Abstract
Cerebral cortical neurons arise from radial glia (direct neurogenesis) or from intermediate progenitors (indirect neurogenesis); intriguingly, the sizes of intermediate progenitor populations and the cortices they generate correlate across species. The generation of intermediate progenitors is regulated by the transcription factor Tbr2, whose expression marks these cells. We investigated how this mechanism might be controlled. We found that acute blockade of mature microRNA biosynthesis in murine cortical progenitors caused a rapid cell autonomous increase in numbers of Tbr2-expressing cells. Acute microRNA-92b (miR-92b) gain of function caused rapid reductions in numbers of Tbr2-expressing cells and proliferating intermediate progenitors. Acute miR-92b loss of function had opposite effects. These findings indicate that miR-92b limits the production of intermediate cortical progenitors.
Keywords: cortical development, microRNAs
The excitatory neurons of the cerebral cortex are generated from radial glia whose cell bodies populate the ventricular zone (VZ) around the inner surface of the developing anterior end of the neural tube (1–3). Radial glia divide either symmetrically, producing two new radial glial cells and expanding the proliferative population, or asymmetrically to generate a new radial glial cell and either a postmitotic neuron (direct neurogenesis) or a secondary (intermediate) progenitor. Whereas postmitotic neurons migrate to the outer surface of the neural tube to contribute to the cortex, intermediate progenitors move into the subventricular zone (SVZ), where they amplify the production of neurons by undergoing a further, usually symmetric, division to generate a pair of postmitotic cortical neurons (indirect neurogenesis) (4–6). In the mouse, these events occur between embryonic days 11 (E11) and E17 (7–9). The transition from radial glia to intermediate progenitors is controlled by a number of proteins; of these, only Tbr2 (also known as Eomesodermin) has been shown to have a direct effect on the specification of the intermediate progenitors (10–12).
MicroRNAs (miRNAs) are a class of short noncoding RNA molecules involved in posttranscriptional regulation of protein expression. As a class, they have been implicated in corticogenesis, most notably in cortical cell survival, but little is known about the functions of miRNAs in general, let alone those of specific miRNAs, in the regulation of cortical progenitors (13–16). Here, we investigated the possibility that miRNAs contribute to the regulation of indirect neurogenesis.
We first examined possible cell autonomous functions of miRNAs in general by generating acute cortical ablations of the key enzyme involved in the biosynthesis of most miRNAs, Dicer, in relatively small numbers of radial glia by electroporating a cre-recombinase expression vector into mouse embryos that were homozygous for the Dicer1fl allele (17). Acute mosaic ablation of Dicer cell autonomously promoted Tbr2-positive progenitor production. Then, based on an in silico prediction that one particular miRNA, miR-92b, might target the 3′UTR of Tbr2, we used acute gain- and loss-of-function approaches to show that this miRNA does indeed contribute to the regulation of intermediate progenitor cells.
Results
Rapid Expansion of the Tbr2-Expressing Progenitor Population After Dicer Loss.
miRNAs act on many developmental processes, and their prolonged loss throughout an entire tissue is likely to induce many indirect, possibly cell nonautonomous, effects including loss of cell viability (13–15). Our aim, therefore, was to examine cortical progenitors as soon as possible after they had been subject to acute mosaic depletion of miRNAs. We generated mosaic ablation of Dicer in a subset of radial glia in a consistent region of mouse lateral cortex by electroporating cre-recombinase expression vector (CAG-cre-IRES-EGFP) into E13.5 Dicer1fl/fl or Dicer1+/fl (control) embryos (Fig. 1A). We assessed the efficiency of the electroporated cre-recombinase expression vector in ablating functional Dicer and reducing mature miRNA levels in Dicer1fl/fl GFP+ cortical cells (Fig. S1A). Levels of Dicer1 mRNA, measured by quantitative RT-PCR (qRT-PCR) in GFP+ cells sorted using FACS, were significantly reduced in these samples of Dicer1fl/fl GFP+ cells 1 d after electroporation (Fig. S1B). As a further read-out of in vivo deletion, some dissociated cortical cells were plated and fixed immediately, and levels of miRNAs in Dicer1fl/fl or Dicer1+/fl cells were compared quantitatively by image analysis of locked nucleic acid in situ hybridizations (18). This approach confirmed rapid and efficient depletion of miRNAs from almost all GFP+ Dicer1fl/fl cells (Fig. S1 C–P).
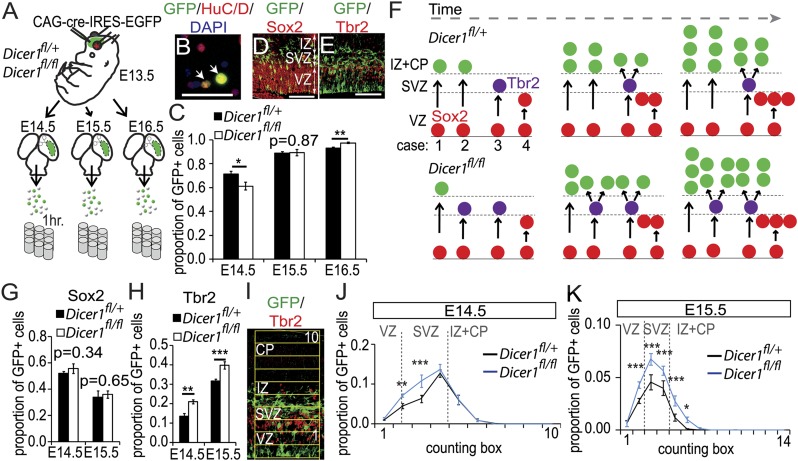
Fig. 1.
