Abstract
Specific stages of the cell cycle are often restricted to particular times of day because of regulation by the circadian clock. In zebrafish, both mitosis (M phase) and DNA synthesis (S phase) are clock-controlled in cell lines and during embryo development. Despite the ubiquitousness of this phenomenon, relatively little is known about the underlying mechanism linking the clock to the cell cycle. In this study, we describe an evolutionarily conserved cell-cycle regulator, cyclin-dependent kinase inhibitor 1d (20 kDa protein, p20), which along with p21, is a strongly rhythmic gene and directly clock-controlled. Both p20 and p21 regulate the G1/S transition of the cell cycle. However, their expression patterns differ, with p20 predominant in developing brain and peak expression occurring 6 h earlier than p21. p20 expression is also p53-independent in contrast to p21 regulation. Such differences provide a unique mechanism whereby S phase is set to different times of day in a tissue-specific manner, depending on the balance of these two inhibitors.
One of the most significant rhythmic processes controlled by the circadian clock is the daily timing of the cell cycle. This process of clock-regulated cell division is observed in most organisms, in tissues and cell lines, as well as during embryo development (1–4). Although gene expression studies in normal mouse organs (5–7) and especially in regenerating liver (3) have begun to elucidate some aspects of the mechanism, remarkably little is known about how the clock controls the cell cycle, particularly regarding the timing of DNA synthesis (S phase) at the cellular level. Disruption of this temporal pattern of cell-cycle regulation has clear implications for cancer initiation and progression (8), but its significance during embryo development is relatively unexplored.
Zebrafish represent a very powerful tool with which to explore this interaction, not least because of the highly decentralized nature of its circadian system. Each cell is believed to contain a complete circadian system, including the means to detect and entrain directly to environmental light signals. Previous studies have shown that both mitosis (M phase) and S phase are clock-regulated and restricted to specific times of the day, not only in cell lines, but also during embryo development (2, 4). S phase, as revealed by BrdU incorporation, shows a particularly robust rhythm, with a peak of DNA replication occurring during the day in developing larvae (2).
Here we demonstrate that a G1/S cell-cycle regulator, cyclin-dependent kinase inhibitor 1d (20 kDa protein, p20), is directly clock-controlled and plays a critical role in cell-cycle timing. Peripheral circadian clocks in zebrafish are set to the same phase, as a consequence of their direct sensitivity to light, a fact that raises issues about how clock outputs are distinctly timed. In the case of the cell cycle, differential expression patterns, both temporally and spatially, between p20 and p21 provide a mechanism to regulate S phase to different times of day in different cell types.
Results and Discussion
Among the most likely candidates to play a role in S-phase regulation is the family of cyclin-dependent kinase (CDK) inhibitors, exemplified by CDK inhibitor 1a (cdkn1a) or p21, a well-established regulator of G1/S cell-cycle progression (9). To explore their role further, we performed a bioinformatic analysis of CDK inhibitors in the zebrafish genome. This led not only to the isolation of the zebrafish p21 coding and regulatory sequences, but also to the discovery of a gene with significant similarity to p21, designated cdkn1d or p20, based on its predicted protein molecular weight. Further analysis led to the identification of p20 in other teleost fish species, such as medaka, stickleback, Fugu, and spotted green pufferfish as well as in birds (chicken and turkey) and reptiles (lizard and turtle). Importantly, we have amplified p20 by RT-PCR from zebrafish, medaka, and chicken embryo RNA, proving that this gene is indeed expressed in these organisms.
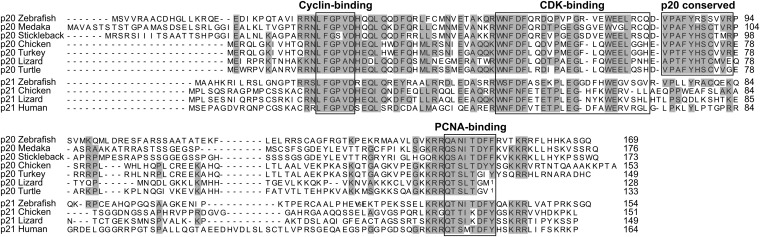
The p20 gene is not just teleost-specific or simply the result of a teleost-specific gene duplication. Its presence in several animal groups implies evolutionary conservation and argues for a significant role in cell-cycle regulation. An evolutionary tree comparing p20 protein sequences with other CDK inhibitors (cdkn1a/p21, cdkn1b/p27, and cdkn1c/p57) in various species reveals that p20 is most closely related to p21. In addition, all p20 proteins cluster together in an independent branch of the tree, suggesting a strong conservation throughout evolution (Fig. S1). Furthermore, p20 and p21 share common domains, which have been previously identified as essential for binding to cyclins (10), CDKs (11), and proliferating cell nuclear antigen (PCNA) (12) (Fig. 1). In particular, the cyclin- and PCNA-binding domains show a high degree of similarity in all p20 and p21 proteins analyzed. The CDK-binding domain, in contrast, is significantly more variable across species, which might suggest differential binding affinities. Interestingly, immediately downstream of this region is an 11-residue domain specific to the p20 proteins. Because of its proximity, this region is likely to modulate p20 interaction with CDKs, or might even be responsible for interaction with other cell-cycle proteins, suggesting that p20 may play a distinct role in cell-cycle regulation.
