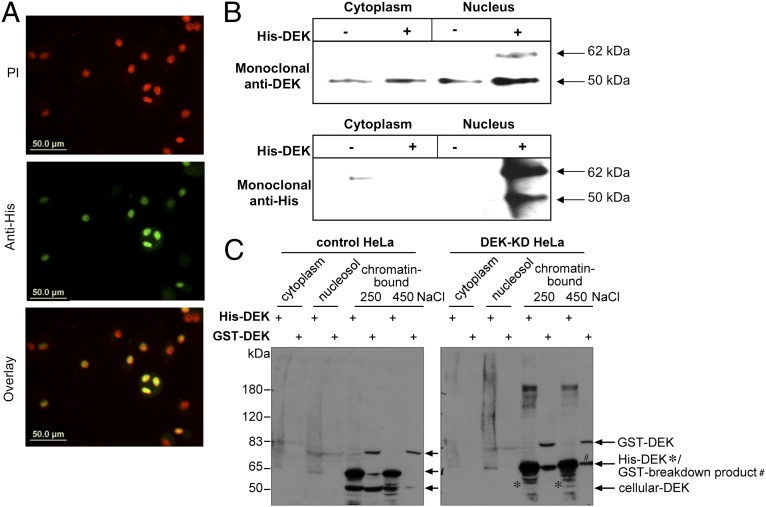
To assess DEK uptake in another biologically relevant cell type, we used activated, primary monocyte-derived macrophages (MDMs) from healthy volunteers, which contain less DEK in their nuclei after differentiation and secretion of their endogenous DEK (28). Indeed, recombinant His-DEK was detected in the nuclei of macrophages incubated with DEK by immunofluorescence (Fig. 3A). These results were confirmed by immunoprecipitation of His-DEK from the nuclear and cytoplasmic fractions. DEK was detected mainly in the nuclear fraction by an anti-His antibody or by a specific monoclonal DEK antibody (Fig. 3B). This indicates that DEK can be taken up by several types of cancer cell lines and by primary cells involved in the immune response.
Fig. 3.
DEK taken up by cells migrates to chromatin. (A) Twelve-day MDMs in 40% human serum were treated overnight with 15 µg of recombinant His-tagged DEK. Cells were stained for DEK with a monoclonal anti-His antibody, and nuclei were stained with propidium iodide (PI). (B) Immunoprecipitation of DEK from cytoplasmic and nuclear fractions of cells treated as in A with a rabbit polyclonal anti-His antibody. The resulting samples were analyzed on an SDS gel and immunoblotted with anti-DEK or anti-His monoclonal antibodies. (C) Control or HeLa DEK-KD cells were incubated with recombinant His-DEK or GST-DEK for 24 h and fractionated into cytosolic, nucleosolic, and chromatin-bound fractions. Proteins from each fraction were precipitated, resolved by SDS/PAGE, and blotted for DEK using a polyclonal antibody.
Endogenous DEK usually localizes to chromatin, where it can reside in euchromatin, as well as in heterochromatin (35). On cellular entry, exogenous DEK localizes mostly to the nucleus of HeLa DEK-KD cells. A fundamental question is whether DEK is functional on internalization. We tested the ability of exogenous DEK to bind to chromatin subsequent to uptake. Both control HeLa cells and HeLa-KD cells were fractionated after 24 h of incubation with either recombinant His-tagged DEK produced in a baculovirus expression system or GST-tagged DEK produced in bacteria. Recombinant DEK, regardless of its respective tag, origin, and phosphorylation status (GST-tagged DEK made in bacteria is not phosphorylated, whereas His-tagged DEK made in baculovirus is phosphorylated), was detected in chromatin fractions extracted by high-salt buffers in both control and DEK-KD cells (Fig. 3C), confirming that exogenous DEK has the ability to bind chromatin on internalization and localizes to fractions in which endogenous DEK is ordinarily found.