Increased generation of intermediate progenitor cells following Dicer1 deletion. (A) Experimental design: electroporated tissue was dissected from Dicer1fl/+ and Dicer1fl/fl brains and dissociated. (B) Dissociated cells were immunolabeled for GFP and a marker of early born neurons HuC/D (arrows). (Scale bar, 50 µm.) (C) Proportions of GFP+ Dicer1−/− cells that were HuC/D-positive were quantified at E14.5, E15.5, and E16.5 (n = 4). (D and E) One day after electroporation, GFP+ cells can be found in the VZ, SVZ, and intermediate zone (IZ). Sox2 is expressed by radial glia and Tbr2 is expressed by intermediate progenitors. (Scale bar, 100 µm.) (F) Neuronal production can be enhanced by an increased incidence of indirect neurogenesis: radial glia (red) can produce neurons directly on cell division in the VZ (cases 1 and 2) or indirectly by generating a Tbr2-positive intermediate progenitor (purple), which will further proliferate at the SVZ to amplify the neuronal output of the radial glia (case 3). In some cases, radial glia proliferate to expand the pool of progenitors (case 4). If more Dicer1−/− progenitors differentiate via the indirect neurogenesis pathway, the proportion of neurons will be initially reduced and later increased, consistent with the observations shown in C. (G and H) Normal proportion of Dicer1−/− GFP+ cells expressed Sox2 at E15.5, whereas a higher proportion of Dicer1−/− GFP+ cells expressed Tbr2 (n = 7–9). (I) Quantification of the distribution of GFP+/Tbr2+ cells used a counting ladder (boxes = 200 × 40 µm). (J and K) Distributions of GFP+ cells double-labeled for Tbr2. Expression of Tbr2 is up-regulated in Dicer1−/− cells (n = 7–11).
Previous studies reported that conditional Dicer deletion in mouse cortex causes rapid-onset large-scale apoptotic cell death (13–15). Interestingly, however, double-immunolabeling of sections through E13.5 electroporated Dicer1+/fl and Dicer1fl/fl cortex at E15.5 and E18.5 for GFP and the apoptotic marker cleaved-caspase-3 revealed no evidence that the incidence of apoptosis was altered when functional Dicer was lost from small subpopulations of cortical progenitors (Fig. S2 A–E). To provide further evidence for the absence of apoptosis in the Dicer-deficient progenitor cell lineages, mice carrying the Dicer1fl allele were crossed to mice carrying the Rosa26R:YFP (R26RYFP) Cre-reporter transgene to generate Dicer1fl/fl;Rosa26RYFP+ or Dicer1+/fl;Rosa26RYFP+ (control) embryos. The progeny of E13.5 electroporated radial glial progenitors were identified at postnatal day 14 using an antibody that recognizes both EGFP and YFP (immunopositive cells are referred to as GFP+). In Dicer1fl/fl P14 electroporated cortices, we found a higher contribution of GFP+ cells than in Dicer1+/fl P14 electroporated cortices (Fig. S2 F–I). This difference indicates that loss of Dicer from a subpopulation of cortical progenitor cells did not result in increased cell-autonomous apoptotic cell death between E13.5 and P14, possibly due to rescue by surrounding WT cells. On the contrary, loss of Dicer caused a long-term increase in the output of affected progenitors.
We examined whether the loss of functional Dicer affects the neuronal output from progenitors. Dissociated cells from cortical areas containing electroporated cells from either Dicer1fl/fl or Dicer1+/fl embryos at E14.5, E15.5, and E16.5 (Fig. 1A) were immunoreacted for GFP and HuC/D, a marker of early postmitotic neurons (Fig. 1B). Quantification showed that 3 d after electroporation, significantly more Dicer1−/− than control GFP+ cells expressed HuC/D (Fig. 1C). This increase in the proportion of early postmitotic neurons was preceded by an earlier phase during which fewer Dicer1−/− than control GFP+ cells were double-labeled for HuC/D (Fig. 1C). One possible explanation for this is that, as Dicer1 levels fall, the progeny of the Sox2+ radial glia (Fig. 1D) enter the indirect neurogenesis pathway at an increased rate and are specified as Tbr2+ intermediate progenitors (Fig. 1E) (10, 12, 19). An early increase in the specification of intermediate progenitors at the expense of neurons could lead to a later boost in neuronal production from the larger-than-normal intermediate progenitor pool, as shown by the diagram in Fig. 1F and in line with our observations on the postnatal effects of Dicer deletion (Fig. S2 F–I).