Fig. 1.
Conservation of functional domains between the newly identified CDK inhibitor p20 and p21. Sequence alignment of p20 and p21 proteins showing the conservation of various domains across species. Alignment was performed using ClustalW, and gray shading indicates identical/conservative amino acids. 1Predicted p20 sequence is incomplete.
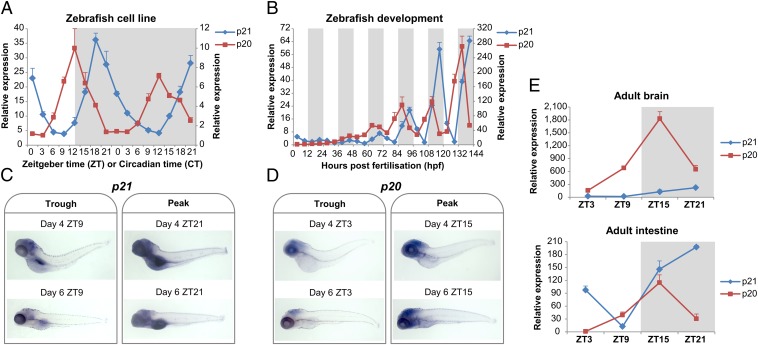
Does p20 as well as p21 show a circadian oscillation in gene expression in zebrafish? To address this question, we examined transcript levels for both genes using quantitative PCR (qPCR) in an embryonic zebrafish cell line (PAC2) (13) under both light-dark (LD) cycle and constant dark (DD) free-running conditions. p21 shows a robust circadian oscillation, peaking in the night at zeitgeber time 18 (ZT18), in which ZT0 refers to lights on (Fig. 2A). Interestingly, p20 is also rhythmically expressed in these cells; however, peak expression occurs ∼6 h earlier at ZT12, during the light to dark transition. Both genes maintain their rhythmicity in DD, showing clear circadian clock regulation. The phase difference between p21 and p20 is also observed during zebrafish development, in particular from 72 h post fertilization (hpf) onward (Fig. 2B). p21 peaks at ZT21, whereas p20 peaks at ZT15, maintaining a phase difference of ∼6 h. It is important to note that the absolute levels of p21 and p20 also vary in cell lines and larvae. Although p21 is clearly more abundant than p20 in the cell line, the exact opposite is true in the zebrafish embryo (note different y axes in Fig. 2 A and B).
Fig. 2.
Differential rhythmic expression of p21 and p20. qPCR analysis of p21 and p20 expression in PAC2 cells (A) and during embryo development (B). The left and right axes correspond to relative levels of p21 and p20, respectively. Representative images of whole mount in situ hybridization on 4- and 6-d-old larvae for p21 (C) and p20 (D). Both trough and peak times were analyzed. (E) qPCR analysis of p21 and p20 expression in adult zebrafish tissues (brain and intestine). White and gray backgrounds represent light and dark phases, respectively.
To determine where these genes are expressed during zebrafish development, we performed whole-mount in situ hybridization on 4- and 6-d-old larvae, selecting both trough and peak times for p21 and p20 expression. p21 is expressed in the brain, but also very strongly in the gut of developing zebrafish (Fig. 2C). In contrast, p20 expression is restricted primarily to the developing brain (Fig. 2D). An examination of expression levels in adult tissues also reveals differences between the two genes. p20 is dramatically more abundant in adult brain than p21, whereas p20 and p21 levels are expressed at similar levels in adult intestine (Fig. 2E).
Both genes show strong circadian oscillations, with distinct timing in their peaks of expression. This might suggest, along with particular sequence differences, that they regulate different stages of the cell cycle, an idea we explore in this article. Rhythmic expression of p20 and p21 is also robust during embryo development and corresponds to the timing of the onset of clock regulation of S phase, adding further support to the idea that one or both of these genes are critical for clock–cell-cycle interactions. Not only is the timing of expression consistently different between p20 and p21, but also the tissue specificity of their expression. The p20 transcript is dramatically more abundant than p21 in the zebrafish adult brain (by 14-fold when comparing peak levels), a fact that suggests this gene may play a critical role in the regulation of the neuronal cell cycle.