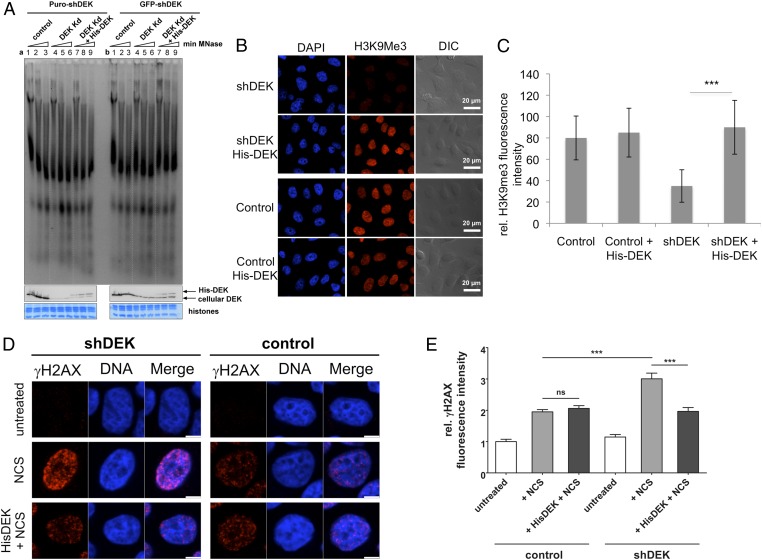
We next asked whether exogenously added DEK that is taken up by cells exhibits biological activity. In mammalian cells and in Drosophila, DEK is vital to global heterochromatin integrity, an activity not dependent on phosphorylation (3). Specifically, DEK augments the binding of heterochromatin protein 1α (HP1α) to the repressive epigenetic mark Histone H3 tri-methylated at lysine 9 (H3K9Me3). Elevated expression of DEK thus results in enhanced H3K9Me3 levels and a more closed chromatin structure (3). Suppression of DEK expression in turn leads to a reduction in H3K9Me3 levels, resulting in a less-condensed chromatin structure. Thus, we tested whether or not internalized DEK could complement this profound DEK-deficient phenotype. Impressively, micrococcal nuclease (MNase) treatment of nuclei from two HeLa DEK-KD cell lines revealed that the addition of exogenous DEK reversed the accelerated digestion of chromatin seen in DEK-KD cells, suggesting that global heterochromatin integrity was restored when exogenous DEK was taken up into the nucleus (Fig. 4A).
Fig. 4.
Exogenous DEK is bioactive on uptake into cells. Control or HeLa DEK-KD cells were incubated with recombinant His-DEK or buffer alone for 48 h and then subjected to functional analysis. (A) Cellular nuclei from control or HeLa DEK-KD, selected either by puromycin (a) or by FACS for GFP expression (b), were isolated, adjusted to 500 μg DNA/mL, and subjected to micrococcal nuclease treatment (5 U/50 μg DNA) of increasing duration (1 min in lanes 1, 4, and 7; 2 min in lanes 2, 5, and 8; 4 min in lanes 3, 6, and 9). The DNA fragments thus produced were analyzed directly by agarose gel electrophoresis and ethidium bromide staining. Equal aliquots of the individual samples were further analyzed by immunoblotting using a DEK-specific antibody, confirming DEK knockdown (a and b, lanes 4–6) and the presence of His-DEK (a and b, lanes 7–9). Protein staining of core histones served as a loading control (histones). (B) Control or HeLa-KD (shDEK) cells were fixed and stained with DAPI (blue) or an antibody that recognizes the H3K9Me3 heterochromatin mark (red) in the presence or absence of exogenously added His-DEK. Shown are representative magnifications of tile scans of three biological replicates (Fig. S4). DIC, differential interference contrast. (C) H3K9Me3 intensities in the individual samples were analyzed using the KNIME image processing software and a newly developed workflow (Figs. S3 and S4). One out of three replicates is shown. On average, 1,500–2,000 cells were analyzed per sample. Datasets were compared using the unpaired Student t test. ***P = 0.0004. (D) Control or HeLa-KD (shDEK) cells were treated with NCS for 15 min to induce DNA double-strand breaks, fixed, and stained with DAPI (blue) or an antibody specific for γH2AX (red), a marker for DNA double-stranded breaks. (E) Graphical representation of γH2AX-specific fluorescence intensity in the nucleus. The fluorescence intensity of γH2AX-positive nuclei was significantly reduced in NCS-treated HeLa-KD cells incubated with exogenous His-DEK. At least 150 nuclei were analyzed per experimental condition. Datasets were compared using the unpaired Student t test. ***P = 0.0001.
