Abstract
Wild-living chimpanzees and gorillas harbor a multitude of Plasmodium species, including six of the subgenus Laverania, one of which served as the progenitor of Plasmodium falciparum. Despite the magnitude of this reservoir, it is unknown whether apes represent a source of human infections. Here, we used Plasmodium species-specific PCR, single-genome amplification, and 454 sequencing to screen humans from remote areas of southern Cameroon for ape Laverania infections. Among 1,402 blood samples, we found 1,000 to be Plasmodium mitochondrial DNA (mtDNA) positive, all of which contained human parasites as determined by sequencing and/or restriction enzyme digestion. To exclude low-abundance infections, we subjected 514 of these samples to 454 sequencing, targeting a region of the mtDNA genome that distinguishes ape from human Laverania species. Using algorithms specifically developed to differentiate rare Plasmodium variants from 454-sequencing error, we identified single and mixed-species infections with P. falciparum, Plasmodium malariae, and/or Plasmodium ovale. However, none of the human samples contained ape Laverania parasites, including the gorilla precursor of P. falciparum. To characterize further the diversity of P. falciparum in Cameroon, we used single-genome amplification to amplify 3.4-kb mtDNA fragments from 229 infected humans. Phylogenetic analysis identified 62 new variants, all of which clustered with extant P. falciparum, providing further evidence that P. falciparum emerged following a single gorilla-to-human transmission. Thus, unlike Plasmodium knowlesi-infected macaques in southeast Asia, African apes harboring Laverania parasites do not seem to serve as a recurrent source of human malaria, a finding of import to ongoing control and eradication measures.
Keywords: diagnostic Laverania PCR, great apes, nextgen sequencing, Plasmodium coinfections, Plasmodium diversity
Malaria is one of the most devastating infectious diseases of humans worldwide, with hundreds of millions of cases of clinical illness and over 650,000 deaths occurring annually (1). Given this enormous health burden, efforts to control and potentially eradicate this disease have become an urgent public health priority (2, 3). Effective control and elimination measures require a clear understanding of parasite, vector, human, and environmental factors that sustain malaria transmission. This includes a systematic evaluation of potential zoonotic reservoirs and the risk that they may pose for humans. Recently, close genetic relatives of the human malaria parasites Plasmodium falciparum, Plasmodium ovale, Plasmodium malariae, and Plasmodium vivax have been identified in wild-living apes in sub-Saharan Africa (4–8). These parasites were tentatively classified on the basis of their sequence relationships into a number of different species, six of which were closely related to human P. falciparum and placed into a separate Plasmodium subgenus, termed Laverania (4, 7, 9, 10). Of these six Laverania species, Plasmodium reichenowi, Plasmodium gaboni, and Plasmodium billcollinsi were identified only in chimpanzees, whereas Plasmodium adleri, Plasmodium blacklocki, and Plasmodium praefalciparum were only found in gorillas. Moreover, P. praefalciparum was shown to be the immediate precursor of human P. falciparum (4). Because candidate Anopheles vectors have been identified that may transmit both ape and human parasites (11), the fact that a large fraction of wild-living apes is endemically Plasmodium infected has raised concerns that they might represent a source of recurring human infections (4, 5, 9, 12, 13).
In this study, we tested humans who live in remote rural areas of southern Cameroon for ape Plasmodium zoonoses. We specifically screened for Laverania infections, because these are the most abundant and widespread in resident ape populations, and because one of them, P. praefalciparum, has crossed the species barrier from gorillas to humans already once (4). Moreover, ape Laverania parasites have been studied extensively at the molecular level, with numerous mitochondrial, apicoplast, and nuclear sequences available for analyses. To detect zoonotic infections, we (i) developed a Plasmodium species-specific diagnostic PCR, (ii) used ultra-deep sequencing to search for low-abundance ape parasites in mixed Plasmodium species infections, and (iii) used single-genome amplification (SGA) to characterize the genetic diversity of human P. falciparum in southern Cameroon. Our study systematically searched for Plasmodium zoonoses in west central Africa, thus providing insight into the host range of human and great ape parasites.
Results
Genetic Analysis of Human Plasmodium Infections in Rural Cameroon.
Cameroon is an area of high malaria endemicity, with nearly 100% of clinical cases believed to be caused by P. falciparum (1). However, few of these infections have been molecularly characterized and the extent of parasite diversity, both at the intraspecies and interspecies level, is largely unknown. Studying the epidemiology and natural history of HIV type 1 (HIV-1) infections in Cameroon, we previously collected large numbers of buffy coat samples, which represent thin layers of leukocytes on the surface of sedimented erythrocytes (14). These samples also contain Plasmodium DNA, because parasite-infected red blood cells concentrate immediately below the buffy coat layer and are thus harvested together with the leukocytes (15). To characterize the Plasmodium species that commonly infect humans in rural Cameroon, we selected samples from 318 inhabitants of seven remote villages (Fig. 1 and SI Appendix, Table S1). These study sites were chosen because of the high Laverania prevalence rates in chimpanzee and gorilla populations in adjacent forest regions (Fig. 1A). All sampled individuals lived in close proximity to ape habitats (Fig. 1B) and included forest dwellers, hunters, members of local pygmy tribes, and individuals who worked at logging concessions. Because many of these subjects spent numerous hours and days in the forest, we reasoned that at least some of them were exposed to ape Plasmodium-infected Anopheles mosquitoes. To examine whether such exposures had resulted in parasite transmission, we screened buffy coat DNA for ape parasites by diagnostic PCR. Using primers previously shown to amplify ape Laverania parasites with high sensitivity and specificity (4), we targeted a 939-bp region spanning most of the cytochrome b (cytb) gene of the Plasmodium mitochondrial DNA (mtDNA) genome (Fig. 2). This analysis identified 194 of the 318 blood samples to be PCR positive (61%), all of which contained human parasites as determined by direct sequencing: 181 samples contained P. falciparum, 12 samples contained P. ovale, and 1 sample contained P. malariae as the predominant Plasmodium species (SI Appendix, Table S1). From this experiment, we concluded that zoonotic Laverania infections, if they indeed occurred, were rare and unlikely to represent monoinfections.