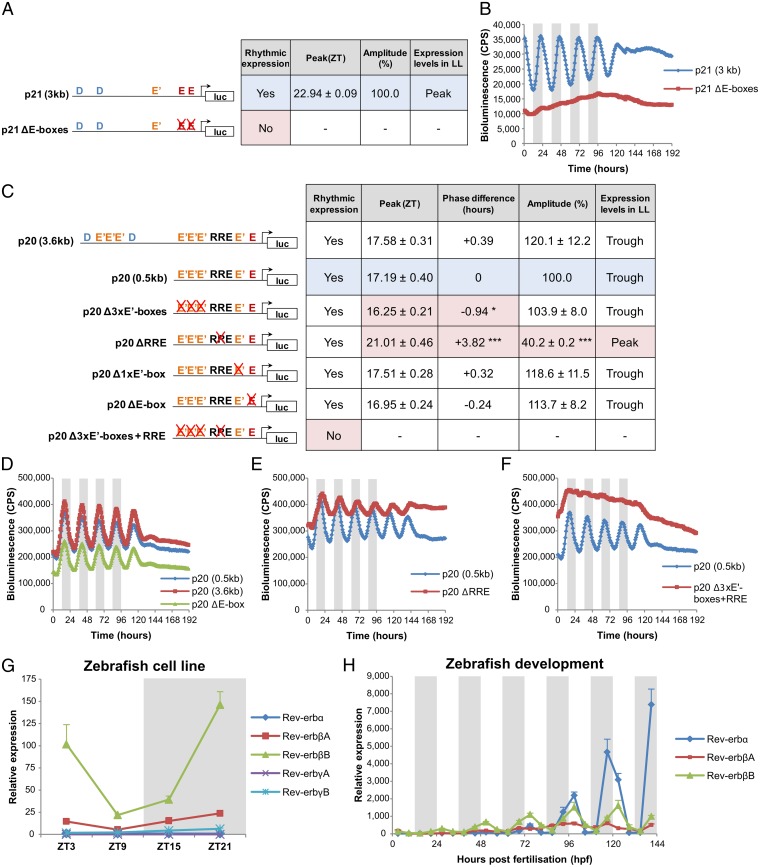
How then does the circadian clock regulate the expression of these two cell-cycle inhibitors? To address this question, we analyzed the promoters of both genes and determined the presence of known clock-controlled regulatory elements, including D boxes (TTA[T/C]GTAA), E (CACGTG)/E′ boxes (CACGTT), and Rev-Erb/retinoid-related orphan receptor response elements (RREs) ([A/T]A[A/T]NT[A/G]GGTCA) (14). A 3.0-kb DNA fragment upstream of the p21 transcriptional start sites (TSS) contains a number of these sequences, including two D boxes, one E′ box, and two E boxes. We fused this promoter region to the firefly luciferase gene and generated a stable luminescent reporter cell line. When exposed to an LD cycle, these cells exhibited a clear circadian rhythm (Fig. 3 A and B), demonstrating that the critical regulatory elements responsible for p21 rhythmicity are contained within this promoter fragment. When these cells were transferred to constant light (LL), a condition known to repress clock function in zebrafish (15), the p21 rhythm was abolished and expression levels were maintained at close to peak values. The deletion of the two perfect E boxes in the p21 promoter resulted in a complete loss of rhythmicity as well as a significant reduction in basal expression levels (Fig. 3 A and B). These results show that p21 regulation requires two essential E boxes, which are most likely activated by direct binding of the central clock components, CLOCK and BMAL.
Fig. 3.
p21 and p20 are direct clock-controlled genes. (A) Summary table of relevant bioluminescent parameters exhibited by the two different p21-luciferase cell lines analyzed. (B) Bioluminescent traces of p21-luciferase cell lines on an LD cycle and then transferred to LL. (C) Summary table of relevant bioluminescent parameters exhibited by the seven different p20-luciferase cell lines analyzed. (D–F) Bioluminescent traces of p20-luciferase cell lines on an LD cycle and then transferred to LL. The blue-shaded row in each table corresponds to the cell line used as a reference for comparison. Red-shaded cells correspond to significant differences relative to reference values. *P < 0.05, ***P < 0.001. qPCR analysis of Rev-erb gene expression in PAC2 cells (G) and during embryo development (H). White and gray backgrounds represent light and dark phases, respectively. luc, luciferase.
