Abstract
Background
Non-coding small RNAs, ranging from 20 to 30 nucleotides in length, mediate the regulation of gene expression and play important roles in many biological processes. One class of small RNAs, microRNAs (miRNAs), are highly conserved across taxa and mediate the regulation of the chromatin state and the post-transcriptional regulation of messenger RNA (mRNA). Another class of small RNAs is the Piwi-interacting RNAs, which play important roles in the silencing of transposons and other functional genes. Although the biological functions of the different small RNAs have been elucidated in several laboratory animals, little is known regarding naturally occurring variation in small RNA transcriptomes among closely related species.
Results
We employed next-generation sequencing technology to compare the expression profiles of brain small RNAs between sympatric species of the Japanese threespine stickleback (Gasterosteus aculeatus). We identified several small RNAs that were differentially expressed between sympatric Pacific Ocean and Japan Sea sticklebacks. Potential targets of several small RNAs were identified as repetitive sequences. Female-biased miRNA expression from the old X chromosome was also observed, and it was attributed to the degeneration of the Y chromosome.
Conclusions
Our results suggest that expression patterns of small RNA can differ between incipient species and may be a potential mechanism underlying differential mRNA expression and transposon activity.
Keywords: Stickleback, Speciation, Variation, miRNA, piRNA, Ecology, Variation
Background
Recent progress in the development of genomic techniques, including next generation sequencers, has greatly facilitated transcriptome analysis of ecologically important animals to reveal variations in mRNA expression patterns among closely related species and ecotypes within species [1-3]. Divergence in mRNA expression patterns is known to contribute to phenotypic evolution [4,5], although amino acid alterations in proteins are also important [6]. While a great deal is known about variation in mRNA expression profiles, information regarding naturally occurring variation in the expression patterns of small RNAs is limited, except for a few cases in plants [7,8] and cichlids [9].
Non-coding small RNAs, ranging from 20 to 30 nucleotides in length, mediate the regulation of gene expression [10-13]. The members of one class of small RNAs, microRNAs (miRNAs), are typically 20–24 nucleotides long and are highly conserved across diverse taxa [11,12]. miRNA post-transcriptionally regulates messenger RNA (mRNA). A miRNA interacts with ten to hundreds of target mRNAs to induce degradation or suppress translation [12]. Another function of miRNA is epigenetic modification of genomic DNA: miRNAs interact with target DNAs to alter the chromatin state and suppress mRNA transcription [14]. miRNAs comprise more than 1% of animal genes [15,16], suggesting that they play important roles in many biological processes. Recent functional studies in laboratory model animals such as mice, flies, and nematodes have demonstrated that miRNAs are important for regulating development, growth, pathogen resistance, and neural functions [11,12,17-19].
Another class of small RNAs is the Piwi-interacting RNAs (piRNAs), which are typically 24–32 nucleotides long and interact with Piwi proteins to suppress the expression of transposons and other functional genes [13,20]. piRNAs often possess uridine at the 5’-end (5′U) [13,20]. piRNAs are expressed from intergenic repetitive elements, active transposons, and piRNA clusters. Importantly, piRNAs may contribute to hybrid dysgenesis [21,22]. For example, some Drosophila strains contain transposons as well as piRNAs that inhibit transposon activity, whereas other strains lack both transposons and inhibitory piRNAs. Because piRNAs are maternally transmitted, hybrid progeny resulting from a cross between a mother lacking both transposons and piRNAs and a father possessing both will inherit the transposons, but not the inhibitory piRNAs. This abnormal activity of transposons in the germ line is likely to result in sterility [21,22]. Thus, maternally transmitted piRNAs can explain why hybrid abnormalities are observed in only one direction of the inter-strain crosses. piRNAs are expressed not only in the gonads, but also in the brain, and they may be involved in the regulation of neuronal functions [23-25]. Compared with miRNAs, piRNAs are less well conserved across taxa. Yet another class of small RNAs, endogenous small interfering RNAs (endo-siRNAs), are usually 21 nucleotides and have been found in some taxa, including nematodes [26], flies [27-29], and mammals [30,31], but it has not been well characterized in other animals.
Evolutionary genetic studies examining small RNAs are important for several reasons. First, genome-wide allele-specific mRNA expression analyses have revealed that both cis- and trans-regulatory changes contribute to differential expression of mRNAs among closely related species [32-34]. Small RNAs can act as trans-regulatory factors, which contribute to differential mRNA expression [35]. Additionally, cis-regulatory changes may include mutations at the target sites of small RNAs [36]; for example, SNPs and insertion-deletion polymorphisms were identified within miRNA-binding sites of 3’-untranslated regions [37,38]. Variations in small RNA transcriptomes and sequences were found to be associated with phenotypic variation in humans and laboratory animals. For example, miRNA and miRNA target site polymorphisms and mutations have been found in humans and are associated with disease susceptibility [39-42]. Polymorphism in a miRNA target site is associated with variation of muscularity in pigs [43]. Second, small RNAs regulate translation of mRNAs. Therefore, transcriptome studies of mRNA alone can overlook the divergence in the total outcome of gene expression among species. Third, piRNAs may contribute to hybrid abnormalities (see above), but generalities regarding the roles of piRNA in different types of hybrid abnormalities remain unclear.
In the present study, we compared brain small RNA transcriptomes between incipient species of the threespine stickleback (Gasterosteus aculeatus). The threespine stickleback is a good model for linking ecological and genetic studies of adaptive evolution and speciation [44-52]. The threespine stickleback has undergone tremendous diversification over the past few million years [44,45,49]. Evolutionary diversification within the stickleback species complex led to a speciation continuum, which ranges from populations with interspecific phenotypic polymorphism to strong divergence with near-complete reproductive isolation [44,53]. Recent genetic studies have revealed that differences in the expression of genes involved in morphological development [54,55], physiology [56,57], and immune function [58] may underlie adaptive divergence among populations or species. Sex bias of the mRNA transcriptome has also been investigated, and genes located on sex chromosomes were found to be female-biased, likely owing to Y-chromosome degeneration and lack of dosage compensation [59]. However, transcriptome analysis of small RNAs has not yet been conducted in any stickleback system.
This study focused on Japanese threespine stickleback species pairs, including a Pacific Ocean form and a Japan Sea form. These sticklebacks diverged during a period of geographical isolation between the Sea of Japan and the Pacific Ocean approximately 1.5–2 million years ago [60,61]. Currently, they are sympatric in eastern Hokkaido, but they are reproductively isolated with a very low level of hybridization [60-62]. In the Japan Sea form, a chromosomal fusion occurred between linkage group (LG) 9 and the ancestral Y chromosome (LG 19), resulting in the evolution of the X1X2Y multiple sex chromosome system [62]. In contrast, the Pacific Ocean form has a simple XY sex chromosome system [62]. Previously, we found that the Pacific Ocean and Japan Sea forms diverge in male courtship behaviors and female mate choice behaviors, contributing to behavioral isolation between these two forms [60,62,63]. Furthermore, we found that divergence in the intensity of courtship behaviour, which is important for mate choice, mapped to a neo-sex chromosome (LG 9).