To test this, we quantified the proportions of cells that expressed Sox2 and Tbr2. Normal proportions of Dicer1−/− GFP+ cells expressed Sox2 at E14.5 and E15.5 (Fig. 1G), indicating no effect on the radial glial population, but the proportions of Dicer1−/− GFP+ cells expressing Tbr2 were increased (Fig. 1H). The distributions of Tbr2-expressing Dicer1−/− and control cells were compared by analyzing proportions of GFP and Tbr2 double-positive cells in counting bins through the cortex (Fig. 1I). This quantification showed increased Tbr2 expression among Dicer1−/− GFP+ cells in the abventricular portion of the VZ and lower part of the SVZ at E14.5 (Fig. 1J) and throughout the normal Tbr2 expression domain at E15.5 (Fig. 1K). These results are in excellent agreement with the proposed model (Fig. 1F).
MiR-92b Overexpression Causes a Rapid Reduction of Intermediate Progenitors.
The model proposed in Fig. 1F suggests that miRNAs might be important for tuning the proportions of cells transiting from Tbr2− radial glia to Tbr2+ intermediate progenitors. However, which miRNAs might be implicated and how might they work? Although the answers to these questions are likely to be complex, we based the next set of experiments on the supposition that, because Tbr2 expression not only marks intermediate progenitors but is implicated in controlling their numbers (10–12), miRNAs targeting Tbr2 might be particularly good candidates. TargetScan (www.targetscan.org) and microRNA.org (www.microrna.org) predicted that about 200 miRNAs might bind Tbr2. From this list, we searched for any that were known to be expressed in cortical progenitors in a complementary pattern to Tbr2. Neural progenitor cells were known to express miR-92b and, although its physiological target was not known (20–22), one publication highlighted it as an interesting candidate because its expression decreases as Tbr2 expression increases during development (20). TargetScan (www.targetscan.org) and microRNA.org both predicted the presence of a single highly conserved site complementary to the seed region of miR-92b in the 3′UTR of the mouse Tbr2 mRNA between nucleotides 2707–2714 (Fig. S3; Fig. 2C). We tested the possible involvement of this miRNA in intermediate progenitor regulation.
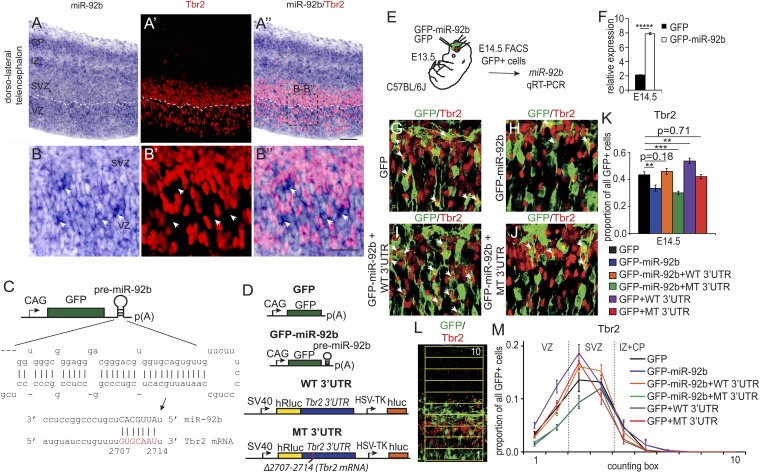
Fig. 2.
miR-92b targets Tbr2 in vivo. (A and B) In situ hybridization for miR-92b at E14.5, (A′ and B′) immunostaining for Tbr2, and (A″ and B″) overlays. Arrows indicate some of the hotspots of miR-92b labeling. (Scale bars: A–A″, 100 µm; B–B″, 50 µm.) (C) miR-92b expression construct used in this study. Response element in the 3′UTR of Tbr2 mRNA is highlighted in red. (D) Constructs generated to test the ability of miR-92b to interact with the 3′UTR. GFP expression plasmid was used as a control. (E) Expression vectors were delivered by electroporation. (F) Overexpression of mature miR-92b was confirmed by qPCR (n = 3). (G–J) Sections through the E14.5 telencephalic wall electroporated at E13.5 with vectors listed in D. Arrows indicate GFP and Tbr2 double-labeled cells. (Scale bar, 50 µm.) (K) Quantification of GFP and Tbr2 double-positive cells. Overexpression of miR-92b results in down-regulation of Tbr2 protein expression (blue) compared with control GFP-vector expression (black), which can be rescued by overexpressing the full length 3′UTR of Tbr2 (orange) but not when the miR-92b response element is mutated (green). When the action of endogenous miRNAs was competed out with the full-length 3′UTR, the Tbr2 expression increased (purple) but not when the 3′UTR lacked the miR-92b response element (red) (n = 8–11). (L) Method of quantification of the distribution of GFP+/Tbr2+ cells at E14.5 (200 × 40-µm boxes). (M) Distribution of GFP/Tbr2 double-positive cells manipulated as in K shows that changes in Tbr2 expression took place mainly in the abventricular portion of the VZ and the SVZ.
In situ hybridization staining for mature miR-92b was present throughout the cortex at E14.5 (Fig. 2 A and B) but was not of uniform intensity. In the VZ, there were many hotspots of high-intensity staining for miR-92b (Fig. 1 A and B, arrows show examples). In these areas, cells were Tbr2− (arrows in Fig. 2 B–B″), whereas cells that were Tbr2+ tended to be located between the hotspots. In the SVZ, there were fewer and smaller miR-92b hotspots, and this corresponded with higher densities of Tbr2+ cells (although, as in the VZ, cells at the hotspots were generally Tbr2−, e.g., see top of Fig. 2B″). This pattern suggests that there is a negative relationship between miR-92b expression and Tbr2 expression in areas of the proliferative zone where miR-92b concentration is above a certain level.