In contrast, the p20 promoter appears much more complex and contains two D boxes, seven E′ boxes, one E box, and one RRE in a 3.6-kb fragment upstream of the TSS. A 0.5-kb fragment, lacking both D boxes and three E′ boxes, showed an identical rhythm to the 3.6-kb fragment, both in terms of phase and amplitude (Fig. 3 C and D), proving that all essential regulatory elements are present in this shorter DNA fragment. A single deletion in the 0.5-kb fragment of either the E box or the E′ box closest to the TSS had no effect on the phase or amplitude of the rhythm. However, the E box deletion did reduce global luminescence levels, showing that this element is important for basal expression (Fig. 3 C and D). Of all of the promoter mutations, the most interesting was deletion of the RRE, which produced a significant reduction in rhythm amplitude (∼60%) and a phase delay of almost 4 h (Fig. 3 C and E). The resulting peak in expression, now ZT21, is much closer to the phase of p21 expression (ZT23), arguing that the RRE is responsible for the phase difference observed between p20 and p21. It is interesting that this one regulatory element may underlie the differential timing in expression of these two cell-cycle genes. This implies that RRE regulation is playing a key role in controlling the precise phase of cell-cycle events.
The significance of the RRE on p20 expression is even greater than just phase regulation because RRE deletion completely reverses the response of this gene to sustained light exposure. Expression of p20 is normally repressed to minimal levels in LL but in the absence of the RRE, light now increases p20 expression to near-maximal levels (Fig. 3 C and E). Furthermore, when the RRE mutation is combined with deletion of the three E′ boxes, rhythmicity is completely abolished and an increase in the basal expression levels is observed (Fig. 3 C and F). This is interesting because deletion of the E′ boxes or RRE alone does not abolish rhythmicity, a result that strongly supports the idea that a critical interaction occurs between the proteins that bind to these clock-controlled elements to generate p20 oscillations. Regulation of the RRE in the p20 promoter not only distinguishes its regulation from that of p21, but also clearly places this regulatory step at the heart of cell-cycle control by this rhythmic factor.
So what upstream proteins could be binding to and regulating this key promoter element? The RRE is a DNA sequence recognized by both transcriptional activators (retinoid-related orphan receptors) and transcriptional repressors (Rev-Erbs). The RRE deletion study shows a loss of repression for p20 expression (Fig. 3E), which suggests that the RRE is primarily acting as a binding site for transcriptional repressors, strongly implicating a role for Rev-Erbs in this process. Consequently, we examined the expression levels for all five zebrafish Rev-Erbs (α, βA, βB, γA, and γB) in our cell line system. Among the five Rev-Erbs analyzed, only Rev-ErbβA and Rev-ErbβB were rhythmically expressed, with the latter being the most abundant (Fig. 3G). In contrast, in the developing zebrafish embryo, Rev-Erbα is the most strongly rhythmic as well as the most abundant transcript from day 4 of development onward (Fig. 3H). Importantly, it is also at this developmental stage when the p21 and p20 phase difference becomes apparent (Fig. 2B), which supports the idea that Rev-Erbα is the essential factor for p20 repression and circadian timing in zebrafish larvae. Furthermore, electrophoretic mobility shift assays show that mouse Rev-Erbα binds directly to a p20 promoter fragment containing the RRE, but not one lacking this sequence (Fig. S2). Therefore, circadian rhythms in Rev-Erb expression, acting through the RRE in the p20 promoter, appear to be essential for the specific, timed regulation of this unique cell-cycle regulator.
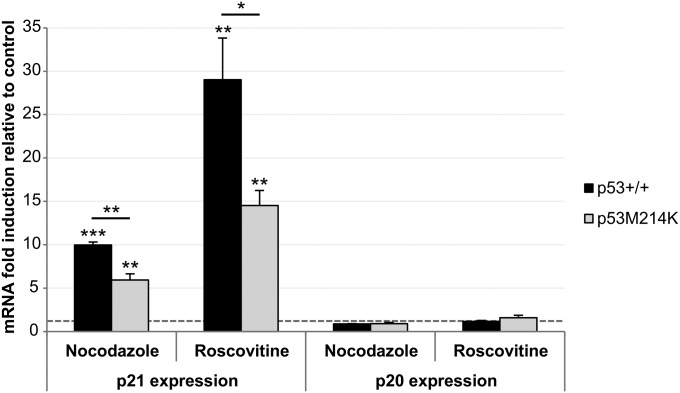
Transcriptional regulation of p20 differs from that of p21, and consequently, the daily timing of its peak expression is also different. However, are there other key aspects of their regulation that differ between these two cell-cycle factors? One important regulator of p21 is the tumor suppressor protein p53, which activates p21 expression in response to cellular stresses (16). Is p20 similarly regulated by p53? To examine this issue, we treated zebrafish embryos with cell cycle–arresting drugs (nocodazole and roscovitine) known to induce p53 expression (17, 18) and assessed p21 and p20 expression levels. A 16-h treatment during zebrafish development led to a significant induction of p53 expression by both drugs (Fig. S3). As expected, p21 expression is also markedly induced by both treatments, and this induction is, at least in part, dependent on p53, given that induction in p53 mutant zebrafish (19) is significantly reduced (Fig. 4). Remarkably, however, p20 levels did not change during either drug treatment (Fig. 4), showing that, unlike p21, the regulation of p20 expression is p53-independent. This result not only distinguishes the regulation of p20 from p21 even further, but also suggests that certain aspects of their function are likely to differ. It would appear from these results that, in contrast to p21, p20 does not play a role in stress-regulated pathways that influence the cell cycle.