Fig. 1.
Screening of humans in rural Cameroon for zoonotic Plasmodium infections. (A) Location of human study sites (red stars). Eight rural villages were selected for molecular epidemiological studies because of their proximity to wild-living chimpanzee (yellow circles) and gorilla (yellow hexagons) populations known to harbor Laverania infections at high prevalence rates. Previously estimated infection rates (4) are shown for the most proximal field sites (denoted by a two-letter code). Country borders, major rivers, and the capital city of Yaoundé (red triangle) are also shown. A red star with asterisk highlights the location of five closely spaced villages (Mboumo, Eboumetoum, Aviation, Nkonzu, and Kompia). (B) View of one rural village, depicting the close proximity of human residences and sleeping quarters to the surrounding forest.
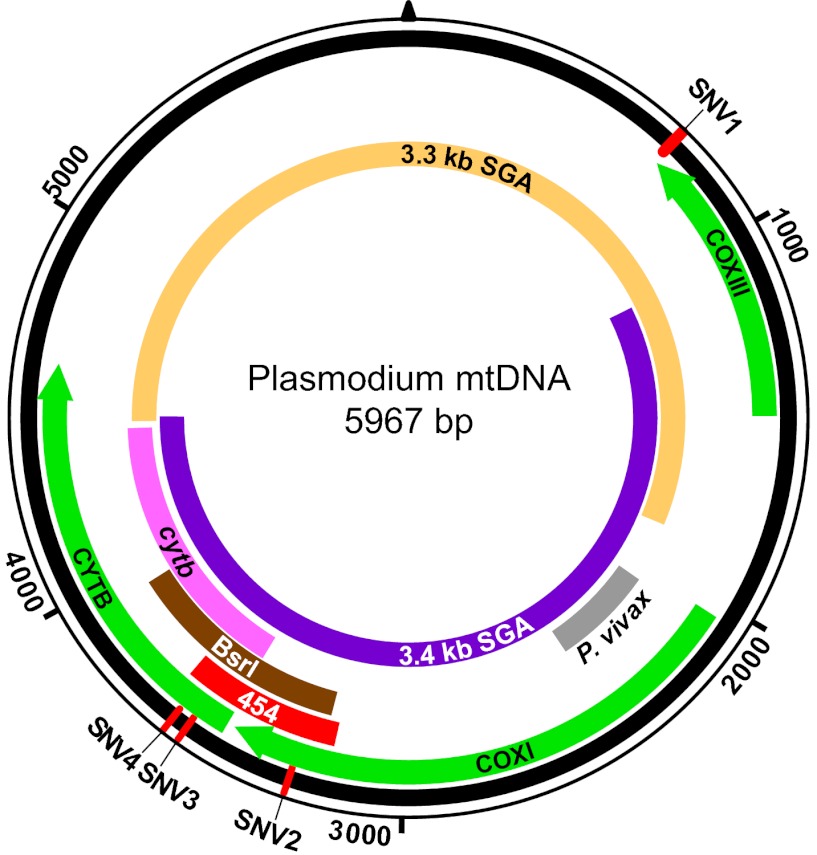
Fig. 2.
Schematic representation of the Plasmodium mitochondrial genome. DNA fragments amplified for diagnostic purposes (cytb, BsrI, P. vivax), 454 sequencing (454), and SGA (mtDNA-3.3 kb; mtDNA-3.4 kb) are shown in relation to cytochrome b (cytb), cytochrome c oxidase subunit I (coxI), and cytochrome c oxidase subunit III (coxIII) coding regions, respectively. The positions of four SNVs (SNV1–SNV4), which distinguish human P. falciparum from ape Laverania parasites, are shown in red.
Diagnostic PCR Capable of Differentiating Human and Ape Laverania Species.
To screen a larger number of individuals, we developed a Plasmodium species-specific PCR. Aligning several hundred ape and human Plasmodium mitochondrial genomes, we had previously noted four single-nucleotide variants (SNVs) that distinguished all known P. falciparum strains from the six ape Laverania species (4). One of these (SNV4) comprised a BsrI restriction enzyme site (ACTGGN) that was present in 134 of 135 ape Laverania sequences, but was absent from all of 859 human Plasmodium sequences in the database (Fig. 2). To determine whether PCR amplification followed by BsrI cleavage could be used to screen human blood samples for ape Laverania infections, we designed primers for a ∼700-bp DNA fragment that spanned the diagnostic SNV4 site (Figs. 2 and 3). Testing these primers on fecal samples from Plasmodium-infected apes, we obtained PCR products that were cleaved by BsrI and yielded the expected fragments for the respective ape Plasmodium species (Fig. 3 A and B). In contrast, amplicons from human P. falciparum, P. malariae, and P. vivax reference strains were not cleaved by BsrI, and although P. ovale amplicons were cleaved once, the resulting fragments were readily distinguishable from those of the ape parasites. BsrI cleavage products were also visible in mixtures of human and ape parasite DNAs, including in preparations that contained P. falciparum at a 10-fold excess. These data indicated that PCR amplification, followed by BsrI cleavage, represented a viable screening approach for zoonotic Laverania infections, even when ape and human parasites were present in mixed-species infections.