We next analyzed whether the restoration of heterochromatin by the addition of exogenous DEK is due to elevated H3K9Me3 levels, based on the finding that endogenous DEK controls heterochromatin integrity by facilitating the recruitment of HP1α and, in turn, the lysine methyltransferase 1 (KMT1) A/B to histones (3). We incubated control HeLa or DEK-KD HeLa cells with recombinant His-DEK for 48 h and then analyzed H3K9Me3 by immunofluorescence. To avoid bias, image analysis was performed automatically using a Konstanz Information Miner (KNIME)-based image processing workflow (36) (Fig. S3). Analysis of tile scans from three biological replicates (Fig. 4 B and C and Fig. S4) showed that internalized DEK indeed induced a significant increase in H3K9Me3 in DEK-KD HeLa cells (P = 0.0004). This effect was absent in control cells, likely owing to the high levels of DEK already present in HeLa cells.
We and others have shown that DEK-KD cells are deficient in their ability to repair DNA double-strand breaks, as measured by the increased persistence of γH2AX-positive foci when cells are subjected to neocarzinostatin (NCS) treatment (9, 10), which induces double-strand breaks (9). As part of the repair mechanism for DNA damage, the histone H2AX is phosphorylated, generating γH2AX, a marker for DNA double-strand breaks (37). Incubation with exogenous DEK before exposure to NCS reverses the increased sensitivity toward NCS-induced DNA double-strand breaks in DEK-KD cells, consistent with the observed restoration of heterochromatin reported above and also with the observation that heterochromatin is more refractory to developing γH2AX foci after DNA damage (Fig. 4 D and E). We treated both control HeLa cells and HeLa-KD cells with NCS with or without previous incubation with recombinant His-DEK, and then fixed and stained these cells for γH2AX. We found a significant reduction (P = 0.0001) in γH2AX-positive foci when the HeLa-KD cells treated with NCS were supplemented with recombinant DEK, as revealed by quantifying nuclear fluorescence signals on mean intensity projections of confocal z-stacks (Fig. 4 D and E). We observed no significant difference in susceptibility to DNA damage in control HeLa cells, again most likely related to the significant levels of endogenous DEK.
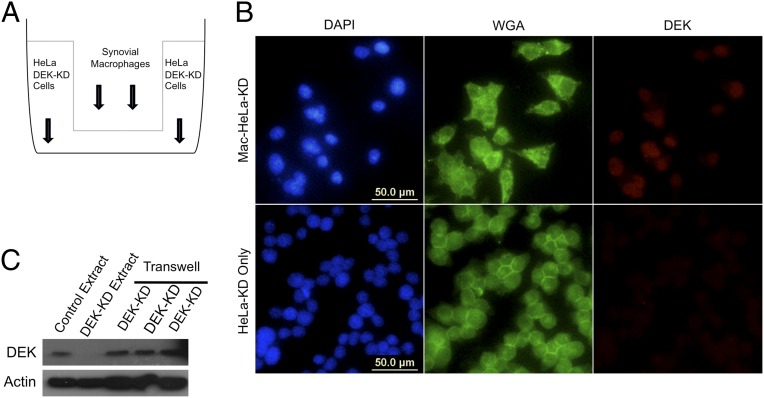
Having shown that recombinant DEK can be internalized by at least three different cell types, we next investigated whether naturally occurring DEK could indeed be secreted by one cell and taken up by another cell. Macrophages isolated from the synovial fluids of patients with JIA actively secrete abundant amounts of DEK, which can subsequently form immune complexes with anti-DEK antibodies (27). We used the ability of these macrophages to secrete DEK to study whether secreted DEK can be internalized by another cell type. We cocultured HeLa DEK-KD cells with synovial macrophages for 3–5 d in a Transwell cell culture apparatus (Fig. 5A). Using immunofluorescence (Fig. 5B) and immunoblot analysis (Fig. 5C), we detected synovial macrophage-derived DEK in HeLa DEK-KD cells, localizing mainly to the nucleus of these cells, consistent with DEK’s preferred endogenous localization. These findings indicate that WT DEK indeed can be transmitted from one cell to another and migrate to the nucleus of the second cell.