Fig. 4.
p20 expression is p53-independent. qPCR analysis of p21 and p20 expression in 52 hpf p53+/+ (AB) and p53M214K embryos treated for 16-h with nocodazole or roscovitine. Bars represent fold-change in expression compared with untreated controls (horizontal dashed line). *P < 0.05, **P < 0.01, ***P < 0.001.
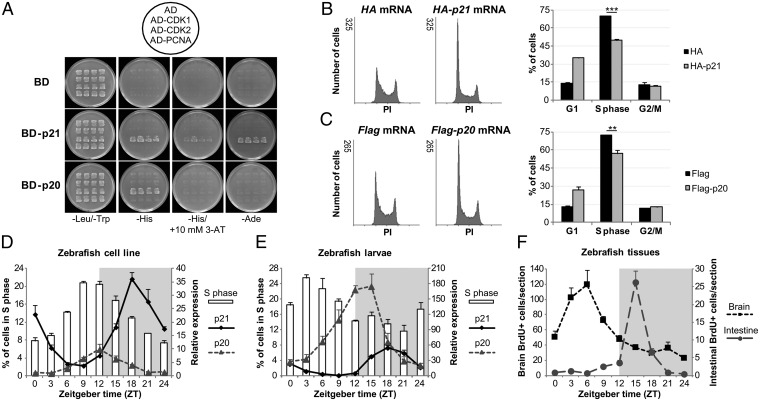
What then is the function of p20 in cell-cycle regulation? The CDK inhibitors function through key protein interactions with other cell-cycle proteins (20), such as CDK1 and CDK2, involved in the regulation of G2/M and G1/S transitions, respectively, and PCNA, an essential DNA replication factor. To explore the protein interactions of both p20 and p21, we performed a series of yeast two-hybrid assays. Neither protein showed significant binding to either CDK1 or PCNA, as revealed by the absence of yeast growth in selective media (Fig. 5A). Conversely, both p21 and p20 interacted with CDK2, although with different affinities. Colony growth was observed for yeast transformed with p21/CDK2 interaction plasmids in all three selective media, whereas p20/CDK2 transformants did not grow on the two most restrictive conditions (−histidine + 3-aminotriazole and −adenine) (Fig. 5A). These results indicate a stronger interaction between p21 and CDK2 than between p20 and CDK2, which could be due to differences within and/or near the CDK-binding domains of p20 and p21 proteins (Fig. 1).
Fig. 5.
p20 regulates circadian cell-cycle timing by inhibiting the G1/S transition. (A) Yeast two-hybrid analysis of p21 and p20 interactions with CDK1, CDK2, and PCNA. Four independent transformants from each plasmid combination were selected on −Leu/−Trp media. Protein interactions were assayed on –His, −His/+10 mM 3-AT, and –Ade media. (B) Representative histograms of DNA content from embryos at 70–80% epiboly injected with 60 pg/embryo of HA or HA-p21 mRNA at the single-cell stage (Left). Cell-cycle distribution of the same embryo samples determined by the Watson Pragmatic Model (Right). (C) Representative histograms of DNA content from embryos at 70–80% epiboly injected with 40 pg/embryo of Flag or Flag-p20 mRNA at the single-cell stage (Left). Cell-cycle distribution of the same embryo samples determined by the Watson Pragmatic Model (Right). **P < 0.01, ***P < 0.001. qPCR analysis of p21 and p20 expression plotted against the percentage of cells in S phase in PAC2 cells (D) and in 6-d-old larvae (E). (F) Quantification of BrdU-positive cells present in cross-sections of the hindbrain and the intestinal bulb of 6-d-old larvae. For each time point, three independent larvae were analyzed for a total of 12 sections. White and gray backgrounds represent light and dark phases, respectively.