To better understand the genetic mechanisms affecting behavioral differences between this Japanese stickleback species pair, it is essential to understand divergence in small RNA transcriptomes of the brain. Both miRNAs and piRNAs play important roles in a diverse array of neuronal functions such as neuronal differentiation, neural stem cell renewal, neuronal outgrowth, and dendritic spine morphogenesis [23,24,64]. Furthermore, variation in miRNA expression patterns in the brain may contribute to behavioral differences among laboratory mouse strains [65]. Additionally, in the Japanese stickleback system, courtship dysfunction may exist in hybrids because a substantial number of hybrids did not perform any courtship behavior in a previous experiment (Supplementary data in [61]). Therefore, it is necessary to examine whether small RNAs, especially piRNAs, which may be regulating transposon activity in the brain, affect hybrid behavior. Here, we characterized the divergence in small RNA transcriptomes in the brain between the species pairs of Japanese threespine stickleback.
Results and discussion
miRNA transcriptome
We conducted small RNA sequencing of four males and four females from both the Pacific Ocean and the Japan Sea forms using the Illumina Genome Analyzer IIx. After quality control of the sequence reads, data was collected from 26.2 ± 3.3 (mean ± SD) million reads per fish (Additional file 1: Table S1). Most of these reads (23.7 ± 2.9 million reads; 86.9–94.6% of the total reads) were located in the Ensembl stickleback miRNA database. More than 50% of these small RNAs were 22 nucleotides in length (Figure 1). In total, 1300 isoforms of miRNA were detected in the brain (924 in the Pacific Ocean males, 924 in the Pacific Ocean females, 916 in the Japan Sea males, and 884 in the Japan Sea females). miRNA expression profiles demonstrated that miRNAs homologous to mir21, mir100, let7, mir101, mir143, and mir9 were the most abundant in the stickleback brain, regardless of the species or sex (Figure 2). Most other miRNAs were expressed at relatively low levels (less than 3% of the annotated reads).
Figure 1.
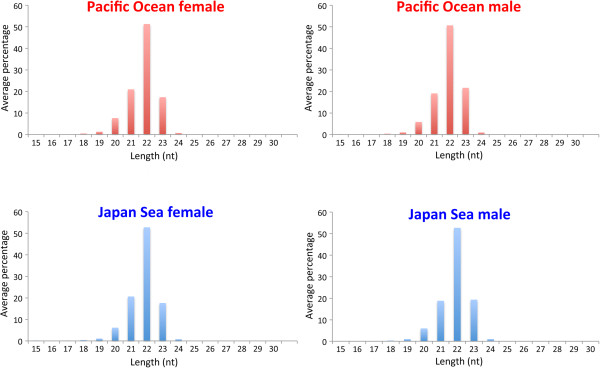
Size distribution of stickleback brain miRNAs. The average of four individuals is shown for each group.
Figure 2.
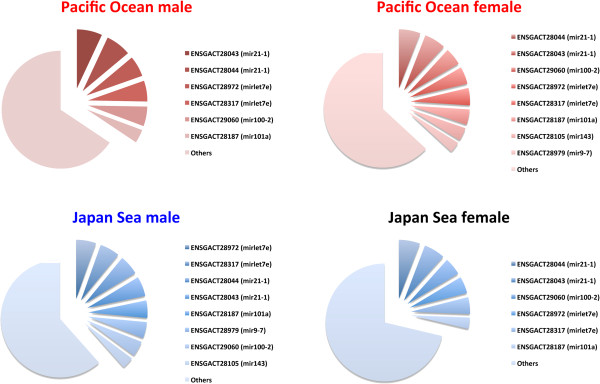
miRNA expression profile in threespine stickleback brains. The area indicates the fraction of read numbers of particular miRNAs among the total read number of annotated miRNAs. Only miRNAs whose expression is higher than 3% of all annotated reads are shown. The average of four individuals is shown for each group. Homologous zebrafish miRNA (blast, E < 10-3) are shown in parentheses.
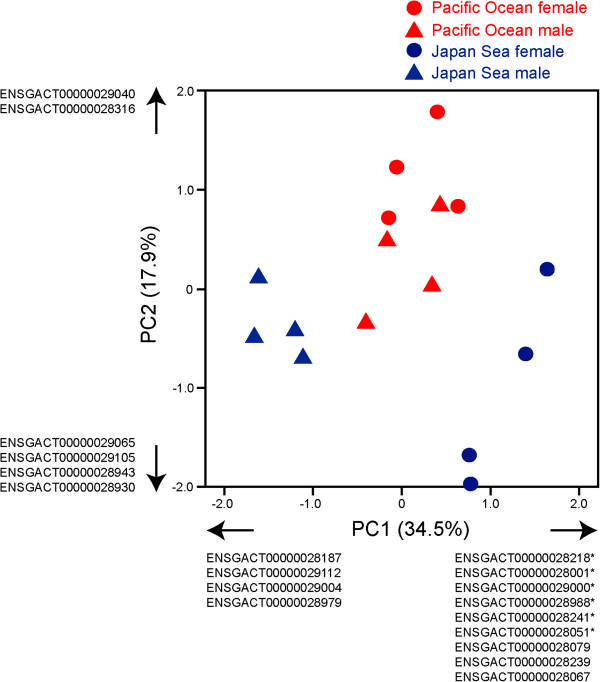
To elucidate the variation in the miRNA transcriptomes, we conducted principal component analysis (Figure 3; Additional file 2: Table S2). The miRNA transcriptomes of the Pacific Ocean and the Japan Sea forms make distinct clusters. Interestingly, the Japan Sea males and females make distinct clusters, whereas the miRNA transcriptomes of the Pacific Ocean males and females overlapped. These data suggest that the magnitude of sex differences of miRNA expression levels might differ between species.
Figure 3.
Principal component analysis (PCA). The first PC (PC1) and second PC (PC2) explained 34.5 and 17.9% of the variances, respectively. The loading components are shown in Table S2. The transcripts with loading component values larger than 0.75 and 0.70 for PC1 and PC2, respectively, are shown in the figure. The asterisks indicate the transcripts that were differentially expressed between the sexes by ANOVA with Bonferroni correction (Table 2).
We identified several miRNAs that were differentially expressed between species (Bonferroni correction of analysis of variance [ANOVA]; Tables 1 and 2). Although quantitative trait loci (QTL) mapping revealed that LG9 contained a courtship display QTL, no miRNAs expressed from LG9 showed significantly different expression levels between species after Bonferroni correction (Table 1). We identified a miRNA homologous to the zebrafish mir7 that was differentially expressed between the stickleback species. In mammals, mir7 expression levels in the brain can change after hyperosmolar stimuli [66] and regulate growth factor signalling pathways [67]. Another miRNA differentially expressed between species, mir30, may be involved in axon guidance [68].