Fig. 5.
Secreted DEK is taken up by adjacent cells. (A) Diagram of the Transwell apparatus used in these experiments. Synovial macrophages were seeded on the top layer of the Transwell apparatus with 0.4-μm pores. HeLa-KD cells were then seeded on the bottom layer 2 d later. Cells were incubated for 3 d, and HeLa extracts were analyzed by immunofluorescence imaging and immunoblot analysis. (B) Immunofluorescence images of HeLa-KD cells with (Mac-HeLa-KD) or without (HeLa-KD only) synovial macrophages on the top layer of the Transwell apparatus. Cells were stained with DAPI (blue), membrane-specific WGA (green), and a polyclonal DEK antibody (red). (C) Western blot of whole cell extracts from control HeLa cells, HeLa DEK-KD cells without synovial macrophages on the top layer of the Transwell apparatus, and HeLa DEK-KD cells with synovial macrophages on the top layer of the Transwell (last three lanes, representing three biological replicates).
The extracellular and intracellular aspects of DEK biology have not previously been considered interdependent processes. Here we have shown that DEK can be taken up by multiple cell lines and primary cell types, including HeLa cells, HAP1 cells, and human MDMs. DEK uptake is HSPG-dependent, as demonstrated by the findings that treatment with heparin inhibited cellular uptake of DEK and genetic depletion of HSPGs blocked DEK internalization. DEK internalization led to nuclear localization, in which, remarkably, exogenous DEK corrected the heterochromatin deficits and the DNA-damage repair aberrations characteristic of DEK-KD cells. Importantly, DEK secreted from activated synovial macrophages derived from a patient with JIA was taken up by neighboring cells. Thus, the secretion and uptake of biologically active DEK may represent an important pathway through which this biochemically distinct protein plays a role in chromatin biology and perhaps in autoimmunity and cancer as well. The discovery of a pathway that shuttles a protein vital for heterochromatin integrity in and out of cells suggests an unexpected new layer of epigenetic regulation.
Materials and Methods
Antibodies.
Detection of DEK was achieved using either a mouse monoclonal antibody (BD Biosciences) or an affinity-purified polyclonal antibody (8). Immunoprecipitation and immunoblotting of His-DEK required the use of a monoclonal anti-polyHistidine antibody (Sigma-Aldrich). Staining for H3K9Me3 (Abcam) and γH2AX (clone JBW301; Millipore) was accomplished using their respective polyclonal or monoclonal antibodies.
Cell Culture Procedures.
HeLa cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS (Atlanta Biologicals) and 100 U/mL penicillin-streptomycin (Gibco/Invitrogen). HeLa DEK-KD cells were cultured as above with the addition of puromycin at a concentration of 1 µg/mL. HAP1 cells were maintained in IMDM supplemented with 10% FBS, 100 U/mL penicillin-streptomycin, and 1.5 µg/mL puromycin. B4GALT7 and SLC35B2 were disrupted in HAP1 cells by retrovirus-mediated insertional mutagenesis. Cells were then reconstituted via lentiviral transduction with either empty vector or functional B4GALT7 or SLC35B2 and selected by puromycin resistance (33). MDMs were isolated from the blood of healthy donors and synovial macrophages derived from patients with JIA as described previously (27, 28). Monocytes were cultured in X-Vivo 15 media with gentamicin (Lonza) supplemented with 25% human serum (Lonza). All cell culture procedures were conducted in a humidified atmosphere at 37 °C and 5% (vol/vol) CO2.
shRNA Procedures.
Stable DEK knockdown in HeLa cells was achieved as described previously (3).
Immunofluorescence Microscopy.