The yeast two-hybrid results suggest that p20 might be involved in the regulation of G1/S transition, the same as the expected role for p21. To test this hypothesis, we injected p20 or p21 mRNA into single-cell stage zebrafish embryos, dissociated them at 70–80% epiboly and stained the cells with propidium iodide (PI) for cell-cycle analysis based on DNA content. The advantage of working at this early developmental time is that most cells in the embryo are actively dividing, a fact that allows us to better evaluate p20 function during the cell cycle. As expected for a G1/S check point regulator, p21-injected embryos exhibited a significant increase in the percentage of cells in G1 and a concomitant decrease in S-phase cells relative to control-injected embryos. No change was registered in the percentage of G2/M cells (Fig. 5B). Significantly, the p20-injected embryos showed a similar DNA profile to p21-injected embryos (Fig. 5C), demonstrating that p20 is inhibiting the G1/S transition. This result, of course, fits well with p20 protein binding to CDK2, which has a critical role in the G1/S transition. Nevertheless, it is not the result we were expecting based on the differential timing of p20 and p21 expression. The 6-h phase difference in gene expression had led us to speculate that p20 and p21 might act at different points in the cell cycle, which does not seem to be the case. So how are p20 and p21 acting in a clock-dependent way to regulate cell-cycle timing?
Both cell-cycle regulators, p20 and p21, are direct clock-controlled inhibitors of the G1/S transition. However, they possess very clear differences in the nature of their transcriptional regulation, which leads to a clear time-of-day difference in their expression pattern. The action of Rev-Erb on RRE elements in the p20 promoter generates a clear, 6-h difference in peak timing. However, not only are they temporally differentially expressed, but also show differences in their tissue- or cell-type level of expression. For example, p21 is the predominant transcript in the PAC2 zebrafish cell line; consequently, S phase is timed to occur relative to the expression of this gene, with a peak at ZT9–12, corresponding to the trough of p21 expression (Fig. 5D). In contrast, in the developing zebrafish embryo, p20 is the predominant transcript, which then controls the peak in S phase to occur 6 h earlier (ZT3), the time corresponding to the phase advance of p20 expression relative to p21 (Fig. 5E). This provides an interesting mechanism to regulate the circadian timing of the cell cycle differentially within different tissues of the organism. In fact, when we examined the cell-cycle profile in specific larval tissues, we found a clear phase difference in the timing of S phase in the brain compared with the intestine (Fig. 5F and Fig. S4). Importantly, these circadian rhythms in DNA synthesis in brain and intestine show an inverse correlation with the expression levels of p20 and p21, respectively. Thus, to restrict S phase to the early morning, p20 will be the “regulator of choice.” However, if S phase is more desirable in the late day/early evening, then it will be regulated by p21. In mammals, the cell cycle, and S phase in particular, is also set to occur at different times of day in different tissues (21), but at this time, nothing is known about the possible mechanism that may underlie this process. In zebrafish, the problem of timing the cell cycle to specific times of day in different tissues or cell types has been solved by using multiple, directly clock-controlled cell-cycle regulators p20 and p21.
Materials and Methods
Bioinformatic and Phylogenetic Analysis.
The zebrafish cdkn1a/p21 gene (Ensembl ID ENSDARG00000076554) was used in a bioinformatic search for orthologous sequences in Ensembl, particularly within regulatory regions, using Regulatory Sequence Analysis Tools (http://rsat.ulb.ac.be). Among the DNA sequences initially retrieved were those of a cdkn1 gene in stickleback (Ensembl ID ENSGACG00000018951), medaka (Ensembl ID ENSORLG00000011013), and chicken (Ensembl ID ENSGALG00000019256), often incorrectly annotated as cdkn1a. A zebrafish ortholog was then identified in Ensembl (Ensembl ID ENSDARG00000088020) and designated cdkn1d or p20 (NCBI RefSeq XR_082680). DNA sequences encompassing the putative coding regions of both p20 and p21 were amplified by RT-PCR from zebrafish, medaka, and chicken embryo RNA. These sequences have been deposited into GenBank [accession nos. KC818433 (zebrafish p20), KC818434 (zebrafish p21), KC818435 (medaka p20), KC818436 (medaka p21), KC818437 (chicken p20), and KC818438 (chicken p21)] (SI Materials and Methods).
Zebrafish Cell Lines.
Zebrafish PAC2 cells were cultured in Leibovitz’s L-15 medium (Gibco) containing 15% (vol/vol) FBS (Biochrom AG), 50 U/mL penicillin/streptomycin (Gibco), and 50 µg/mL gentamicin (Gibco) as previously described (13). Cells were plated (250,000 cells per well) in triplicate wells of a six-well dish (Greiner) and incubated in a large volume water bath at 28 °C on an LD cycle (12L:12D) for 6 d and then transferred into DD for the seventh day.