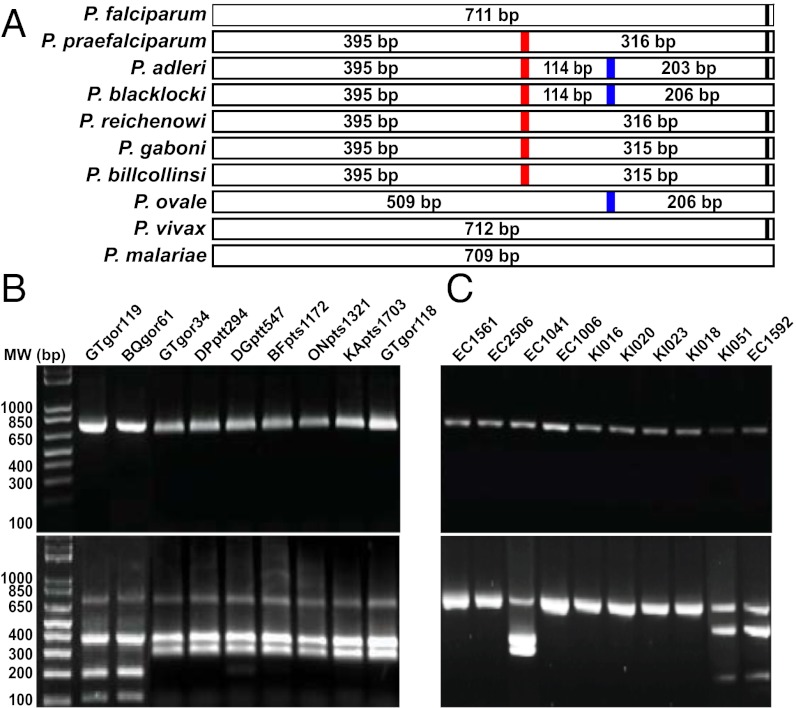
Fig. 3.
A Plasmodium species-specific PCR assay capable of differentiating human and ape Laverania parasites. (A) Predicted BsrI cleavage products for different Plasmodium species infecting humans and apes. A red vertical line highlights a BsrI site unique to all ape Laverania parasites. A second BsrI site found only in P. adleri, P. blacklocki, and P. ovale is highlighted in blue. (B) Diagnostic PCR of ape Laverania infections. Laverania-positive ape fecal samples were PCR positive (Upper) and yielded appropriately sized fragments upon BsrI cleavage (Lower). (C) Diagnostic PCR of human Plasmodium infections. Plasmodium-positive human samples yielded amplicons of predicted size (Upper), with BsrI cleavage products observed for three (Lower).
Using this Plasmodium species-specific PCR assay, we screened 1,165 buffy coat samples from villagers native to southeastern Cameroon (Fig. 1 and SI Appendix, Fig. S1). For control, we also analyzed 85 samples from HIV-1–infected individuals in the capital city Yaoundé. Testing a total of 1,250 samples, we amplified BsrI-specific fragments from 872 of them (SI Appendix, Table S1), three of which were cleaved by BsrI (Fig. 3C). Two of these samples (KI051 and EC1592) yielded PCR cleavage products consistent with P. ovale infection, which was confirmed by sequence analysis. The third sample (EC1041), from a child in Mboumo, yielded the ape-specific PCR cleavage products of 395 and 316 bp, respectively (Fig. 3C), suggestive of ape Laverania infection. However, sequence analysis of the BsrI fragment failed to confirm this diagnosis, identifying instead a P. falciparum variant that exhibited a single point mutation at the SNV4 site. This was confirmed by sequencing the entire mitochondrial genome of this variant (amplified as two partially overlapping 3.4- and 3.3-kb fragments; Fig. 2), which contained the SNV4 point mutation, but lacked additional ape Plasmodium-specific signatures. Thus, the BsrI diagnostic PCR had uncovered a rare P. falciparum variant whose mitochondrial sequence was identical to that of other P. falciparum strains, except for a single (ape-like) back mutation at the SNV4 site.
Molecular Characterization of Human Plasmodium Infections by 454 Deep Sequencing.
The cytb and BsrI PCR results indicated that the vast majority of villagers in rural Cameroon harbored human Plasmodium parasites, with P. falciparum representing the predominant species. We thus reasoned that ape Laverania parasites—if they were transmitted to humans—would likely replicate less efficiently and represent only a minor fraction of the total parasite burden within an infected individual. To increase the likelihood of detecting such variants, we selected an ultra–deep-sequencing approach, which generates tens of thousands of sequences of the same genetic locus and can thus detect low-abundance variants with great sensitivity (16–18). Specifically, we used the 454 GS FLX Titanium chemistry to sequence a 405-bp fragment of the Plasmodium mtDNA genome that included three of the four diagnostic SNVs (Fig. 2) and thus differentiated even the closest human and ape parasites (SI Appendix, Fig. S2).