We generated a miR-92b overexpression vector (GFP-miR-92b) by cloning the genomic sequence containing the pre-miR-92b stem loop sequence downstream of a stop codon in a GFP expression vector (Fig. 2C). The GFP expression vector lacking the premiR-92b stem loop was used as a control (Fig. 2D). Overexpression of miR-92b in HEK-293 cells reduced Renilla luciferase reporter activity when the reporter sequence was joined to the full-length WT 3′UTR of Tbr2 (construct WT 3′UTR) but not when the response element between nucleotides 2707–2714 of the full-length Tbr2 3′UTR was deleted using site-directed mutagenesis (construct MT 3′UTR; Fig. 2D; Fig. S4).
The ability of miR-92b to regulate the expression of Tbr2 in vivo was assessed by electroporating the GFP-miR-92b vector in utero into the cortex of WT E13.5 embryos. Overexpression was confirmed at E14.5 by qRT-PCR using RNA extracted from GFP+ cells isolated by FACS (Fig. 2 E and F). Electroporation of this miR-92b overexpression vector resulted, within a day, in fewer GFP+ cells that were double-positive for Tbr2 than electroporation of control GFP-only vector (Fig. 2 G and H; compare black and blue bars in K). This phenotype was rescued when GFP-miR-92b was coelectroporated with the WT-3′UTR vector (Fig. 2D), which expresses the WT 3′UTR of Tbr2 to act as a competitor with the endogenous 3′UTR (Fig. 2I; orange bar in K). Conversely, the MT-3′UTR vector (Fig. 2D) expressing the 3′UTR of Tbr2 mRNA lacking the miR-92b putative binding site failed to rescue the phenotype (Fig. 2J; green bar in K). Analysis of the laminar distribution of GFP+/Tbr2+ double-positive cells revealed that the reduction in the number of Tbr2+ cells at E14.5 after miR-92b overexpression was greatest in the abventricular portion of the VZ and lower SVZ (counting boxes 2 and 3 in Fig. 2 L and M).
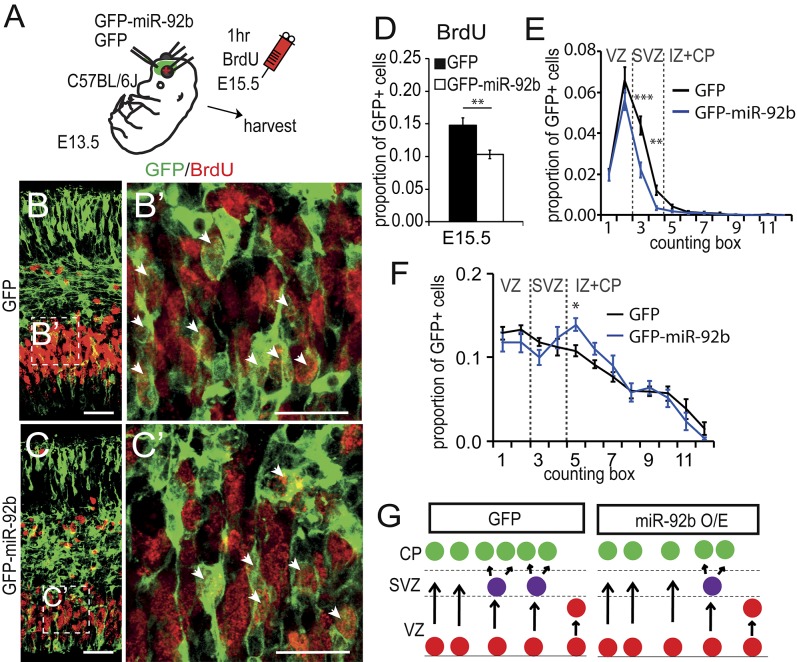
To examine the effect of miR-92b on the proliferation of cortical progenitors, we electroporated WT E13.5 embryos with either the GFP-miR-92b overexpression vector or the GFP control vector (Fig. 3A). The embryos were exposed to BrdU for 1 h on E15.5 to label cells in S-phase (Fig. 3 A–C′). The result of overexpression of miR-92b was to significantly reduce by about 30% the overall proportion of GFP+ cells that incorporated BrdU (Fig. 3D), as a consequence of reductions specifically in the SVZ and not in other layers (Fig. 3E). Overexpression also caused an increase in the relative proportion of GFP+ cells (most of which did not contain BrdU) in the intermediate zone (Fig. 3F), suggesting that the decrease in the proliferative population in the SVZ was accompanied by a corresponding increase in the exit of cells from the proliferative zones toward the cortical plate. Our data strongly suggest that miR-92b contributes to the regulation of neuronal output from the radial glia, by controlling the balance between the intermediate progenitor and the postmitotic states (see schematic in Fig. 3G).
Fig. 3.