Zebrafish Lines.
Zebrafish lines AB/TL, AB, and p53M214K were used in this study and maintained under standard conditions in the University College London fish facility at 28.5 °C on an LD cycle (14L:10D). All experiments have been conducted in accordance with the United Kingdom Animals (Scientific Procedures) Act 1986. Zebrafish embryos were collected and transferred into 25-cm2 tissue culture flasks (Greiner) (20–50 embryos each) in E3 embryo medium. Flasks of embryos were incubated in a large-volume water bath at 28 °C on an LD cycle (12L:12D) for up to 6 d. The AB/TL line was used for all experiments unless otherwise stated.
Quantitative PCR.
Zebrafish PAC2 cells, embryos/larvae, or adult tissues were harvested at the indicated ZT or circadian time in TRIzol (Ambion). Total RNA was isolated following the manufacturer’s instructions (Ambion), and cDNA was synthesized from 1 to 2 µg of RNA using SuperScript II Reverse Transcriptase (Invitrogen). qPCR was carried out as described (4). ΔCt was calculated using ribosomal protein L13α (RPL13α) or elongation factor 1α (ef1α) as a reference gene. Relative expression levels were then plotted by determining ΔΔCt by normalizing to a single sample with a high ΔCt value. Primer sequences and amplification efficiencies are listed in Table S1.
Whole Mount in Situ Hybridization.
In situ hybridization was performed according to standard protocols (22), with minor modifications (SI Materials and Methods).
Promoter Analysis.
Stable luminescent zebrafish cell lines were generated for promoter analysis. WT and mutant p21 and p20 promoters were fused to the firefly luciferase gene and plasmids were electroporated into PAC2 cells using the Neon transfection system (Invitrogen) (SI Materials and Methods).
Bioluminescence Assays.
Luminescent cell lines were plated in medium supplemented with 0.5 mM beetle luciferin (Promega). Bioluminescence was monitored on a Packard TopCount NXT scintillation counter (SI Materials and Methods).
Drug Treatments.
Zebrafish p53+/+ (AB line) and p53M214K (19) embryos were treated with 0.5 µg/mL nocodazole (Sigma-Aldrich) or 17.75 µg/mL roscovitine (Sigma-Aldrich) for 16 h (SI Materials and Methods).
Yeast Two-Hybrid Assays.
The full-length coding regions of zebrafish p21, p20, CDK1, CDK2, and PCNA were used for yeast two-hybrid assays (SI Materials and Methods).
Microinjections.
The zebrafish p21 and p20 coding regions were subcloned in-frame into plasmids pCS2HA and pCS2Flag (Addgene), respectively. Capped mRNA was synthesized with the SP6 mMessage mMachine (Ambion). One-cell stage embryos were injected with the indicated amounts of mRNA. Embryos at 70–80% epiboly were dissociated with 0.05% trypsin-EDTA (Gibco), and the cell suspension filtered through a 70-µm cell strainer (BD Biosciences).
Flow Cytometry.
For cell-cycle analysis, PAC2 cells or dissociated embryos/larvae were fixed in cold 70% (vol/vol) ethanol, treated with 100 µg/mL RNase A (Sigma-Aldrich), and stained with 50 µg/mL PI (Sigma-Aldrich). Samples were then analyzed on a FACSCalibur (BD Biosciences) in the FACS facility at Cancer Research U.K. (Lincoln’s Inn Fields, London, U.K.). Cell-cycle distribution was analyzed using FlowJo (Tree Star Inc.) by fitting the histogram of DNA content to the Watson Pragmatic Model (23).
BrdU Labeling.
Day 6 larvae were incubated at the indicated ZT for 40 min in a BrdU solution [10 mM BrdU and 5% (vol/vol) DMSO in E3 medium] at 28 °C. Larvae were fixed in 4% (wt/vol) paraformaldehyde (PFA) immediately after the BrdU exposure and cryoprotected in 30% (wt/vol) sucrose in 0.1 M phosphate buffer at 4 °C overnight. Then, larvae were embedded in Tissue-Tek OCT Compound (Sakura) and rapidly frozen on dry ice. 5 µm-thick cross-sections were mounted in SuperFrost Plus slides (Thermo Scientific) and stored at –80 °C. BrdU immunocytochemistry was performed as previously described (24), with rat anti-BrdU (Abcam) and goat anti-rat conjugated to Alexa 488 (Molecular Probes) antibodies. BrdU-positive cells were quantified on a Leica DMLB fluorescence microscope (Leica Microsystems).