To explore the utility of the 454-sequencing approach, we initially pooled amplicons from 77 Plasmodium positive human buffy coat samples, which yielded 465,391 high-quality reads. Each read was classified by determining its minimum edit distance to a large set of Plasmodium reference sequences (SI Appendix, Table S2). This approach identified 458,676 reads (98.56%) to represent P. falciparum, 76 reads (0.02%) to represent P. ovale, and 6,266 reads (1.35%) to represent P. malariae (SI Appendix, Fig. S3 A–C), the classification of which was confirmed by phylogenetic analyses (SI Appendix, Fig. S4). One single read was classified as P. praefalciparum (SI Appendix, Fig. S3D); however, closer inspection of its sequence revealed multiple indels that caused inactivating frameshift mutations as well as a substitution that was not found in any other P. praefalciparum strain. Moreover, this read differed from the closest P. praefalciparum reference by five mutations, but from the closest P. falciparum reference by six mutations, and contained only two of the three ape-specific SNVs. We thus concluded that this read was erroneously classified as P. praefalciparum due to PCR and/or 454 process errors, and that there was no evidence of ape Laverania infection in any of the 77 sequenced individuals (SI Appendix, Results and Analysis).
Identification of Plasmodium Multispecies Infections by 454 Deep Sequencing.
To extend the search for zoonotic Laverania infections, we selected an additional 437 samples for 454 sequencing (SI Appendix, Fig. S1) but improved the methodology. First, we inserted a 12-mer barcode into the sequencing primer to permit the computational sorting of individual samples (19). Second, we reversed the sequencing direction to increase the number of reads that covered at least two diagnostic SNVs (SI Appendix, Fig. S2). Third, we amplified samples using the lowest possible number of cycles to reduce PCR-introduced errors (SI Appendix, Table S3). Finally, we included cloned fragments (3.4 kb) of the P. falciparum, P. malariae, and P. ovale mitochondrial genome as controls, which allowed us to perform a formal error calculation for each pyrosequencing run (SI Appendix, Results and Analysis). The resulting pyrosequencing reads were sorted by sample and analyzed.
The identification of rare ape Plasmodium parasites necessitated a method that could differentiate true sequence changes from 454-sequencing error. We thus used a maximum-likelihood based approach to determine which and how many different Plasmodium species were present in each barcoded human sample (SI Appendix, Results and Analysis). For each sample, we generated pairwise alignments of all reads with all Plasmodium reference sequences and then used a model of empirically determined 454-sequencing error (SI Appendix, Table S4) to calculate the probability that a read was derived from a particular reference. Using this approach, we determined the Plasmodium species composition in all barcoded human samples. Of 437 samples, 349 contained only P. falciparum, one contained only P. malariae, and 4 contained only P. ovale sequences (Table 1). A further 61 samples contained both P. falciparum and P. malariae, 13 samples contained both P. falciparum and P. ovale, and 9 samples contained all three species (Table 1). Importantly, none of the human blood samples contained any of the six ape Laverania species, including P. praefalciparum. Moreover, none of the samples contained P. vivax sequences.
Table 1.
Species composition of human Plasmodium infections in Cameroon as determined by 454 sequencing
| Identified species* | No. of samples |
| P. falciparum | 349 |
| P. falciparum and P. malariae | 61 |
| P. falciparum and P. ovale | 13 |
| P. falciparum, P. malariae, and P. ovale | 9 |
| P. ovale | 4 |
| P. malariae | 1 |
| Total | 437 |
See SI Appendix, Fig. S6, for a breakdown of the parasite composition of individual samples.
To be certain that our inability to find ape Plasmodium zoonoses was not due to technical limitations, we used the identical 454 methodology to deep sequence Plasmodium parasites from fecal samples of infected apes. Analysis of 37,644 filtered reads from two western lowland gorillas (Gorilla gorilla gorilla), three central chimpanzees (Pan troglodytes troglodytes), and one eastern chimpanzee (Pan troglodytes schweinfurthii) confirmed the presence of all six ape Laverania species as well as P. vivax-like parasites (SI Appendix, Fig. S5). We also characterized the proportion of humans who harbored multiple P. falciparum variants (SI Appendix, Results and Analysis). Multiple variant infections were detected in 10% of all subjects, with a maximum of four variants per person (SI Appendix, Table S5 and Fig. S6). Importantly, minor variants could be identified at levels as low as 0.006% of the total parasite burden, thus providing direct evidence that our deep-sequencing approach was capable of identifying very low-abundance Plasmodium variants.
Genetic Diversity of P. falciparum in Rural Cameroon.