Cells incubated with recombinant His-DEK were fixed with 3.7% (wt/vol) paraformaldehyde in PBS for 10 min at room temperature on poly-L-lysine–treated slides (LabScientific). For membrane visualization, slides were treated with 5 μg/mL WGA (Molecular Probes/Invitrogen) at room temperature for 10 min. Cells were washed in PBS and permeabilized with 0.2% Triton-X 100 for 15 min at room temperature, then washed again with PBS and blocked with 1% BSA in PBS for 30 min. Primary antibodies were diluted in blocking buffer (10% goat serum in PBS) and incubated with cells for 1 h at room temperature. After more washes, cells were incubated with 2 μg/mL goat anti-mouse or anti-rabbit antibody (Molecular Probes/Invitrogen) in blocking buffer for 1 h. Subsequently, cells were again fixed with 3.7% paraformaldehyde for 10 min at room temperature and washed with distilled water. Nuclei were stained with either DAPI (Molecular Probes/Invitrogen) or propidium iodide (Sigma-Aldrich). DEK-positive foci were visualized by laser scanning confocal microscopy (Olympus FV-500).
Cell Fractionation, Nuclease Digestion, and Immunoprecipitation.
Cell fractionations and nuclease digestions were carried out as described by Kappes et al. (3). Cells prepared for immunoprecipitation were washed and incubated in lysis buffer [50 mM Tris (pH 7.5), 150 mM sodium chloride, 1% Nonidet P-40, 0.1% SDS, 0.5% sodium deoxycholate, and complete EDTA-free protease inhibitor mixture] for 15 min on ice. The lysate was centrifuged at 12,000 × g for 20 min at 4 °C. The supernatant was then recovered, and the protein extract was incubated with a polyclonal rabbit anti-His antibody. Immunoprecipitation samples were separated by SDS/PAGE and then immunoblotted.
Cell Extract Preparation and Immunoblot Analysis.
Cells were pelleted and dissolved in 2% (vol/vol) SDS and heated in a boiling water bath for 15 min. Extracts were supplemented with beta-mercaptoethanol and heated for an additional 2 min at 95 °C. Samples were then separated using SDS/PAGE, transferred to PVDF membranes, and immunoblotted.
Transwell Cell Culture System.
Macrophages from the synovial fluids of patients with JIA were isolated as described previously (27) and plated in X-Vivo-15 supplemented with 10% human serum. Nonadherent cells were plated on the top layer of a Transwell cell culture-permeable system with 0.4-µM pores (Corning). Two days later, HeLa DEK-KD cells were plated on the bottom layer of the Transwell system. Cells were incubated for an additional 3 d and then harvested, and cell extracts were prepared for immunofluorescence microscopy and immunoblotting as described above using a specific polyclonal DEK antibody (8).
DNA Damage Susceptibility Assay.
HeLa Puro-C (control) and HeLa DEK-1165 (shDEK) cells were seeded in 12-well plates containing glass coverslips at a density of 5 × 104 cells/mL using medium without puromycin. After 24 h, the medium was supplemented with 1.6 μg/mL His-DEK and incubated for an additional 48 h. For DNA damage induction, NCS was added to a final concentration of 75 ng/mL. Cells were fixed and stained as outlined above. γH2AX-positive foci were visualized by spinning disk confocal microscopy using a Zeiss Cell Observer SD microscope. Stacks consisting of 12 z-slices at 0.46-μm intervals were recorded. Image analysis was performed on mean intensity projections using ImageJ. The mean signal intensity of single nuclei was determined for at least 150 nuclei in each experimental condition. Statistical analysis was performed using an unpaired Student t test.
KNIME Image Analysis and Statistical Analysis.