MiR-92b regulates intermediate progenitor cell specification. (A) Electroporation of GFP or GFP-miR-92b constructs followed by BrdU labeling of cells in S-phase. (B and C) Example images showing some GFP+ cells in the telencephalon incorporate BrdU. High-magnification examples are shown in B′ and C′ (arrows). (D) Fewer GFP-miR-92b–expressing cells incorporated BrdU (n = 12). (E) Fewer GFP-miR-92b cells incorporated BrdU on E15.5 in the SVZ but not the VZ (n = 12). (F) Relative distribution of GFP+ cells. More cells were found in the IZ following miR-92b overexpression. (G) We propose that miR-92b overexpression (O/E) results in fewer cells entering the indirect neurogenesis pathway.
MiR-92b Inhibition Causes a Rapid Increase in Tbr2-Expressing Cells.
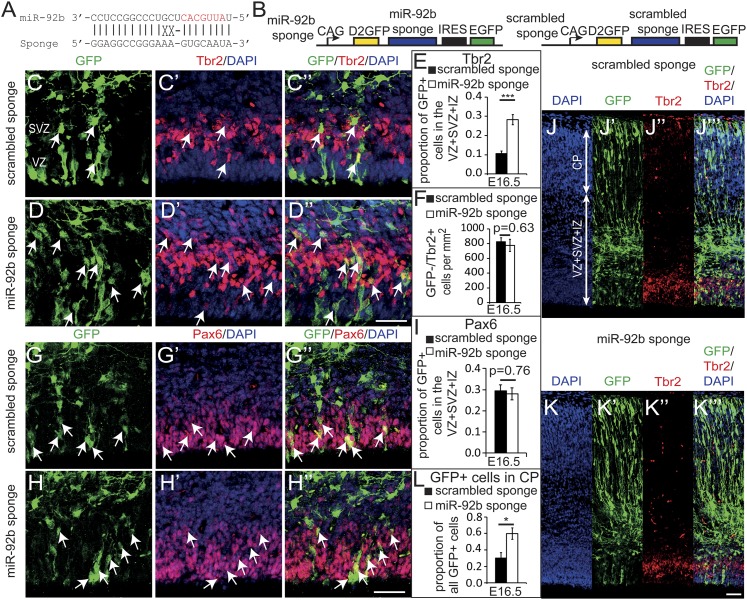
We used two approaches to test whether endogenous miR-92b can limit the numbers of Tbr2+ cells in vivo. First, we coelectroporated the GFP and WT-3′UTR vectors (Fig. 2D) at E13.5, because competition between WT-3′UTR and the endogenous binding site in the 3′UTR of Tbr2 mRNA should lower the levels, and hence the function, of miR-92b at the endogenous site. This manipulation resulted in an increased proportion of cells expressing GFP and Tbr2 compared with cells electroporated only with the GFP expression vector by E14.5 (Fig. 2 K and M, purple bar and line). Conversely, the MT 3′UTR vector (Fig. 2D) lacking the miR-92b binding site did not produce the same effect (Fig. 2 K and M, red bar and line). Second, we used a miRNA sponge (SI Materials and Methods) designed to outcompete endogenous transcripts binding to miR-92b (Fig. 4 A and B). Vectors containing the miR-92b sponge or scrambled control were electroporated at E13.5, and cortices were collected at E16.5. Higher proportions of GFP+ cells expressed Tbr2 in the proliferative zones following electroporation with the miR-92b sponge (Fig. 4 D–D″ and E) rather than with scrambled control sponge (Fig. 4 C–C″ and E). Densities of GFP−/Tbr2+ cells were no different between control and experimental groups, indicating that the effect of miR-92b inhibition on the generation of Tbr2+ cells was cell autonomous (Fig. 4F). There were no differences in the proportions of GFP+ cells that expressed Pax6, a marker of radial glial cells, in agreement with the effects being specifically on the intermediate progenitor population (Fig. 4 G–I). Our findings indicate that miR-92b is an important physiological inhibitor of the generation of Tbr2-expressing intermediate cortical progenitor cells.
Fig. 4.
Endogenous miR-92b controls neuronal output of the progenitor population. (A) A sponge was designed to provide binding sites for endogenous miR-92b. (B) Constructs containing miR-92b sponge or scrambled sponge (control). (C–D″) VZ and SVZ at E16.5 electroporated with scrambled sponge or miR-92b sponge immunostained for GFP and Tbr2, with DAPI nuclear counterstain. Arrows indicate GFP and Tbr2 double-positive cells. (E) Quantification of the proportion of GFP-positive cells in the VZ+SVZ+IZ-expressing Tbr2 (n = 5). (F) Quantification of the proportion of GFP-negative cells in the VZ+SVZ+IZ-expressing Tbr2 (n = 5). (G–H″) E16.5 VZ and SVZ electroporated with scrambled sponge or miR-92b sponge and immunostained for GFP and Pax6, with DAPI counterstain. Arrows indicate GFP and Pax6 double-positive cells. (Scale bars in C-H″, 50 µm.) (I) Quantification of the proportion of GFP-positive cells in the VZ+SVZ+IZ-expressing Pax6 (n = 5). (J–K″′) Cross sections through E16.5 cortex electroporated with scrambled sponge (J–J″′) or miR-92b sponge (K–K″′), counterstained with DAPI, and immunostained for GFP and Tbr2. (Scale bar, 50 µm.) (L) Quantification showing an increased proportion of GFP-positive cells in the CP following miR-92b sponge electroporation compared with control (n = 5).