Although there are over a hundred near–full-length P. falciparum mitochondrial DNA sequences in the database, little is known about the extent of genetic diversity of this parasite in central Africa. In particular, there are no molecularly characterized human strains from areas where wild-living apes are endemically infected with Laverania parasites. To characterize the P. falciparum variants prevalent in rural Cameroon, we selected Plasmodium-positive samples from seven different locations (SI Appendix, Table S1) and subjected them to SGA, targeting the region of the mitochondrial genome (3.4 kb) known to exhibit the greatest diversity between ape and human Laverania lineages (Fig. 2). We selected SGA rather than conventional PCR, because this method eliminates Taq polymerase-induced recombination as well as nucleotide misincorporations in finished sequences, and thus ensures an accurate representation of parasite variants as they exist in vivo (4, 20). Sequencing between one and eight SGA amplicons per sample, we generated a total of 684 half-genome mtDNA sequences. Phylogenetic analyses revealed that these represented 69 unique P. falciparum haplotypes, 62 of which had not previously been reported. Despite this diversity, all variants grouped with previously identified P. falciparum sequences, forming a single well-supported clade within the radiation of P. praefalciparum from gorilla (Fig. 4). This was the case even after inclusion of a P. falciparum variant (EC1041; Fig. 3) that contained one of the three ape-specific SNVs at the BsrI cleavage site (Fig. 4). These results failed to uncover additional cross-species transmissions, including human-to-ape transfers, and thus confirmed that extant P. falciparum emerged in humans following a single introduction of a gorilla parasite.
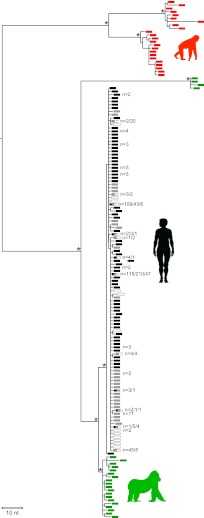
Fig. 4.
Phylogeny of P. falciparum strains from rural Cameroon. Newly derived P. falciparum sequences from humans living in Cameroon (black) are shown in relation to P. falciparum sequences from GenBank (white) and the Sanger Institute (gray), as well as to P. praefalciparum and P. reichenowi sequences from gorillas (green) and chimpanzees (red), respectively. The tree includes 684 new SGA-derived 3.4-kb mitochondrial sequences from 229 infected humans, including one that contained a back mutation at the ape-specific SNV4 site (highlighted by arrow; Fig. 3C). Numbers at tips indicate the number of times that a sequence was found in Cameroon (black), the Sanger dataset (gray), or GenBank (white; for sequences present in multiple datasets, numbers are listed in sequence). The tree was inferred using maximum-likelihood methods. The asterisks indicate posterior probabilities above 0.9. (The scale bar represents 10-nt substitutions.)
Absence of P. vivax in Humans from Southern Cameroon.
Although the great majority of individuals in Cameroon are Duffy negative (21), it has been proposed that P. vivax persists in west central human populations at a very low frequency (22). Because deep sequencing failed to identify evidence of P. vivax infection in 514 individuals, we considered the possibility that the 454 primers were less efficient in amplifying P. vivax compared with the other Plasmodium species. We thus designed P. vivax-specific primers in the mtDNA cox1 gene (Fig. 2) and used these to screen an additional 558 human samples (SI Appendix, Fig. S1). Although 47 samples yielded a visible amplification product (SI Appendix, Table S6), none of these represented P. vivax or P. vivax-like infections as determined by sequence analysis of the corresponding amplicon. Instead, 37 of the PCR-positive samples contained P. malariae, whereas the remaining 10 contained P. ovale. Moreover, sequence analysis of the Duffy promoter region from 90 human samples confirmed a Duffy-negative phenotype in all of them (SI Appendix, Table S6). Thus, using both conventional PCR and 454-sequencing approaches, we found no evidence of P. vivax infections in individuals living in rural Cameroon.
Discussion
Chimpanzees and gorillas harbor at least 10 different Plasmodium species, including six of the subgenus Laverania that are closely related to P. falciparum (4–8). The discovery of this previously unrecognized reservoir has prompted concerns that wild-living apes might constitute a source of recurrent human infection (4, 5, 9, 11–13). In this study, we set out to examine this possibility for several reasons. First, the ape reservoir is substantial, both in terms of geographic distribution and complexity of Plasmodium species. Second, both western gorillas and chimpanzees are infected at high endemicity throughout their habitat. Third, Plasmodium zoonoses can have significant public health impact. A case in point is Plasmodium knowlesi, a macaque parasite that has been shown to cause hundreds of cases of human malaria every year (23–25). Finally, Plasmodium zoonoses have been misdiagnosed in the past: P. knowlesi was initially mistaken for P. malariae, ultimately requiring the development of molecular tools to facilitate its detection (24). Given that malaria infections in central Africa are rarely genetically characterized, we considered the possibility that ape Plasmodium zoonoses might have also been overlooked. To test this, we developed diagnostic PCR assays and next-generation sequencing approaches that permitted the detection of rare Plasmodium variants, even when they occurred in the context of mixed-species infection with P. falciparum, which is known to reach very high blood titers. Using these approaches to test 1,400 blood samples from individuals native to rural Cameroon, we failed to detect previously unknown human Plasmodium infections (SI Appendix, Table S1). There was no evidence for zoonotic infection with any of the six ape-specific Laverania species or non-Laverania parasites identified only in wild apes (4). Instead, we detected P. falciparum, P. ovale, and P. malariae in a large fraction of individuals, both as monospecies and mixed-species infections (Table 1 and SI Appendix, Fig. S6). From these data, we conclude that ape Laverania zoonoses can be ruled out as an ongoing threat to public health in this region of west central Africa.