Table 1.
miRNA differentially expressed between species
|
Transcript ID |
LG |
Start position |
Median reads per million (RPM) ± S.D. |
Zebrafish homolog |
|||
|---|---|---|---|---|---|---|---|
| Pacific Ocean males | Pacific Ocean females | Japan Sea males | Japan Sea females | ||||
| ENSGACT00000029029 |
I |
7684495 |
2852.3 (227.8) |
4343.0 (1716.7) |
10218.3 (1270.0) |
4026.2 (860.8) |
mir22a-1 |
| ENSGACT00000028961 |
III |
7312510 |
563.3 (78.4) |
1250.9 (684.9) |
3683.1 (408.8) |
1115.0 (176.9) |
mir7a-3 |
| ENSGACT00000028970 |
VI |
4231011 |
2465.3 (127.5) |
2270.1 (62.9) |
1858.6 (244.0) |
1966.8 (141.3) |
mir30c |
| ENSGACT00000029035 |
XI |
1952924 |
9700.4 (165.6) |
11004.3 (1503.8) |
14871.2 (475.2) |
12259.0 (332.2) |
mir152 |
| ENSGACT00000028984 |
XIII |
7256736 |
609.2 (80.5) |
1294.3 (679.4) |
3601.1 (393.0) |
1134.8 (168.2) |
mir7b |
| ENSGACT00000029072 |
XIX |
2936579 |
547.6 (75.4) |
1228.1 (681.4) |
3503.5 (381.2) |
1073.8 (169.6) |
mir7a-1 |
| ENSGACT00000029039 | XX | 15698389 | 568.3 (73.6) | 1245.5 (680.7) | 3560.3 (396.7) | 1099.6 (167.8) | mir7a-2 |
LG: linkage group.
Table 2.
ANOVA of miRNA
|
Transcript ID |
LG |
Species |
Sex |
Species X Sex |
|||
|---|---|---|---|---|---|---|---|
| F | P | F | P | F | P | ||
| ENSGACT00000029029 |
I |
34.2 |
<0.001 |
5.9 |
0.032 |
46.1 |
<0.001 |
| ENSGACT00000028961 |
III |
44.5 |
<0.001 |
8.8 |
0.012 |
39.6 |
<0.001 |
| ENSGACT00000028970 |
VI |
25.9 |
<0.001 |
0.1 |
0.782 |
2.9 |
0.115 |
| ENSGACT00000029035 |
XI |
57.6 |
<0.001 |
0.9 |
0.353 |
23.8 |
<0.001 |
| ENSGACT00000028984 |
XIII |
42.6 |
<0.001 |
8.7 |
0.012 |
39.8 |
<0.001 |
| ENSGACT00000029072 |
XIX |
41.2 |
<0.001 |
8.0 |
0.015 |
38.2 |
<0.001 |
| ENSGACT00000029039 |
XX |
42.1 |
<0.001 |
8.4 |
0.013 |
38.2 |
<0.001 |
| ENSGACT00000029075 |
IV |
4.6 |
0.053 |
50.3 |
<0.001 |
0.0 |
0.926 |
| ENSGACT00000028051 |
IV |
0 |
0.971 |
84 |
<0.001 |
29.2 |
<0.001 |
| ENSGACT00000028218 |
VII |
0 |
0.852 |
77.9 |
<0.001 |
26.1 |
<0.001 |
| ENSGACT00000028988 |
XII |
0 |
0.97 |
83.1 |
<0.001 |
28.8 |
<0.001 |
| ENSGACT00000028001 |
XVII |
0 |
0.956 |
76.5 |
<0.001 |
27.9 |
<0.001 |
| ENSGACT00000029000 |
XVII |
0 |
0.966 |
83.2 |
<0.001 |
28.9 |
<0.001 |
| ENSGACT00000029064 |
XIX |
5.1 |
0.043 |
51 |
<0.001 |
0.0 |
0.963 |
| ENSGACT00000028241 | XIX | 0 | 0.976 | 84.3 | <0.001 | 29.1 | <0.001 |
LG: linkage group.
Bold letters indicate significance even after Bonferroni correction (P < 0.0003).
Sex differences in the expression levels were identified for several miRNAs (Tables 2 and 3). Interestingly, all sex-biased miRNAs belonged to the let-7 family (Table 3) [69]. miRNAs of the let-7 family are highly conserved across taxa and are important during development [70,71]. Two sexually dimorphic miRNAs (ENSGACT00000028241 and ENSGACT00000029064) were expressed from LG19; both of these were female-biased. In species with an XY-sex chromosome system, suppression of recombination can lead to degeneration of the Y chromosome [72,73]. Unless dosage compensation mechanisms evolve, expression of genes located on the X-specific region becomes female-biased [74]. Therefore, we investigated the relationship between Y-degeneration and sex differences in miRNA expression levels. We found that expression of all miRNAs derived from the X-specific region (i.e. the counterpart region of the Y is likely degenerated) were female-biased, whereas expression of miRNA derived from the pseudoautosomal region was not necessarily female-biased (Figure 4), and log2 (fold difference between male and female) significantly differed between the pseudo-autosomal and the X-specific regions (Mann–Whitney U test, U = 7, Z = 814.5, P < 0.001, N = 17). These results suggest that Y chromosome degeneration may have a substantial impact not only on mRNA expression [59], but also on miRNA expression.
Table 3.
miRNA differentially expressed between males and females
|
Transcript ID |
LG |
Start position |
Median reads per million (RPM) ± S.D. |
Zebrafish homolog |
|||
|---|---|---|---|---|---|---|---|
| Pacific Ocean males | Pacific Ocean females | Japan Sea males | Japan Sea females | ||||
| ENSGACT00000029075 |
IV |
20044816 |
1606.0 (181.4) |
2616.6 (434.6) |
1421.9 (60.4) |
2339.2 (396.8) |
mirlet7i |
| ENSGACT00000028051 |
IV |
19861309 |
20839.3 (1126.5) |
22453.2 (1081.8) |
18010.6 (611.4) |
25558.9 (1427.5) |
mirlet7a-3 |
| ENSGACT00000028218 |
VII |
18314432 |
19933.6 (1139.6) |
21367.4 (1035.2) |
17101.4 (549.5) |
24133.6 (1426.5) |
mirlet7a-4 |
| ENSGACT00000028988 |
XII |
12165589 |
20768.4 (1137.2) |
22362.8 (1085.0) |
17924.8 (600.1) |
25445.9 (1434.9) |
mirlet7a-6 |
| ENSGACT00000028001 |
XVII |
11962085 |
20520.0 (1120.3) |
21925.0 (1071.1) |
17736.6 (610.0) |
24909.7 (1419.4) |
mirlet7a-1 |
| ENSGACT00000029000 |
XVII |
3393574 |
20777.3 (1135.5) |
22362.4 (1084.3) |
17935.0 (602.4) |
25456.6 (1431.4) |
mirlet7a-6 |
| ENSGACT00000029064 |
XIX |
4582643 |
1608.3 (184.2) |
2633.7 (436.6) |
1411.8 (66.8) |
2327.6 (395.7) |
mirlet7i |
| ENSGACT00000028241 | XIX | 4785031 | 20810.0 (1125.6) | 22433.4 (1083.4) | 17991.0 (609.0) | 25531.1 (1426.0) | mirlet7a-3 |
LG: linkage group.
Figure 4.