We predicted that increased generation of Tbr2+ cells induced by the miR-92b sponge might translate into an increased output of cortical plate cells, as occurred following Dicer deletion (Fig. S2 F–I). Indeed, we observed that proportions of GFP+ cells in the cortical plate 3 d after electroporation were higher with the miR-92b sponge (Fig. 4 K–K″′ and L) than the scrambled control (Fig. 4 J–J″′ and L ). A previous study showed that Tbr2 gain of function in cortical progenitors at the onset of corticogenesis increased the proportions of their descendants fated to the superficial cortical layers and decreased the proportions fated to deep layers (12). We tested for a similar trend following long-term inhibition of miR-92b by injecting E12.5 WT embryos with retroviruses expressing GFP and the miR-92b sponge or, as controls, with retroviruses expressing GFP and the scrambled sponge. Animals were allowed to develop to P14, and we quantified the positions of the neurons in the resulting clones. Similar to the previous report (12), we found that about 30% of labeled neurons in controls were in superficial layers (I–III), with about 70% in deep layers (IV–VI; Fig. S5). Infection with the miR-92b sponge–expressing retrovirus resulted in significant increases in the proportions of labeled neurons in superficial layers and corresponding decreases in the proportions in deep layers (Fig. S5). These changes are consistent with our hypothesis that inhibition of miR-92b results in increased specification of Tbr2+ intermediate progenitor cells and with the proposed role of Tbr2 in superficial layer neurogenesis (10, 12).
Discussion
During normal corticogenesis, most cells expressing the transcription factor Tbr2 are located in the SVZ, where intermediate cortical progenitors reside. It is also expressed by some cells in the VZ, thought to be the daughters of asymmetric divisions of radial glia. Previous studies investigating the role of Tbr2 in cortical development suggested that, far from being simply a marker of the intermediate progenitor population, Tbr2 drives VZ cells toward an intermediate progenitor fate (10–12). Tbr2 loss-of-function experiments have shown that Tbr2 is required for the generation of normal numbers of intermediate progenitors, whereas Tbr2 overexpression in radial glial progenitors causes the overproduction of intermediate progenitors and the cortical neurons that they generate (10–12). It is likely, therefore, that mechanisms that regulate the levels of Tbr2 in cells newly generated by the radial glia will impact on the production of intermediate progenitors. Limiting Tbr2 levels among the progeny of the radial glia is likely to bias against intermediate progenitor production, whereas enhancing Tbr2 levels is likely to favor production. Our experiments suggest that one such mechanism might involve miR-92b.
In our histological work, we observed that hotspots containing the highest levels of miR-92b in the VZ coincided with areas where cells were Tbr2 negative, whereas regions with lower miR-92b levels were Tbr2 positive. These observations are compatible with the possibility that miR-92b is involved in controlling levels of Tbr2 in VZ cells. To test this hypothesis, we raised or lowered miR-92b levels experimentally. Our results showed that miRNAs, including miR-92b and possibly others, normally restrict cell autonomously the production of Tbr2+ intermediate cortical progenitor cells. Both acute depletion of miRNAs in general and up- or down-regulation of specifically miR-92b levels caused rapid-onset changes in the intermediate progenitor population. We suggest, therefore, that miR-92b is a significant component of a process determining the proportions of newly generated VZ cells entering the intermediate progenitor cell state as opposed to adopting other fates, for example reentering the radial glial cell cycle. Given that miR-92b appears able to target Tbr2 directly, a parsimonious hypothesis is that miR-92b’s actions on intermediate progenitor cell numbers is mediated at least in part by its modulation of the levels of Tbr2 within newborn daughters of radial glia. Thus, in those newly generated cells in which such a mechanism keeps levels of Tbr2 mRNA below a critical threshold, entry into the intermediate progenitor state would be prevented or inhibited.
Although this proposal offers a straightforward way of drawing together several strands of evidence, it is likely that miR-92b also operates through other pathways. The mechanisms controlling cell states are complex and even more so where miRNAs are involved. Individual miRNAs probably have numerous targets. For example, TargetScan, which takes evolutionary conservation into account, lists about 700 predicted targets of miR-92b, although none of those that score higher than Tbr2 are implicated in the regulation of cortical intermediate progenitors. Other studies have shown, for example, that miR-92b targets mRNAs for p57 in mouse embryonic stem cells (23) and for Dickkopf-3 in neuroblastoma cells (24) and is likely to target a similar repertoire of transcripts as miR92a (25). Furthermore, although the requirement for Tbr2 in promoting intermediate progenitor cell development is well established, ablation of Tbr2 may not cause complete loss of all intermediate progenitors (10, 12), suggesting that other factors could promote their development. There is strong evidence that the intermediate progenitor pool size is affected by the function of Pax6, Cdc42, Tlx, Id4, Ccnd2, Lrp6, or Ngn2 (11, 26), although some of these effects may be indirect. Our bioinformatic searches predicted single binding sites to miR-92b in the 3′UTR elements of Cdc42, Id4, Pax6, and Ccnd2, although numbers of Pax6-expressing cells were not affected by miR-92b manipulation in our experiments. Overall, our work implicating miRNAs, and miR-92b in particular, in the development of intermediate progenitors should be seen as just one important element of a network of regulation that is likely to be complex.