Although we failed to find ape Laverania infections in humans, our data cannot exclude the possibility of very rare transmission events (4). Depending on the host, parasite, and/or vector properties required for successful transmission, a much larger number of individuals from geographically more diverse regions will have to be screened to exclude rare spillovers. Because the Anopheles species that transmit Plasmodium parasites among wild apes are only beginning to be characterized (11), it is conceivable that the ecology, distribution, and feeding preferences of these vectors play a much greater role in determining the likelihood of zoonotic transmission than the mere geographic proximity of human habitations to infected ape populations. Nonetheless, it seems unlikely that the absence of zoonotic Laverania infections in rural Cameroon is solely due to a lack of human exposure. This is because even among endemically infected chimpanzees and gorillas, there is no evidence that Laverania parasites cross between the two ape species (4). This remarkable host specificity suggests a restriction at the parasite–host interface, which is supported by comparisons of P. falciparum and P. reichenowi gene sequences. It is known that the genes involved in erythrocyte invasion are evolving rapidly between Laverania parasites (26). Moreover, erythrocyte invasion of P. falciparum is absolutely dependent on the interaction of its PfRh5 ligand with the human Ok blood group antigen basigin (27). Human, chimpanzee, and gorilla homologs of basigin are highly divergent, suggesting that ape Laverania species have to overcome significant adaptive hurdles before they can spread efficiently in a different host.
Given these restrictions, the question arises how the gorilla precursor of P. falciparum managed to colonize humans. One possibility is that P. praefalciparum underwent a very specific mutation in a host-compatibility factor that changed its host preference from gorilla to human. Because P. falciparum emerged only once (Fig. 4), this mutation must have been difficult to generate and/or must have arisen under exceedingly favorable transmission conditions. Another possibility is that the immediate precursor of P. falciparum was the product of a rare recombination event. Regardless of the circumstances, it seems clear that the generation of an ape Laverania strain that is capable of spreading in humans is an extremely rare event, which may explain why we failed to detect such variants.
In addition to Laverania species, wild-living chimpanzees and gorillas also harbor P. vivax, P. malariae, and P. ovale-like infections. Because very few of these ape parasites have been molecularly characterized, it remains unknown whether they represent members of the same or different Plasmodium species as their human counterparts. Transmission studies conducted nearly a hundred years ago demonstrated that non-Laverania species cross between apes and humans more readily than Laverania species (28–32). Two species, Plasmodium schwetzi and Plasmodium rodhaini, have been experimentally transmitted to humans in the past (29, 30, 32). Because neither species has been molecularly characterized, it is unknown whether they represent the P. vivax, P. malariae, and/or P. ovale-like infections that have more recently been identified in wild apes. It will thus be important to characterize more of these ape parasites to understand their evolutionary history and characterize their zoonotic potential.
The genetic diversity of P. falciparum in central Africa is largely unknown because only very few strains from this geographic region have been molecularly characterized. It has thus been argued that this lack of sequence information has confounded previous evolutionary analyses and led to erroneous conclusions concerning the origin of P. falciparum (33). In particular, it has been proposed that P. praefalciparum is more likely the result of a human-to-gorilla transmissions of P. falciparum than the other way around (33). To examine this possibility, we amplified mitochondrial half-genome sequences from 229 P. falciparum-positive samples collected in remote rural areas of Cameroon. We found that P. falciparum strains from rural Cameroon were indeed genetically more diverse than previously appreciated. Analyzing 684 single template-derived sequences, we identified 69 unique P. falciparum variants, 62 of which had not previously been reported. However, none of these variants changed previous conclusions concerning the evolutionary history of P. falciparum. Phylogenetic analysis revealed that all newly characterized variants grouped with previously reported P. falciparum strains, forming a well-supported clade within the P. praefalciparum radiation (Fig. 4). An additional 354 sequences of cosmopolitan P. falciparum strains from the Sanger Institute supported this conclusion (34), indicating that all human P. falciparum sequences coalesced to a single common ancestor. Thus, the addition of over 1,000 new P. falciparum sequences, including over 600 from individuals living near wild-ape populations, confirmed that P. falciparum is of gorilla origin and emerged in humans following a single cross-species transmission event (4).
P. vivax is extremely rare in humans in west and central Africa due to the near fixation of the Duffy-negative phenotype, which confers resistance to this parasite (21). Nonetheless, P. vivax seems to be able to cause clinical infection of Duffy-negative individuals in Madagascar (35) and P. vivax-specific antibodies have been reported in 13% of humans living in Pointe-Noire, a city in the Republic of Congo (36). To examine whether P. vivax is maintained in west central African human populations, we screened nearly 700 blood samples from Cameroonian villagers for P. vivax mitochondrial sequences (SI Appendix, Fig. S1). Using both PCR and 454-sequencing approaches, we failed to identify P. vivax infection in inhabitants from seven different rural villages. Finally, all human samples tested were Duffy negative, suggesting that the fraction of Duffy-positive individuals in rural areas of west central Africa is exceedingly low. These data differ from results of others who have reported the presence of P. vivax in Equatorial Guinea and Angola. In these studies, the P. vivax-infected individuals were either Duffy positive (37) or diagnosed solely based on P. vivax-specific PCR products without sequence verification (38). Given the frequency of off-target amplification even with P. vivax-specific primers (SI Appendix, Table S6), any P. vivax diagnosis in central Africa should be confirmed by sequence analysis. Until this is done, the presence of P. vivax in rural west central Africa remains questionable.
Methods
Sample Collections.
Human buffy coat samples (n = 1,402) were selected from anonymized sample collections previously obtained for molecular epidemiological studies of HIV-1 in Cameroon (14). Fecal samples from wild-living apes known to contain Laverania and non-Laverania parasites served as positive controls (4). The study was reviewed and approved by the Institutional Review Board of the University of Pennsylvania.