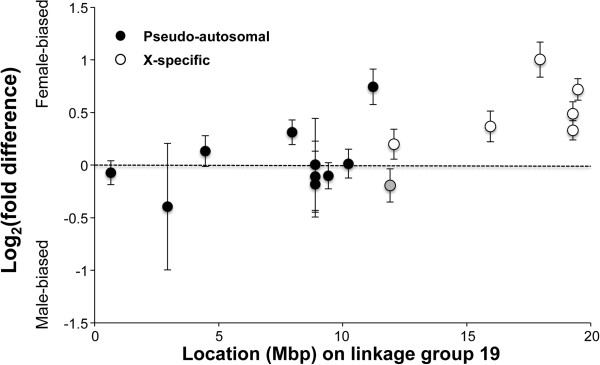
Female-biased expression of miRNA expressed from the non-recombining region of the X chromosome (linkage group 19). Black circles indicate small RNAs on the pseudoautosomal region, whereas white circles indicate small RNAs from the X-specific region. A small RNA, for which it was not clear whether the RNA was located on the pseudoautosomal or X-specific region, is indicated by a grey circle. For statistical analysis, the grey circle was excluded. Data from the Japan Sea and Pacific Ocean fish were pooled. Because the order of the LG19 sequence assembly on the ensembl is inverted after 3.822 Mbp, physical locations on LG19 followed [62]. Error bars indicate S.E.
Because miRNAs can target many mRNAs [75,76], divergence in miRNA expression patterns may have substantial effects on the expression patterns of many mRNAs. Further experimental studies examining the roles of small RNAs in fish will be necessary to understand the functional effects of the miRNA transcriptome variation. This would be possible by developing either transgenic fish specifically overexpressing small RNAs or small RNA-deficient knockout fish [77-81].
Small RNAs homologous to repetitive sequences
Small RNAs with no matches in the stickleback non-coding RNA database were further analyzed. A histogram of the read length of these small RNAs revealed two peaks, with one peak at 22 nucleotides and the other at 27–29 nucleotides (Figure 5). The fraction of large-sized small RNAs may contain piRNAs, whereas the other fraction may correspond to novel miRNAs and/or endo-siRNAs. A histogram of reads per million (RPM) of the unidentified small RNAs revealed that most were expressed at low levels, with only a few expressed at high levels (Figure 6). Thirty-one novel small RNAs expressed at relatively high abundance (mean RPM > 50 in at least one of the four groups) could be classified into 17 isoforms on the basis of sequence identity (Additional file 3: Table S3).
Figure 5.
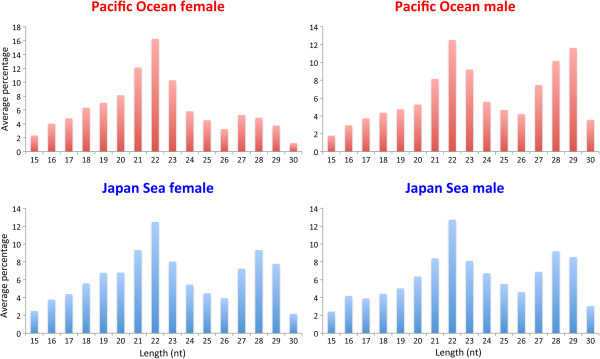
Size distribution of non-annotated small RNAs. The average of four individuals is shown for each group.
Figure 6.

Histogram of reads per million (RPM) of non-annotated small RNAs. The average of four individuals is shown for each group.
A homology search against the piRNABank database revealed that some of these were similar to previously reported piRNAs (Table 4). Additionally, seven isoforms contained 5′U (T in Table 4), which is often found in previously reported piRNAs. However, compared with miRNAs, piRNAs are less conserved across taxa. Therefore, we examined whether these 17 small RNAs showed homology to repetitive sequences such as transposons. For all 17 isoforms, multiple homologous sites were identified in the stickleback genome (Table 5). Most of these potential small RNA target sites overlapped with repetitive sequences (Tables 5 and Additional file 4: Table S4). Four isoforms (iso-smRNA6, 9, 12, and 13) showed a high level of homology to the non-long terminal repeat (non-LTR) retrotransposon. One isoform (iso-smRNA5) was homologous to the LTR retrotransposon and two isoforms (iso-smRNA11 and 17) were homologous to ERV1-type retrovirus genes. One (iso-smRNA8) was similar to a DNA transposon. Because we did not confirm that these sequences actually bind to Piwi proteins, we could not exclude the possibility that the identified sequences are not piRNAs. However, all of these were longer than 24 nucleotides, and some of them contained the 5′U. Recent studies have demonstrated that retrotransposons are active in the adult mammalian brain and are thought to increase neuronal function diversity [82]. Therefore, regardless of whether these sequences are piRNAs, it is interesting that small RNAs highly homologous to transposons are expressed in the stickleback brain.
Table 4.
Small RNAs with nucleotide lengths larger than 25 nt
| smallRNA ID | Sequence | Length (nt) | Hit in the piRNABank | E-value |
|---|---|---|---|---|
| iso_smRNA1 |
CCCTCGGTTCTGGCGTCAAGCGGGCCGGC |
29 |
No hit |
- |
| iso_smRNA2 |
GCATGTGGTTCAGTGGTAGAATTCTCG |
27 |
hsa_piR_018570 |
0.0053 |
| iso_smRNA3 |
GCATTGGTGGTTCAGTGGTAGAATTCTCGC |
30 |
dr_piR_0029993 |
0.0000065 |
| iso_smRNA4 |
GCATTGTGGTTCAGTGGTAGAATTCTCGCC |
30 |
hsa_piR_018570 |
0.00065 |
| iso_smRNA5 |
GCCCGGCTAGCTCAGTCGGTAGAGCATGA |
29 |
hsa_piR_000794 |
0.000017 |
| iso_smRNA6 |
GGGTTCGATTCCCGGTCAGGGAACCA |
26 |
No hit |
- |
| iso_smRNA7 |
GGTTCCATGGTGTAATGGTTAGCACTCTG |
29 |
hsa_piR_020582 |
0.000019 |
| iso_smRNA8 |
GGTTCTATGGTGTAATGGTTAGCACTCTG |
29 |
hsa_piR_020582 |
0.00011 |
| iso_smRNA9 |
GTTGTCGTGGCCGAGTGGTTAAGGCAATG |
29 |
hsa_piR_015249 |
0.0054 |
| iso_smRNA10 |
GTTTCCGTAGTGTAGTGGTTATCACGTTCG |
30 |
rno_piR_005901 |
0.0000065 |
| iso_smRNA11 |
TCCCATATGGTCTAGCGGTTAGGATTCCT |
29 |
dr_piR_0027014 |
0.018 |
| iso_smRNA12 |
TCCCTGGTGGTCTAGTGGTTAGGATTCGGC |
30 |
ona_piR_166322 |
0.0000065 |
| iso_smRNA13 |
TCCCTGTGGTCTAGTGGTTAGGATTCGGCG |
30 |
ona_piR_166322 |
0.00049 |
| iso_smRNA14 |
TCCTCGTATAGTGGACAGTATCTCCGCC |
28 |
No hit |
- |
| iso_smRNA15 |
TGAAAGACAACTCTTAGCGGTGGATC |
26 |
No hit |
- |
| iso_smRNA16 |
TGCGACCTCAGATCAGACGAGACAACCC |
28 |
dr_piR_0026826 |
0.0056 |
| iso_smRNA17 | TGGCTTCCTAAGCCAGGGATTGTGGG | 26 | No hit | - |
E-value smaller than 0.05 is shown here.
Table 5.