For analysis of H3K9Me3 intensities in control and DEK-KD cells with or without added His-DEK, cells were treated with His-DEK (1.6 μg/mL medium) or with the corresponding buffer-only control for 48 h. Cells were then washed with PBS, fixed with 3.7% paraformaldehyde for 20 min, and stained for H3K9Me3, as outlined by Kappes et al. (3). To allow for quantitative comparison of H3K9Me3 levels between samples, laser intensities were adjusted to the sample with the highest signal. All samples were scanned (tile scan: 4 × 4 optical fields; Fig. S4) in a timely fashion on a Zeiss LSM 710 confocal laser scanning microscope with identical settings. For subsequent unbiased and fully automated high-throughput image analysis, we used a newly created workflow on the open-source platform KNIME (http://tech.knime.org/community/image-processing; workflow Measure Intensity version of 03.05.2012). The individual steps of this workflow are depicted in Fig. S3. On average, 1,500–2,000 cells were analyzed per sample and tile scan. We used a modified version of this workflow to analyze the DEK-specific signals shown in Fig. 2B.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of our wondrous young colleague and friend Amruta Mundade (“Amruta ki divy atma ke prati hardik shraddhanjali”). We thank Jen Lewis for assisting with manuscript preparation, the staff of the University of Michigan’s Microscopy Core for helping with image production and analysis, and Christian Dietz (University of Konstanz) for developing the intensity measurement workflow using the KNIME high-throughput, open-source image analysis platform. F.K. is supported by a William D. Robinson Fellowship from the Arthritis Foundation/Michigan Chapter and by the START program of the Faculty of Medicine, RWTH Aachen, and is the recipient of an Arthritis Foundation postdoctoral fellowship. A.D. is supported by the German Research Foundation through Research Training Group 1331. N.M.-V. is supported by National Institutes of Health Grants R03 AR-056748-01 and K01 AR-055620, University of Michigan Tumor Immunology Training Program Grant T32-CA-88784-03, University of Michigan Rheumatic Disease Core Center Grant 5-P30-AR-048310-07, and University of Michigan Postdoctoral Translational Scholars Program Grant UL1-RR-024986. Work in E.F.-M.’s laboratory is funded by the Excellence Initiative of the German federal and state governments. Work in D.M.M.’s laboratory is supported by National Institutes of Health Grants R01 AI062248 and R01 AI087128 and a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
2Deceased February 17, 2009.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220751110/-/DCSupplemental.
References
- 1.von Lindern M, Poustka A, Lerach H, Grosveld G. The (6;9) chromosome translocation, associated with a specific subtype of acute nonlymphocytic leukemia, leads to aberrant transcription of a target gene on 9q34. Mol Cell Biol. 1990;10(8):4016–4026. doi: 10.1128/mcb.10.8.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Lindern M, et al. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12(4):1687–1697. doi: 10.1128/mcb.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kappes F, et al. The DEK oncoprotein is a Su(var) that is essential to heterochromatin integrity. Genes Dev. 2011;25(7):673–678. doi: 10.1101/gad.2036411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGarvey T, et al. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J Cell Biol. 2000;150(2):309–320. doi: 10.1083/jcb.150.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soares LM, Zanier K, Mackereth C, Sattler M, Valcárcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312(5782):1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 6.Fu GK, Grosveld G, Markovitz DM. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc Natl Acad Sci USA. 1997;94(5):1811–1815. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams BS, et al. DEK binding to class II MHC Y-box sequences is gene- and allele-specific. Arthritis Res Ther. 2003;5(4):R226–R233. doi: 10.1186/ar774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khodadoust MS, et al. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69(16):6405–6413. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kappes F, et al. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28(10):3245–3257. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kavanaugh GM, et al. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res. 2011;39(17):7465–7476. doi: 10.1093/nar/gkr454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexiadis V, et al. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 2000;14(11):1308–1312. [PMC free article] [PubMed] [Google Scholar]