Detection of Plasmodium Infections.
Human buffy coat samples were screened for Plasmodium sequences using a diagnostic (cytb) PCR (4) followed by direct amplicon sequencing. Samples were also screened using a newly developed Plasmodium species-specific (BsrI) PCR assay (SI Appendix, Materials and Methods, and Fig. S1).
Pyrosequencing.
A 405-bp fragment of the mitochondrial genome spanning three SNVs unique to ape Laverania parasites (Fig. 2) was amplified and sequenced on a Genome Sequencer FLX Titanium Series (Roche) (SI Appendix, Materials and Methods).
SGA and Sequencing.
To derive Plasmodium mitochondrial sequences devoid of PCR-induced substitutions and/or recombination, a 3.4-kb fragment was amplified and sequenced from 229 cytb PCR-positive human samples as described (4). DNA was endpoint diluted such that single templates were amplified, thus excluding PCR-induced substitution and recombination errors (SI Appendix, Materials and Methods).
Phylogenetic Analyses.
SGA-derived 3.4-kb mitochondrial sequences were aligned with human and simian reference sequences. Trees were inferred using maximum-likelihood and Bayesian methods (SI Appendix, Materials and Methods).
Detection of P. vivax Infection.
Human samples were screened for P. vivax infections by nested PCR. Primers were specifically designed to avoid off-target amplification of P. falciparum or other Laverania species, and were shown to amplify ape P. vivax-like parasites as well as human P. vivax with high sensitivity and specificity (SI Appendix, Materials and Methods).
Duffy Phenotype.
Buffy coat DNA samples were used to amplify the Duffy promoter region by nested PCR analysis. The Duffy phenotype was determined by direct amplicon sequencing (SI Appendix, Materials and Methods).
Supplementary Material
Acknowledgments
We thank Avelin Aghokeng, Eugenie Etam Ebong, Arrah Atem Tamba, Celine Montavon, Julius Chia, Nathalie Nkue, Mireille Mpoudi, Géraldine Manirakiza, Marie Bourgeois, Audrey Gleize, Justin Wadi, and Anke Bourgeois for field work and logistics in Cameroon; the Ministry of Public Health (Cameroon) for authorizations and logistical support; Dan Soppet and Claudia Stewart in the Laboratory of Molecular Technology at the Frederick National Laboratory for Cancer Research for 454 sequencing; Aubrey Bailey and Nirav Malani for maintenance and organization of the 454-sequencing datasets; Rohini Sinha for contributing various deep-sequencing analysis tools; Xiaowen Zhang for assistance with laboratory techniques; Patricia Crystal for artwork and manuscript preparation; and Christian Hoffman, Robert W. Doms, Dustin C. Brisson, David S. Roos, and Sarah A. Tishkoff for helpful discussions. This work was supported by grants from National Institutes of Health (R37 AI050529, R01 AI091595), the National Cancer Institute under Contract HHSN261200800001E, the Agence Nationale de Recherche (Programme Blanc, Sciences de la Vie, de la Santé et des Ecosystèmes and ANR 11 BSV3 021 01), the Wellcome Trust (098051), the University of Pennsylvania Centers for AIDS Research (P30 045008), and the Penn Genome Frontiers Institute through a grant from the Pennsylvania Department of Health. S.A.S. was supported by National Institutes of Health Training Grant T32 AI007532.
Footnotes
The authors declare no conflict of interest.
Data deposition: All newly derived Plasmodium sequences have been deposited in the GenBank database (accession nos. KC175306–KC175322 and KC203521–KC203587). The 454 pyrosequencing read data have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA), www.ncbi.nlm.nih.gov/sra (accession no. SRP019191).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305201110/-/DCSupplemental.
References
- 1.World Health Organization 2011. World Malaria Report 2011 (World Health Organization, Geneva). Available at www.who.int/entity/malaria/world_malaria_report_2011/9789241564403_eng.pdf. Accessed November 11, 2012.