Our findings are also potentially interesting from an evolutionary perspective. Recently, it has been proposed that the evolutionary expansion of the neocortex is due largely to expansion of the SVZ and its intermediate progenitor population, thereby amplifying the output of the radial glia by indirect neurogenesis (27). The miR-92b response element in the 3′UTR of Tbr2 mRNA is conserved in mammalian species, as is most of the miR-92b sequence (Fig. S3), making it likely that an interaction between miR-92b and the 3′UTR of Tbr2 mRNA is also conserved. There are, however, nucleotide differences between rodent and primate miR-92b, particularly in the unpaired regions of the hairpin (Fig. S3C) that could affect the processing and stability of the premiRNA hairpin and hence the miRNA’s function (28). It is conceivable that these or other changes affecting miRNA function could have contributed to the evolution of the cerebral cortex.
Materials and Methods
Animals.
Mice homozygous for Dicer1fl allele were purchased from Jackson Laboratories. Rosa26R:YFP (R26RYFP) cre-reporter transgenic mice were described before (29). For miRNA studies, WT C57BL/6J mice were used. In utero surgery was performed as previously described (30). Anesthesia was maintained using inhaled isoflurane. Cre expression vector was injected at a concentration of 1.35 mg/mL. For coelectroporation, plasmids were injected at a concentration of 2.5 mg/mL. BrdU injections were performed i.p. into pregnant dams at 50 µg/g body weight. Pregnant females were killed by cervical dislocation, and embryos were dissected and decapitated. Tissues were fixed in 4% (wt/vol) paraformaldehyde (PFA) in PBS. Tissue was cryoprotected with 15% (wt/vol) sucrose before freezing in 15% (wt/vol) sucrose in PBS/optimum cutting temperature (OCT) (Fisher). Cryosections were cut at 20 µm. The license authorizing this work was approved by the University of Edinburgh's Ethical Review Committee on September 22, 2008 (Application PL35-08) and by the Home Office on November 6, 2008.
Cell Dissociation.
Cortical tissue containing GFP-positive cells were dissected in ice-cold Eagle’s balanced salt solution, dissociated with trypsin (Gibco), and triturated to obtain single-cell suspension. Cells were plated at 103–104 cells per glass coverslip (9 mm diameter; VWR) precoated with 100 µg/mL of poly-l-lysine (Sigma), and allowed to adhere for 1 h before fixation in PFA for 20 min. For FACS sorting, dissociated cells were resuspended in 1% (vol/vol) FCS/PBS and sorted using BD FacsAria II.
Quantitative PCR Analysis.
Total RNA and miRNA were extracted from FACS-sorted GFP+ cells using the miRNeasy Mini Kit (Qiagen). Total RNA was reverse-transcribed using the Super-Script III First Strand Synthesis System (Invitrogen). MiRNA92b was reverse transcribed using the miScript II RT Kit (Qiagen) and a miScript Primer Assay (Qiagen). Quantitative real-time PCR for Dicer1 mRNA expression was performed using the Quantitect SYBR Green PCR Mix (Qiagen) in a DNA Engine Opticon Real-Time Thermal Cycler (MJ Research). Relative expression was compared with Gapdh mRNA. Four control and three mutant embryos were analyzed. Quantitative real-time PCR for miR-92b was performed using the miScript SYBR Green PCR Kit (Qiagen) and compared with snRNA U6 (Qiagen). Twelve control embryos and nine embryos electroporated with miR-92b were analyzed.
Luciferase Assay.
Dual luciferase reporter assay was performed in HEK-293 cells cultured in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS (Gibco). Luciferase expression vectors, wild type (WT) 3′UTR or mutant (MT) 3′UTR, and GFP or GFP-miR-92b were cotransfected using Lipofectamine 2000 (Invitrogen) in 96-well plates. Luciferase activity was assayed 48 h later using the dual luciferase reporter system (Promega) and the GloMax Luminometer (Promega).
In Situ Hybridization and Immunohistochemistry.
In situ hybridization was performed as described previously (31). Immunohistochemistry followed standard protocols with heat-induced antigen retrieval achieved by microwaving the slides in 10 mM sodium citrate, pH = 6.0. Primary antibodies are listed in SI Materials and Methods. Binding was revealed using an appropriate flurophore-conjugated secondary AlexaFluor antibody raised in donkey (1:400; Invitrogen). Nuclear counterstain DAPI (Vector) was applied in PBS. Slides were mounted in Vectashield hard-set (Vector). All images were collected using a Leica DFC480 or DFC 360 FX camera or a Zeiss LSM 150 confocal system. Image analysis was performed either in ImageJ or MatlabR2009a (Mathworks).
Quantification of Immunopositive Cells.