Characterization of small RNAs with high homology to repetitive sequences
|
smallRNA ID |
Length (nt) |
Blast hits (E <10-4) |
Potential target repeat |
Description |
Median reads per million (RPM) ± S.D of the longest isoform |
|||
|---|---|---|---|---|---|---|---|---|
| Pacific Ocean male | Pacific Ocean female | Japan Sea male | Japan Sea female | |||||
| iso_smRNA1 |
29 |
scaffolds(7) |
LSU-rRNA_Mfr |
rRNA |
72.6 (17.5) |
56.9 (43.6) |
58.9 (11.4) |
22.6 (9.8) |
| iso_smRNA2 |
27 |
LG1(12) |
tRNAGlyGGC_CB |
tRNA |
64.2 (20.9) |
51.9 (71.8) |
56.9 (101.1) |
25.5 (13.7) |
| iso_smRNA3 |
30 |
LG1(12), LG12(1) |
tRNAGlyGGC_CB |
tRNA |
52.6 (7.5) |
34.4 (143.5) |
91.1 (32.1) |
18.1 (8.7) |
| iso_smRNA4 |
30 |
LG1(12), LG12(1) |
tRNAGlyGGC_CB |
tRNA |
308.0 (161.0) |
131.8 (611.1) |
729.2 (366.5) |
179.7 (105.6) |
| iso_smRNA5 |
29 |
LG1(2), LG7(2), LG8(3), LG10(1), LG11(7), LG15(1), LG16(1), LG17(1), scaffolds(86) |
Gypsy-14_DAn-I |
LTR retrotransposon |
304.6 (162.0) |
120.7 (572.5) |
51.5 (266.1) |
22.0 (30.8) |
| iso_smRNA6 |
26 |
LG3(1), LG7(1), LG11(2), LG12(1), LG17(5), LG18(1), LG19(1), LG20(1) |
SINE2-1_EC |
Non-LTR Retrotransposon |
79.0 (29.0) |
67.7 (28.7) |
83.7 (30.3) |
62.3 (23.4) |
| iso_smRNA7 |
29 |
LG3(54), LG5(1), LG9(1), LG11(2), LG12(1), LG13(1), LG17(1), LG19(1), LG20(32), scaffolds(30) |
tRNA-Val-GTA |
tRNA |
86.9 (22.0) |
57.2 (123.0) |
25.7 (16.6) |
12.7 (10.8) |
| iso_smRNA8 |
29 |
LG5(1), LG9(1), LG11(2), LG12(1), LG13(1), LG17(1) |
DNA-TTAA-5_NV |
DNA transposon |
219.9 (50.9) |
138.8 (312.8) |
53.7 (47.9) |
41.5 (23.3) |
| iso_smRNA9 |
29 |
LG8(1), LG9(1), LG11(1), LG12(1), LG13(1), LG16(1), scaffolds(5) |
SINE2-8_SP |
Non-LTR Retrotransposon |
72.0 (10.9) |
58.4 (61.2) |
15.2 (17.9) |
9.9 (12.2) |
| iso_smRNA10 |
30 |
LG7(6), LG18(10), scaffolds(44) |
tRNA-Val-GTA |
tRNA |
309.1 (78.0) |
150.1 (701.4) |
581.7 (417.1) |
153.0 (99.2) |
| iso_smRNA11 |
29 |
LG1(1), LG4(1), LG7(1), LG12(1), scaffold(1) |
LTR10A2_SS |
ERV1-type endogenous retrovirus |
292.2 (76.3) |
177.0 (434.9) |
364.8 (124.6) |
155.6 (74.5) |
| iso_smRNA12 |
30 |
LG3(1), LG7(1), LG11(2), LG12(1), LG17(5), LG18(1), LG19(1), LG20(1) |
SINE2-1_EC |
Non-LTR Retrotransposon |
94.4 (21.8) |
29.9 (90.1) |
51.3 (19.5) |
19.9 (7.5) |
| iso_smRNA13 |
30 |
LG3(1), LG7(1), LG11(2), LG12(1), LG17(5), LG18(1), LG19(1), LG20(1) |
SINE2-1_EC |
Non-LTR Retrotransposon |
135.5 (47.2) |
43.5 (124.1) |
101.0 (54.8) |
35.9 (12.3) |
| iso_smRNA14 |
28 |
LG7(7), LG12(1) |
tRNA-Asp-GAY |
tRNA |
86.0 (16.3) |
68.4 (51.3) |
140.7 (166.9) |
74.8 (25.0) |
| iso_smRNA15 |
26 |
scaffolds(10) |
LSU-rRNA_Mfr |
rRNA |
142.1 (40.6) |
122.1 (42.0) |
193.3 (64.9) |
69.9 (27.3) |
| iso_smRNA16 |
28 |
scaffolds(10) |
LSU-rRNA_Mfr |
rRNA |
140.7 (52.4) |
170.6 (82.9) |
140.1 (29.1) |
74.3 (26.3) |
| iso_smRNA17 | 26 | LG12(1) | LTR41_SS | ERV1-type endogenous retrovirus | 76.8 (3.3) | 57.2 (20.0) | 64.1 (35.9) | 41.0 (5.8) |
The numbers in parentheses in the Blast hits indicate the number of hits on that linkage group.
The remaining six and three isoforms overlapped with repetitive sequences homologous to tRNA and rRNA, respectively. Previous studies also identified a number of tRNA-derived small RNAs in humans [83,84], Giardia lamblia[85], and zebrafish [86]. These tRNA-derived small RNAs may contribute to gene regulation [84], although little is yet known about their functions. Interestingly, some transposons are derived from tRNAs [87,88], so there appears to be an interesting link between tRNA and small RNAs.
Finally, we found that the iso-smRNA9, whose potential targets are predicted to be non-LTR transposons, was more highly expressed in the Japan Sea sticklebacks than in the Pacific Ocean sticklebacks (ANOVA, F1, 13 = 14.5, P = 0.002). Although expression levels of some other piRNAs may differ between different species and sexes, the differences were not significant after Bonferroni correction (Additional file 5: Table S5). None of the non-annotated small RNAs showed sex differences in the expression levels (Additional file 5: Table S5).
Thus, we identified small RNAs homologous to repetitive sequences such as RNA and DNA transposons. Hybrids between species often exhibit courtship dysfunction [89-94]. Abnormal transposon activity in hybrids may cause hybrid courtship dysfunction, but this has not been tested in any organism. Intra- and inter-population variation in the presence and absence of non-LTR retrotransposons has been found in sticklebacks [95]. In addition, our analysis involving whole genome sequence comparisons also revealed that DNA transposon insertion sites diverge between the Pacific Ocean and the Japan Sea sticklebacks (Kitano, unpublished data). Therefore, further studies examining variation in transposon activity between the different species and transposon activity in the hybrids will lead to a better understanding of speciation mechanisms.
Conclusions
Our study demonstrates that closely related species can show divergence in expression patterns of small RNAs, including miRNAs and piRNAs. Some of the sex differences in miRNA expression levels might result from Y-chromosome degeneration. Therefore, variation in small RNA transcriptomes should be examined as a potential mechanism underlying phenotypic divergence between incipient species.