Statistical Analysis.
The data in this study are presented as the mean ± SEM (n ≥ 3). Statistical significance was determined by a two-tailed Student t test.
Supplementary Material
Acknowledgments
We thank D. Davies and the FACS facility at the London Research Institute for their technical assistance and expertise. This work was supported by Biotechnology and Biological Sciences Research Council (BBSRC) and ERASysBio+ Grant BB/I004823/1 and doctoral fellowship SFRH/BD/63863/2009 from Fundação para a Ciência e a Tecnologia (to R.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. KC818433 (zebrafish p20), KC818434 (zebrafish p21), KC818435 (medaka p20), KC818436 (medaka p21), KC818437 (chicken p20), and KC818438 (chicken p21)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217912110/-/DCSupplemental.
References
- 1.Johnson CH. Circadian clocks and cell division: What’s the pacemaker? Cell Cycle. 2010;9(19):3864–3873. doi: 10.4161/cc.9.19.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dekens MP, et al. Light regulates the cell cycle in zebrafish. Curr Biol. 2003;13(23):2051–2057. doi: 10.1016/j.cub.2003.10.022. [DOI] [PubMed] [Google Scholar]
- 3.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302(5643):255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 4.Tamai TK, Young LC, Cox CA, Whitmore D. Light acts on the zebrafish circadian clock to suppress rhythmic mitosis and cell proliferation. J Biol Rhythms. 2012;27(3):226–236. doi: 10.1177/0748730412440861. [DOI] [PubMed] [Google Scholar]
- 5.Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12(7):540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 6.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 7.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 8.Lévi F, Filipski E, Iurisci I, Li XM, Innominato P. Cross-talks between circadian timing system and cell division cycle determine cancer biology and therapeutics. Cold Spring Harb Symp Quant Biol. 2007;72:465–475. doi: 10.1101/sqb.2007.72.030. [DOI] [PubMed] [Google Scholar]
- 9.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 10.Lin J, Reichner C, Wu X, Levine AJ. Analysis of wild-type and mutant p21WAF-1 gene activities. Mol Cell Biol. 1996;16(4):1786–1793. doi: 10.1128/mcb.16.4.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakanishi M, Robetorye RS, Adami GR, Pereira-Smith OM, Smith JR. Identification of the active region of the DNA synthesis inhibitory gene p21Sdi1/CIP1/WAF1. EMBO J. 1995;14(3):555–563. doi: 10.1002/j.1460-2075.1995.tb07031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warbrick E, Lane DP, Glover DM, Cox LS. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol. 1995;5(3):275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- 13.Whitmore D, Foulkes NS, Sassone-Corsi P. Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature. 2000;404(6773):87–91. doi: 10.1038/35003589. [DOI] [PubMed] [Google Scholar]
- 14.Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37(2):187–192. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- 15.Tamai TK, Young LC, Whitmore D. Light signaling to the zebrafish circadian clock by Cryptochrome 1a. Proc Natl Acad Sci USA. 2007;104(37):14712–14717. doi: 10.1073/pnas.0704588104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 17.Tishler RB, Lamppu DM, Park S, Price BD. Microtubule-active drugs taxol, vinblastine, and nocodazole increase the levels of transcriptionally active p53. Cancer Res. 1995;55(24):6021–6025. [PubMed] [Google Scholar]
- 18.Lee KC, et al. Detection of the p53 response in zebrafish embryos using new monoclonal antibodies. Oncogene. 2008;27(5):629–640. doi: 10.1038/sj.onc.1210695. [DOI] [PubMed] [Google Scholar]
- 19.Berghmans S, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA. 2005;102(2):407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: Cell cycle regulators and beyond. Dev Cell. 2008;14(2):159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Scheving LE, Tsai TH, Scheving LA. Chronobiology of the intestinal tract of the mouse. Am J Anat. 1983;168(4):433–465. doi: 10.1002/aja.1001680405. [DOI] [PubMed] [Google Scholar]
- 22.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3(1):59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 23.Watson JV, Chambers SH, Smith PJ. A pragmatic approach to the analysis of DNA histograms with a definable G1 peak. Cytometry. 1987;8(1):1–8. doi: 10.1002/cyto.990080101. [DOI] [PubMed] [Google Scholar]
- 24.Kustermann S, Hildebrandt H, Bolz S, Dengler K, Kohler K. Genesis of rods in the zebrafish retina occurs in a microenvironment provided by polysialic acid-expressing Müller glia. J Comp Neurol. 2010;518(5):636–646. doi: 10.1002/cne.22232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.