For each experiment using dissociated cells, 50 images of fields containing GFP-expressing cells were taken from five coverslips. Cells were assessed from four experiments per age, using at least three embryos per experiment for each genotype. Areas were selected based only on the GFP staining to image as many cells as possible. Cells were then counted manually. Quantification of immunopositive cells in brain sections is described in SI Materials and Methods. Statistical analysis using either two-way unpaired Student t test or Tukey’s test was performed using Microsoft Office Excel and Sigmastat (SYSTAT Software). Error bars represent SEM in all figures. In all graphs, *P < 0.05, **P < 10−2, ***P < 10−3, and *****P < 10−5. The n numbers quoted are the numbers of animals in each group.
Supplementary Material
Acknowledgments
We thank Veronica van Heyningen (Edinburgh University) for helpful comments, Sandro Banfi (Telethon Institute of Genetics and Medicine) for help in the identification of the candidate miRNA, Trudi Gillespie for help with confocal microscopy, Fiona Rossi and Shonna Maccall (Queen’s Medical Research Institute) for help with FACS, and Anjen Chenn (Northwestern University) for providing the cre expression vector. This work was funded by The Wellcome Trust (83213) and the Medical Research Council, UK.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219385110/-/DCSupplemental.
References
- 1.Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31(5):727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 2.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409(6821):714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 3.Malatesta P, Hartfuss E, Götz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127(24):5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi T, Nowakowski RS, Caviness VS., Jr Early ontogeny of the secondary proliferative population of the embryonic murine cerebral wall. J Neurosci. 1995;15(9):6058–6068. doi: 10.1523/JNEUROSCI.15-09-06058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: A major site of neurogenesis. Proc Natl Acad Sci USA. 2004;101(9):3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7(2):136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 7.Caviness VS., Jr Neocortical histogenesis in normal and reeler mice: A developmental study based upon [3H]thymidine autoradiography. Brain Res. 1982;256(3):293–302. doi: 10.1016/0165-3806(82)90141-9. [DOI] [PubMed] [Google Scholar]
- 8.Gillies K, Price DJ. The fates of cells in the developing cerebral cortex of normal and methylazoxymethanol acetate-lesioned mice. Eur J Neurosci. 1993;5(1):73–84. doi: 10.1111/j.1460-9568.1993.tb00207.x. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi T, Nowakowski RS, Caviness VS., Jr The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 1995;15(9):6046–6057. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold SJ, et al. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22(18):2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30(1-3):24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- 12.Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60(1):56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Pietri Tonelli D, et al. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135(23):3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev Dyn. 2009;238(11):2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowakowski TJ, Mysiak KS, Pratt T, Price DJ. Functional dicer is necessary for appropriate specification of radial glia during early development of mouse telencephalon. PLoS ONE. 2011;6(8):e23013. doi: 10.1371/journal.pone.0023013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volvert ML, Rogister F, Moonen G, Malgrange B, Nguyen L. MicroRNAs tune cerebral cortical neurogenesis. Cell Death Differ. 2012;19(10):1573–1581. doi: 10.1038/cdd.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci USA. 2005;102(31):10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RHA. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3(1):27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 19.Ochiai W, et al. Periventricular notch activation and asymmetric Ngn2 and Tbr2 expression in pair-generated neocortical daughter cells. Mol Cell Neurosci. 2009;40(2):225–233. doi: 10.1016/j.mcn.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen JA, Lau P, Maric D, Barker JL, Hudson LD. Integrating microRNA and mRNA expression profiles of neuronal progenitors to identify regulatory networks underlying the onset of cortical neurogenesis. BMC Neurosci. 2009;10:98. doi: 10.1186/1471-2202-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapsimali M, et al. MicroRNAs show a wide diversity of expression profiles in the developing and mature central nervous system. Genome Biol. 2007;8(8):R173. doi: 10.1186/gb-2007-8-8-r173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meza-Sosa KF, Valle-García D, Pedraza-Alva G, Pérez-Martínez L. Role of microRNAs in central nervous system development and pathology. J Neurosci Res. 2012;90(1):1–12. doi: 10.1002/jnr.22701. [DOI] [PubMed] [Google Scholar]
- 23.Sengupta S, et al. MicroRNA 92b controls the G1/S checkpoint gene p57 in human embryonic stem cells. Stem Cells. 2009;27(7):1524–1528. doi: 10.1002/stem.84. [DOI] [PubMed] [Google Scholar]
- 24.Haug BH, et al. MYCN-regulated miRNA-92 inhibits secretion of the tumor suppressor DICKKOPF-3 (DKK3) in neuroblastoma. Carcinogenesis. 2011;32(7):1005–1012. doi: 10.1093/carcin/bgr073. [DOI] [PubMed] [Google Scholar]
- 25.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glickstein SB, Monaghan JA, Koeller HB, Jones TK, Ross ME. Cyclin D2 is critical for intermediate progenitor cell proliferation in the embryonic cortex. J Neurosci. 2009;29(30):9614–9624. doi: 10.1523/JNEUROSCI.2284-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146(1):18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 29.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito T. In vivo electroporation in the embryonic mouse central nervous system. Nat Protoc. 2006;1(3):1552–1558. doi: 10.1038/nprot.2006.276. [DOI] [PubMed] [Google Scholar]
- 31.Wallace VA, Raff MC. A role for Sonic hedgehog in axon-to-astrocyte signalling in the rodent optic nerve. Development. 1999;126(13):2901–2909. doi: 10.1242/dev.126.13.2901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.