Methods
Small RNA sequencing
Sympatric Pacific Ocean and Japan Sea sticklebacks were collected using minnow traps from the Bekanbeushi River System on Hokkaido Island, Japan in June 2007 [60,62]. Fish were brought to a laboratory to examine the brain transcriptome of courting male and spawning female fish, and mating experiments were conducted in June and July 2007, as described previously [60,63,96]. Once the male fish had constructed a nest, a conspecific gravid female fish was placed in the same tank. Immediately after the female fish had inspected the nest, both the male and the female fish were removed from the tank prior to spawning. After immersing the fish in a lethal dose of tricaine methanesulfonate, the brains of each fish were dissected and stored separately at −70°C.
For RNA sequencing, we used four Pacific Ocean male, four Pacific Ocean female, four Japan Sea male, and four Japan Sea female fish (N = 16 in total). Total RNA was isolated using TRIzol Reagent (Life Technologies, Grand Island, NY, USA), and the quality of RNA was evaluated using the BioAnalyzer (Agilent, Santa Clara, CA, USA). RNA Integrity Number (RIN) ranged from 9.3 to 10 with the median of 10. Libraries were constructed using the TruSeq Small RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Small RNAs with 20–30 nucleotides were isolated according to the manufacturer’s instructions. Sequencing was performed using the Genome Analyzer IIx (Illumina, San Diego, CA, USA) at the University of Tokyo. We used 16 lanes of the Genome Analyzer IIx (one fish per one lane).
miRNA analysis
Sequence analyses were conducted using the CLC Genomics Workbench Software (CLC bio, Katrinebjerg, Denmark). First, we discarded reads with a low quality score (quality score on the Phred scale of less than 0.05), very short length (less than 14 bp), or three or more ambiguous nucleotides (Additional file 1: Table S1). Next, identical reads were clustered together to group different types of small RNAs. Next, the sequences were mapped against the Ensembl stickleback miRNA database (http://asia.ensembl.org/info/data/ftp/index.html) with two nucleotides mismatches allowed.
Principal component analysis (PCA) of reads per million (RPM) was conducted on a Pearson correlation matrix. To identify differentially expressed miRNAs between various species and sexes, statistical analyses were conducted on the square-root transformed RPM of each miRNA. Using the statistical package R [97], analysis of variance (ANOVA) was conducted to examine whether the species, sex, and their interactions significantly influenced RPM. Because the patterns of sex differences varied between species (see Figure 3), we included the interaction term for the analysis. The Bonferroni correction was used for multiple comparison correction. Shapiro-Wilk and Bartlett’s tests were used to test for normal distribution of the data and homogeneity of variances, respectively. None of the miRNAs violated the assumptions of the normal distribution or homogeneity of variances after Bonferroni correction. Only miRNAs with a mean RPM higher than 1000 in at least one of the four groups (Pacific Ocean male, Pacific Ocean female, Japan Sea male, or Japan Sea female) were used for PCA and ANOVA.
piRNA analysis
Small RNAs with no match to any sequences in the stickleback miRNA database were mapped against sequences in the Ensembl stickleback non-coding RNA database (http://asia.ensembl.org/info/data/ftp/index.html), which includes transfer RNA (tRNA), ribosomal RNA (rRNA), small cytoplasmic RNA, small nuclear RNA, small nucleolar RNA, microRNA precursors, long intergenic non-coding RNAs, and other miscellaneous RNA. The parameters used were the same as above. Small RNAs with no match to any sequences in the stickleback non-coding RNA database may contain piRNAs; thus, we further analyzed these small RNAs to identify stickleback piRNAs. We first examined non-annotated small RNAs with nucleotide lengths >25 because this fraction is more likely to contain piRNAs. Among these longer small RNAs, we examined small RNAs with a mean RPM higher than 50 in at least one of the four groups (Pacific Ocean male, Pacific Ocean female, Japan Sea male, or Japan Sea female). Although some small RNAs showed differences in length, identical sequences were observed (Additional file 3: Table S3); hence, identical reads of different sizes were considered to be the same isoform.
To examine whether these small RNAs contain homologous sequences against any repetitive sequences, the longest isoforms were blasted against the stickleback genome (BROADS 1.56) using the default parameters (match = 1; mismatch = −3; gap existence = 5; gap extension = 2) of the CLC Genomics Workbench software. Next, flanking sequences (3,000 bp of upstream and 3,000 bp of downstream) were downloaded using Perl script [98]. We then examined whether these regions contained any repetitive sequences using the CENSOR software [99] on the Genetic Information Research Institute website (http://www.girinst.org/). When hits of repetitive sequences were identified in the region, we investigated whether the small RNA sequences overlapped with repetitive sequences. We also blasted these sequences against the piRNABank database [100].
To identify differentially expressed piRNAs between different species and sexes, statistical analysis was conducted on the square-root transformed RPM. Square-root transformed RPM values were subjected to ANOVA, followed by Bonferroni correction. Shapiro-Wilk and Bartlett’s tests were used to test for normal distribution of the data and homogeneity of variances, respectively. Two small RNAs, iso_smRNA3 and iso_smRNA14, did not meet the assumptions of homogeneity of variance (Bartlett’s test, K-squared = 15.2, P = 0.0017) and normal distribution (Shapiro-Wilk test, W = 0.779, P = 0.0014), respectively. For these small RNAs, we also conducted a Mann–Whitney U-test to confirm that these two small RNAs did not exhibit any differences between sexes and species. Because the interaction between the sexes and different species showed no significant effect for any of these small RNAs, the interaction term was excluded.
Additional files
Additional files: Tables S1-S5. All of the short read sequences are deposited in the Sequence Read Archive (DRA) of the DNA Data Bank of Japan (DDBJ): accession number DRA000919.
Abbreviations
miRNA: microRNA; mRNA: messenger RNA; piRNA: Piwi-interacting RNA; LG: linkage group; RPM: reads per million; PCA: principal component analysis; ANOVA: analysis of variance; tRNA: transfer RNA; rRNA: ribosomal RNA.
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
JK carried out most of the experiments and analyses and wrote the manuscript. KY helped with the analyses. YS conducted RNA sequencing. All authors read and approved the final manuscript.
Supplementary Material
Sequence raeds from each fish.
Component loading of principal component analysis.
Non-annotated small.
Repetitive sequences homologous to small RNAs.
ANOVA of non-annotated small RNAs.
Contributor Information
Jun Kitano, Email: jkitano@lab.nig.ac.jp.
Kohta Yoshida, Email: koyoshid@nig.ac.jp.
Yutaka Suzuki, Email: ysuzuki@k.u-tokyo.ac.jp.
Acknowledgements
This research is supported by the JST PRESTO program and Grant-in-Aid for Scientific Research on Innovative Areas ‘Gene Correlative System’ (23113007 and 23113001) and ‘Genome Science’ (221S0002) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Animal use protocols were approved by the Institutional Animal Care and Use Committee of the Fred Hutchinson Cancer Research Center (1575).