- 12.Broxmeyer HE, et al. DEK regulates hematopoietic stem engraftment and progenitor cell proliferation. Stem Cells Dev. 2011;21(9):1449–1454. doi: 10.1089/scd.2011.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koleva RI, et al. C/EBPalpha and DEK coordinately regulate myeloid differentiation. Blood. 2012;119(21):4878–4888. doi: 10.1182/blood-2011-10-383083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wise-Draper TM, et al. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26(20):7506–7519. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wise-Draper TM, et al. The human DEK proto-oncogene is a senescence inhibitor and an up-regulated target of high-risk human papillomavirus E7. J Virol. 2005;79(22):14309–14317. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung TH, et al. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482(7386):524–528. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kondoh N, et al. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59(19):4990–4996. [PubMed] [Google Scholar]
- 18.Kroes RA, et al. The identification of novel therapeutic targets for the treatment of malignant brain tumors. Cancer Lett. 2000;156(2):191–198. doi: 10.1016/s0304-3835(00)00462-6. [DOI] [PubMed] [Google Scholar]
- 19.Kappes F, et al. DEK expression in melanocytic lesions. Hum Pathol. 2011;42(7):932–938. doi: 10.1016/j.humpath.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans AJ, et al. Defining a 0.5-mb region of genomic gain on chromosome 6p22 in bladder cancer by quantitative-multiplex polymerase chain reaction. Am J Pathol. 2004;164(1):285–293. doi: 10.1016/S0002-9440(10)63118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grasemann C, et al. Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene. 2005;24(42):6441–6449. doi: 10.1038/sj.onc.1208792. [DOI] [PubMed] [Google Scholar]
- 22.Orlic M, Spencer CE, Wang L, Gallie BL. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45(1):72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- 23.Privette Vinnedge LM, et al. The human DEK oncogene stimulates β-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene. 2011;30(24):2741–2752. doi: 10.1038/onc.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daibata M, et al. Differential gene-expression profiling in the leukemia cell lines derived from indolent and aggressive phases of CD56+ T-cell large granular lymphocyte leukemia. Int J Cancer. 2004;108(6):845–851. doi: 10.1002/ijc.11647. [DOI] [PubMed] [Google Scholar]
- 25.Babaei-Jadidi R, et al. FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J Exp Med. 2011;208(2):295–312. doi: 10.1084/jem.20100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong X, et al. Autoantibodies to DEK oncoprotein in a patient with systemic lupus erythematosus and sarcoidosis. Arthritis Rheum. 1998;41(8):1505–1510. doi: 10.1002/1529-0131(199808)41:8<1505::AID-ART23>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 27.Mor-Vaknin N, et al. DEK in the synovium of patients with juvenile idiopathic arthritis: Characterization of DEK antibodies and posttranslational modification of the DEK autoantigen. Arthritis Rheum. 2011;63(2):556–567. doi: 10.1002/art.30138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mor-Vaknin N, et al. The DEK nuclear autoantigen is a secreted chemotactic factor. Mol Cell Biol. 2006;26(24):9484–9496. doi: 10.1128/MCB.01030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang D, et al. High mobility group box-1 protein induces the migration and activation of human dendritic cells and acts as an alarmin. J Leukoc Biol. 2007;81(1):59–66. doi: 10.1189/jlb.0306180. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, et al. Cellular uptake of exogenous human PDCD5 protein. J Biol Chem. 2006;281(34):24803–24817. doi: 10.1074/jbc.M600183200. [DOI] [PubMed] [Google Scholar]
- 31.Console S, Marty C, García-Echeverría C, Schwendener R, Ballmer-Hofer K. Antennapedia and HIV transactivator of transcription (TAT) “protein transduction domains” promote endocytosis of high molecular weight cargo upon binding to cell surface glycosaminoglycans. J Biol Chem. 2003;278(37):35109–35114. doi: 10.1074/jbc.M301726200. [DOI] [PubMed] [Google Scholar]
- 32.Cronican JJ, et al. A class of human proteins that deliver functional proteins into mammalian cells in vitro and in vivo. Chem Biol. 2011;18(7):833–838. doi: 10.1016/j.chembiol.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosmarin DM, et al. Attachment of Chlamydia trachomatis L2 to host cells requires sulfation. Proc Natl Acad Sci USA. 2012;109(25):10059–10064. doi: 10.1073/pnas.1120244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477(7364):340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kappes F, Burger K, Baack M, Fackelmayer FO, Gruss C. Subcellular localization of the human proto-oncogene protein DEK. J Biol Chem. 2001;276(28):26317–26323. doi: 10.1074/jbc.M100162200. [DOI] [PubMed] [Google Scholar]
- 36.Preisach C, Burkhardt H, Decker R, Schmidt-Thieme L. Proceedings of the 31st Annual Conference of the Gesellschaft für Klassifikation e.V., Albert-Ludwigs-Universität Freiburg, March 7–9, 2007. Berlin: Springer; 2008. Data analysis, machine learning and applications. [Google Scholar]
- 37.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.