- 2.Alonso PL, et al. A research agenda to underpin malaria eradication. PLoS Med. 2011;8(1):e1000406. doi: 10.1371/journal.pmed.1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. malERA Consultative Group on Basic Science and Enabling Technologies (2011) A research agenda for malaria eradication: Basic science and enabling technologies. PLoS Med 8(1):e1000399. [DOI] [PMC free article] [PubMed]
- 4.Liu W, et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature. 2010;467(7314):420–425. doi: 10.1038/nature09442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prugnolle F, et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci USA. 2010;107(4):1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaiser M, et al. Wild chimpanzees infected with 5 Plasmodium species. Emerg Infect Dis. 2010;16(12):1956–1959. doi: 10.3201/eid1612.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krief S, et al. On the diversity of malaria parasites in African apes and the origin of Plasmodium falciparum from Bonobos. PLoS Pathog. 2010;6(2):e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rich SM, et al. The origin of malignant malaria. Proc Natl Acad Sci USA. 2009;106(35):14902–14907. doi: 10.1073/pnas.0907740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rayner JC, Liu W, Peeters M, Sharp PM, Hahn BH. A plethora of Plasmodium species in wild apes: A source of human infection? Trends Parasitol. 2011;27(5):222–229. doi: 10.1016/j.pt.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duval L, et al. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc Natl Acad Sci USA. 2010;107(23):10561–10566. doi: 10.1073/pnas.1005435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paupy C, et al. Anopheles moucheti and Anopheles vinckei are candidate vectors of ape Plasmodium parasites, including Plasmodium praefalciparum in Gabon. PLoS One. 2013;8(2):e57294. doi: 10.1371/journal.pone.0057294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prugnolle F, et al. A fresh look at the origin of Plasmodium falciparum, the most malignant malaria agent. PLoS Pathog. 2011;7(2):e1001283. doi: 10.1371/journal.ppat.1001283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verhulst NO, Smallegange RC, Takken W. Mosquitoes as potential bridge vectors of malaria parasites from non-human primates to humans. Front Physiol. 2012;3:197. doi: 10.3389/fphys.2012.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laurent C, et al. Commercial logging and HIV epidemic, rural Equatorial Africa. Emerg Infect Dis. 2004;10(11):1953–1956. doi: 10.3201/eid1011.040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spielman A, et al. Malaria diagnosis by direct observation of centrifuged samples of blood. Am J Trop Med Hyg. 1988;39(4):337–342. doi: 10.4269/ajtmh.1988.39.337. [DOI] [PubMed] [Google Scholar]
- 16.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437(7057):376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee R, et al. Switching between raltegravir resistance pathways analyzed by deep sequencing. AIDS. 2011;25(16):1951–1959. doi: 10.1097/QAD.0b013e32834b34de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: Application to HIV-1 drug resistance. Genome Res. 2007;17(8):1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binladen J, et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. PLoS One. 2007;2(2):e197. doi: 10.1371/journal.pone.0000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar-Gonzalez JF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82(8):3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerra CA, et al. The international limits and population at risk of Plasmodium vivax transmission in 2009. PLoS Negl Trop Dis. 2010;4(8):e774. doi: 10.1371/journal.pntd.0000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Culleton RL, Ferreira PE. Duffy phenotype and Plasmodium vivax infections in humans and apes, Africa. Emerg Infect Dis. 2012;18(10):1704–1705. doi: 10.3201/eid1810.120120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox-Singh J, et al. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin Infect Dis. 2008;46(2):165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh B, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363(9414):1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 25.Antinori S, Galimberti L, Milazzo L, Corbellino M. Plasmodium knowlesi: The emerging zoonotic malaria parasite. Acta Trop. 2013;125(2):191–201. doi: 10.1016/j.actatropica.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Rayner JC, Huber CS, Galinski MR, Barnwell JW. Rapid evolution of an erythrocyte invasion gene family: The Plasmodium reichenowi reticulocyte binding like (RBL) genes. Mol Biochem Parasitol. 2004;133(2):287–296. doi: 10.1016/j.molbiopara.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 27.Crosnier C, et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 2011;480(7378):534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coatney GR. Simian malarias in man: Facts, implications, and predictions. Am J Trop Med Hyg. 1968;17(2):147–155. doi: 10.4269/ajtmh.1968.17.147. [DOI] [PubMed] [Google Scholar]
- 29.Rodhain J, Dellaert R. Contribution à l'étude du Pl. schwetzi E. Brumpt. III. L’infection à Plasmodium schwetzi chez l’homme. Ann Soc Belg Med Trop. 1955;35(6):757–775. [PubMed] [Google Scholar]
- 30.Contacos PG, et al. Transmission of Plasmodium schwetzi from the chimpanzee to man by mosquito bite. Am J Trop Med Hyg. 1970;19(2):190–195. doi: 10.4269/ajtmh.1970.19.190. [DOI] [PubMed] [Google Scholar]
- 31.Rodhain J. Contribution à l'étude des Plasmodiums des anthropoïdes africains. Ann Soc Belg Med Trop. 1948;28(1):39–49. [PubMed] [Google Scholar]
- 32.Rodhain J. Les Plasmodiums des anthropoïdes de l'Afrique centrale et leurs relations avec les Plasmodiums humains. Ann Soc Belg Med Trop. 1940;20(4):489–505. [Google Scholar]
- 33.Hughes AL, Verra F. Malaria parasite sequences from chimpanzee support the co-speciation hypothesis for the origin of virulent human malaria (Plasmodium falciparum) Mol Phylogenet Evol. 2010;57(1):135–143. doi: 10.1016/j.ympev.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manske M, et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature. 2012;487(7407):375–379. doi: 10.1038/nature11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ménard D, et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci USA. 2010;107(13):5967–5971. doi: 10.1073/pnas.0912496107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Culleton R, et al. Evidence for the transmission of Plasmodium vivax in the Republic of the Congo, West Central Africa. J Infect Dis. 2009;200(9):1465–1469. doi: 10.1086/644510. [DOI] [PubMed] [Google Scholar]
- 37.Rubio JM, et al. Semi-nested, multiplex polymerase chain reaction for detection of human malaria parasites and evidence of Plasmodium vivax infection in Equatorial Guinea. Am J Trop Med Hyg. 1999;60(2):183–187. doi: 10.4269/ajtmh.1999.60.183. [DOI] [PubMed] [Google Scholar]
- 38.Mendes C, et al. Duffy negative antigen is no longer a barrier to Plasmodium vivax—molecular evidences from the African West Coast (Angola and Equatorial Guinea) PLoS Negl Trop Dis. 2011;5(6):e1192. doi: 10.1371/journal.pntd.0001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.