References
- Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol. 2008;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- Jeukens J, Renaut S, St-Cyr J, Nolte AW, Bernatchez L. The transcriptomics of sympatric dwarf and normal lake whitefish ( Coregonus clupeaformis spp., Salmonidae) divergence as revealed by next-generation sequencing . Mol Ecol. 2010;19:5389–5403. doi: 10.1111/j.1365-294X.2010.04934.x. [DOI] [PubMed] [Google Scholar]
- Gayral PF, Weinert L, Chiari Y, Tsagkogeorga G, Ballenghien M, Galtier N. Next-generation sequencing of transcriptomes: a guide to RNA isolation in nonmodel animals . Mol Ecol Resour. 2011. 11. [DOI] [PubMed]
- Stern DL, Orgogozo V. The loci of evolution: how predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-Devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Zhai J, Liu J, Liu B, Li P, Meyers BC, Chen X, Cao X. Small RNA-directed epigenetic natural variation in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000056. doi: 10.1371/journal.pgen.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Balasubramanian S, Cao J, Ott F, Sureshkumar S, Schneeberger K, Meyer RC, Altmann T, Weigel D. Natural variation in biogenesis efficiency of individual Arabidopsis thaliana microRNAs. Curr Biol. 2012;22:166–170. doi: 10.1016/j.cub.2011.11.060. [DOI] [PubMed] [Google Scholar]
- Loh Y-HE, Yi SV, Streelman JT. Evolution of microRNAs and the diversification of species. Genome Biol Evol. 2011;3:55–65. doi: 10.1093/gbe/evq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nature Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nature Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nature Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Wu L, Zhou H, Zhang Q, Zhang J, Ni F, Liu C, Qi Y. DNA methylation mediated by a microRNA pathway. Mol Cell. 2010;38:465–475. doi: 10.1016/j.molcel.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Bartel DP, Chen C-Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Sun W, Julie Li Y-S, Huang H-D, Shyy JYJ, Chien S. microRNA: a master regulator of cellular processes for bioengineering systems. Annu Rev Biomed Eng. 2010;12:1–27. doi: 10.1146/annurev-bioeng-070909-105314. [DOI] [PubMed] [Google Scholar]
- Lindsay MA. microRNAs and the immune response. Trends Immunol. 2008;29:343–351. doi: 10.1016/j.it.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Im H-I, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsi GA, Joyce EF, Couble P, McKim KS, Loppin B. Drosophila I-R hybrid dysgenesis is associated with catastrophic meiosis and abnormal zygote formation. J Cell Sci. 2010;123:3515–3524. doi: 10.1242/jcs.073890. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149:693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharap A, Nakka VP, Vemuganti R. Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke. 2011;42:1105–1109. doi: 10.1161/STROKEAHA.110.598391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Emerson JJ, Li W-H. The genetic basis of evolutionary change in gene expression levels. Phil Trans Roy Soc B. 2010;365:2581–2590. doi: 10.1098/rstb.2010.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Duff MO, Eipper-Mains J, Graveley BR, Wittkopp PJ. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 2010;20:816–825. doi: 10.1101/gr.102491.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Saunders MA, Liang H, Li W-H. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci USA. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheat CW, Fescemyer HW, Kvist J, Tas EVA, Vera JC, Frilander MJ, Hanski I, Marden JH. Functional genomics of life history variation in a butterfly metapopulation. Mol Ecol. 2011;20:1813–1828. doi: 10.1111/j.1365-294X.2011.05062.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Bartel DP. Allelic imbalance sequencing reveals that single-nucleotide polymorphisms frequently alter microRNA-directed repression. Nature Biotechnol. 2009;27:472–477. doi: 10.1038/nbt.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abelson JF, Kwan KY, O'Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rašin M-R, Gunel M. Sequence variants in SLITRK1 Are associated with Tourette's Syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Carbonell J, Alloza E, Arce P, Borrego S, Santoyo J, Ruiz-Ferrer M, Medina I, Jimenez-Almazan J, Mendez-Vidal C, Gonzalez-del Pozo M. A map of human microRNA variation uncovers unexpectedly high levels of variability. Genome Medicine. 2012;4:62. doi: 10.1186/gm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorc M, Jevsinek Skok D, Godnic I, Calin GA, Horvat S, Jiang Z, Dovc P, Kunej T. Catalog of microRNA seed polymorphisms in vertebrates. PLoS ONE. 2012;7:e30737. doi: 10.1371/journal.pone.0030737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges M, Coppieters W, Charlier C. Polymorphic miRNA-mediated gene regulation: contribution to phenotypic variation and disease. Curr Opin Genet Dev. 2007;17:166–176. doi: 10.1016/j.gde.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen J-M, Eychenne F. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
- McKinnon JS, Rundle HD. Speciation in nature: the threespine stickleback model systems. Trends Ecol Evol. 2002;17:480–488. doi: 10.1016/S0169-5347(02)02579-X. [DOI] [Google Scholar]
- Schluter D. The Ecology of Adaptive Radiation. New York: Oxford University Press; 2000. [Google Scholar]
- Kingsley DM, Peichel CL. In: Biology of the three-spined stickleback. Östlund-Nilsson S, Mayer I, Huntingford FA, editor. Boca Raton: CRC Press; 2007. The molecular genetics of evolutionary changes in sticklebacks; pp. 41–81. [Google Scholar]
- Peichel CL. Fishing for the secrets of vertebrate evolution in threespine sticklebacks. Dev Dynam. 2005;234:815–823. doi: 10.1002/dvdy.20564. [DOI] [PubMed] [Google Scholar]
- Cresko W, McGuigan K, Phillips P, Postlethwait J. Studies of threespine stickleback developmental evolution: progress and promise. Genetica. 2007;129:105–126. doi: 10.1007/s10709-006-0036-z. [DOI] [PubMed] [Google Scholar]
- McPhail JD. In: The evolutionary biology of the threespine stickleback. Bell MA, Foster SA, editor. Oxford: Oxford University Press; 1994. Speciation and the evolution of reproductive isolation; pp. 399–437. [Google Scholar]
- Wootton RJ. The Biology of Sticklebacks. London: Academic Press; 1976. [Google Scholar]
- Wootton RJ. A Functional Biology of Sticklebacks. London: Croom Helm; 1984. [Google Scholar]
- Bell MA, Foster SA. The Evolutionary Biology of the Threespine Stickleback. Oxford: Oxford University Press; 1994. [Google Scholar]
- Hendry AP, Bolnick DI, Berner D, Peichel CL. Along the speciation continuum in sticklebacks. J Fish Biol. 2009;75:2000–2036. doi: 10.1111/j.1095-8649.2009.02419.x. [DOI] [PubMed] [Google Scholar]
- Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. cis-Regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- McCairns RJS, Bernatchez L. Adaptive divergence between freshwater and marine sticklebacks: insights into the role of phenotypic plasticity from an integrated analysis of candidate gene expression. Evolution. 2010;64:1029–1047. doi: 10.1111/j.1558-5646.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- Kitano J, Lema SC, Luckenbach JA, Mori S, Kawagishi Y, Kusakabe M, Swanson P, Peichel CL. Adaptive divergence in the thyroid hormone signaling pathway in the stickleback radiation. Curr Biol. 2010;20:2124–2130. doi: 10.1016/j.cub.2010.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz TL, Eizaguirre C, Rotter B, Kalbe M, Milinski M. Exploring local immunological adaptation of two stickleback ecotypes by experimental infection and transcriptome-wide digital gene expression analysis. Mol Ecol. 2013;22:774–786. doi: 10.1111/j.1365-294X.2012.05756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder EH, Cano JM, Leinonen T, O'Hara RB, Nikinmaa M, Primmer CR, Merilä J. Female-biased expression on the X Chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol Biol Evol. 2010;27:1495–1503. doi: 10.1093/molbev/msq031. [DOI] [PubMed] [Google Scholar]
- Kitano J, Mori S, Peichel CL. Phenotypic divergence and reproductive isolation between sympatric forms of Japanese threespine sticklebacks. Biol J Linn Soc. 2007;91:671–685. doi: 10.1111/j.1095-8312.2007.00824.x. [DOI] [Google Scholar]
- Higuchi M, Goto A. Genetic evidence supporting the existence of two distinct species in the genus Gasterosteus around Japan. Environ Biol Fish. 1996;47:1–16. [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461:1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Mori S, Peichel CL. Divergence of male courtship displays between sympatric forms of anadromous threespine stickleback. Behaviour. 2008;145:443–461. doi: 10.1163/156853908792451430. [DOI] [Google Scholar]
- Kosik KS. The neuronal microRNA system. Nature Rev Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- Parsons M, Grimm C, Paya-Cano J, Fernandes C, Lin L, Philip V, Chesler E, Nietfeld W, Lehrach H, Schalkwyk L. Genetic variation in hippocampal microRNA expression differences in C57BL/6 J X DBA/2 J (BXD) recombinant inbred mouse strains. BMC Genomics. 2012;13:476. doi: 10.1186/1471-2164-13-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-J, Palkovits M, Young WS. miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation . Proc Natl Acad Sci USA. 2006;103:15669–15674. doi: 10.1073/pnas.0605781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S. microRNA-7 Inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in Glioblastoma. Cancer Res. 2008;68:3566–3572. doi: 10.1158/0008-5472.CAN-07-6639. [DOI] [PubMed] [Google Scholar]
- Schonrock N, Ke YD, Humphreys D, Staufenbiel M, Ittner LM, Preiss T, Götz J. Neuronal microRNA deregulation in response to Alzheimer's disease amyloid-β. PLoS One. 2010;5:e11070. doi: 10.1371/journal.pone.0011070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Graves JAM. Sex chromosome dynamics and Y chromosome degeneration. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond Ser B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM. Dosage compensation of the sex chromosomes. Annu Rev Genet. 2011;46:537–560. doi: 10.1146/annurev-genet-110711-155454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruike Y, Ichimura A, Tsuchiya S, Shimizu K, Kunimoto R, Okuno Y, Tsujimoto G. Global correlation analysis for micro-RNA and mRNA expression profiles in human cell lines. J Hum Genet. 2008. [DOI] [PubMed]
- Lu J, Clark AG. Impact of microRNA regulation on variation in human gene expression. Genome Res. 2012;22:1243–1254. doi: 10.1101/gr.132514.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjose A, Zhao X-F. Inhibition of the microRNA pathway in zebrafish by siRNA. RNA Therapeutics. 2010;629:237–253. doi: 10.1007/978-1-60761-657-3_15. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Takeda S, Furutani-Seiki M, Kamei Y, Todo T, Sasado T, Deguchi T, Kondoh H, Mudde J, Yamazoe M. Generation of medaka gene knockout models by target-selected mutagenesis. Genome Biol. 2006;7:1–14. doi: 10.1186/gb-2006-7-12-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlem TJ, Hoshijima K, Jurynec MJ, Gunther D, Starker CG, Locke AS, Weis AM, Voytas DF, Grunwald DJ. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 2012;8:e1002861. doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore FE, Reyon D, Sander JD, Martinez SA, Blackburn JS, Khayter C, Ramirez CL, Joung JK, Langenau DM. Improved somatic mutagenesis in zebrafish using transcription activator-like effector nucleases (TALENs) PLoS One. 2012;7:e37877. doi: 10.1371/journal.pone.0037877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh J-RJ. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012. [DOI] [PMC free article] [PubMed]
- Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JWS, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Luo J, Zhou H, Liao J-Y, Ma L-M, Chen Y-Q, Qu L-H. Stress-induced tRNA-derived RNAs: a novel class of small RNAs in the primitive eukaryote Giardia lamblia. Nucleic Acids Res. 2008;36:6048–6055. doi: 10.1093/nar/gkn596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Salichos L, Wittgrove CM, Rokas A, Patton JG. Transcriptome-wide analysis of small RNA expression in early zebrafish development. RNA. 2012;18:915–929. doi: 10.1261/rna.029090.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurek O, Nikaido M, Boeadi BM, Okada N. Unique mammalian tRNA-derived repetitive elements in Dermopterans: The t-SINE family and its retrotransposition through multiple sources. Mol Biol Evol. 2003;20:1659–1668. doi: 10.1093/molbev/msg187. [DOI] [PubMed] [Google Scholar]
- Kido Y, Aono M, Yamaki T, Matsumoto K, Murata S, Saneyoshi M, Okada N. Shaping and reshaping of salmonid genomes by amplification of tRNA-derived retroposons during evolution. Proc Natl Acad Sci USA. 1991;88:2326–2330. doi: 10.1073/pnas.88.6.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Almendarez Y, Reiland J, Smith KR. The genetics of reproductive isolation and the potential for gene exchange between Drosophila pseudoobscura and D. persimilis via backcross hybrid males. Evolution. 2001;55:512–521. doi: 10.1554/0014-3820(2001)055[0512:TGORIA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Clark ME, O'Hara FP, Chawla A, Werren JH. Behavioral and spermatogenic hybrid male breakdown in Nasonia. Heredity. 2010;104:289–301. doi: 10.1038/hdy.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N, Aiello A, Mallet J, Pomiankowski A, Silberglied RT. Speciation in two neotropical butterflies: extending Haldane's rule. Proc Biol Sci. 1997;264:845–851. doi: 10.1098/rspb.1997.0118. [DOI] [Google Scholar]
- Price DK, Boake CRB. Behavioral reproductive isolation in Drosophila silvestris, D. heteroneura, and their F1 hybrids (Diptera: Drosophilidae) J Insect Behav. 1995;8:595–616. doi: 10.1007/BF01997233. [DOI] [Google Scholar]
- Stratton GE, Uetz GW. The inheritance of courtship behavior and its role as a reproductive isolaing mechanism in two species of Schizocosa wolf spiders (Araneae; Lycosidae) Evolution. 1986;40:129–141. doi: 10.2307/2408610. [DOI] [PubMed] [Google Scholar]
- Lade BI, Thorpe WH. Dove songs as innately coded patterns of specific behaviour. Nature. 1964;202:366–368. doi: 10.1038/202366a0. [DOI] [Google Scholar]
- Blass E, Bell M, Boissinot S. Accumulation and rapid decay of non-LTR retrotransposons in the genome of the three-spine stickleback. Genome Biol Evol. 2012;4:687–702. doi: 10.1093/gbe/evs044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Kawagishi Y, Mori S, Peichel CL, Makino T, Kawata M, Kusakabe M. Divergence in sex steroid hormone signaling between sympatric species of Japanese threespine stickleback. PLoS One. 2011;6:e29253. doi: 10.1371/journal.pone.0029253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria; 2011. [Google Scholar]
- Tisdall JD. Beginning Perl for Bioinformatics. Sebastopol: O'Reilly Media; 2001. [Google Scholar]
- Kohany O, Gentles A, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173–D177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence raeds from each fish.
Component loading of principal component analysis.
Non-annotated small.
Repetitive sequences homologous to small RNAs.
ANOVA of non-annotated small RNAs.