Abstract
Epigenetics acts as an interface between environmental / exogenous factors, cellular responses and pathological processes. Aberrant epigenetic signatures are a hallmark of complex multifactorial diseases, including non-neoplastic disorders (e.g., cardiovascular diseases, hypertension, diabetes mellitus, autoimmune diseases, and some infectious diseases) and neoplasms (e.g., leukemias, lymphomas, sarcomas, and breast, lung, prostate, liver and colorectal cancers). Epigenetic signatures (DNA methylation, mRNA and microRNA expression, etc.) may serve as biomarkers for risk stratification, early detection, and disease classification, as well as targets for therapy and chemoprevention. DNA methylation assays are widely applied to formalin-fixed paraffin-embedded archival tissue specimens as clinical pathology tests. To better understand the interplay between etiologic factors, cellular molecular characteristics, and disease evolution, the field of “Molecular Pathological Epidemiology (MPE)” has emerged as an interdisciplinary integration of “molecular pathology” and “epidemiology”, with a similar conceptual framework to systems biology and network medicine. In contrast to traditional epidemiologic research including genome-wide association studies (GWAS), MPE is founded on the unique disease principle; that is, each disease process results from unique profiles of exposomes, epigenomes, transcriptomes, proteomes, metabolomes, microbiomes, and interactomes in relation to the macro-environment and tissue microenvironment. The widespread application of epigenomics (e.g., methylome) analyses will enhance our understanding of disease heterogeneity, epigenotypes (CpG island methylator phenotype, LINE-1 hypomethylation, etc.), and host-disease interactions. MPE may represent a logical evolution of GWAS, termed “GWAS-MPE approach”. Though epigenome-wide association study attracts increasing attention, currently, it has a fundamental problem in that each cell within one individual has a unique, time-varying epigenome. This article will illustrate increasing contribution of modern pathology to broader public health sciences, which attests pivotal roles of pathologists in the new integrated MPE science towards our ultimate goal of personalized medicine and prevention.
Keywords: molecular pathologic epidemiology, genetics, omics, hypermethylation, hypomethylation, personalized therapy, unique tumor principle, CIMP, long interspersed nucleotide element
Introduction
Epigenetic mechanisms constitute an essential mode of gene regulation and act as an interface between environmental exposures, cellular response and pathological processes. DNA methylation level and its location constitute important gene regulatory mechanisms.1–4 Abnormal epigenetic marks, including DNA methylation alterations, are a hallmark of most human diseases. Importantly, epigenetic modifications are reversible, and represent potential targets for disease prevention and therapy.2–6 There are other epigenetic mechanisms of gene regulations such as non-coding RNA including microRNA.7–13 Gene expression levels are consequences of epigenetic regulation; however, there exists a challenge in accurate quantification of transcript levels in archival tissues.14 Since most studies which have examined host exposures and epigenetic alterations utilized DNA methylation as biomarkers, our discussion on previous data mostly addresses DNA methylation.
Accumulating evidence suggests that epigenetic aberrations induced by environmental, dietary, lifestyle, and microbial factors contribute to specific disease processes.15–22 To examine the complex relationships between etiologic factors, molecular alterations, and disease evolution, “molecular pathology” and “epidemiology” have recently become integrated, generating the interdisciplinary field of “Molecular Pathological Epidemiology (MPE)”.21–23
As clinical molecular pathology testing is becoming more and more common, we anticipate that molecular pathology data can be acumulated into disease registries around the world. This enables MPE to become routine epidemiology and pathology research practice. Thus, a role of pathologists as educators for epidemiologists will increase. We must emphasize pivotal roles of modern pathologists in broader transdisciplinary biomedical and public health sciences, as well as in clinical decision making process.
In this article, we provide an overview of the MPE paradigm, and proceed to illustrate the contribution made by epigenetic research. While we exploit MPE data on neoplastic disorders, MPE approaches and paradigms can conceptually extend to the study of non-neoplastic diseases.
Tissue and Cellular Heterogeneity: Challenges in Epigenetic Research
In many non-neoplastic, non-hematologic, non-dermatologic diseases, access to diseased cells is limited by current technologies.24–27 Even if tissues affected by a non-neoplastic disease (e.g., inflamed liver) can be obtained, those tissues consist of many different cell types with varying epigenomes. Therefore, epigenetic analysis of non-neoplastic diseases faces a fundamental challenge of heterogeneity in tissue, cells, and epigenomes, which is often overlooked or underestimated in research studies and proposals (not limited to MPE).
Notably, the epigenome differs between specific cell-types (even within a single organ). In a single cell, the epigenome changes, as the cell responds to the microenvironmental changes over time. A single organ (or tissue) consists of numerous cell-types with different epigenomes.
A particular human disease process is caused by dysfunction of a specific cell-type, or multiple cell-types (in one organ or across multiple organ systems). Thus, the optimal approach is to analyze molecular changes in the afflicted cell-types specific to the disease process. To study a psychiatric or neuronal disease, it would be best to analyze disordered neurons (within the context of local microenvironment), rather than blood leukocytes or brain tissue as a mixture of different cell-types.28,31,32 Clearly, we must analyze specific cell-types in each tissue in a particular microenvironmental and disease context, using techniques such as laser capture microdissection and flow cytometry.29,30
Epigenomic differences between cell-types may be present in a small part of the genome (with overall similar epigenomic status); however, those minor differences are critical in cell-type specific function and disease pathogenesis, and unlikely inferred from examining the epigenomes of different cell-types. Though many epigenetic studies rely on blood leukocytes as a surrogate for alterations in other diseased cell-types,24–27,31–36 there is very little evidence supporting the validity of inferring epigenetic mechanisms of non-hematologic disorders from epigenetic analysis of blood leukocytes.
In contrast to non-neoplastic diseases, neoplastic diseases are characterized by uncontrolled cellular proliferation, which can provide abundant amounts of diseased cells for epigenetic analyses. We nevertheless should note; 1) that a tumor consists of many different cell-types (transformed neoplastic cells and various non-transformed cells such as fibroblasts, endothelial cells, smooth muscle cells, and inflammatory cells), and 2) that, even within a single tumor, neoplastic cells are heterogeneous.37 While we should be aware of these caveats, neoplastic diseases still give opportunities to study epigenetic alterations in diseased cells, by providing relatively enriched disease cell population.
Basic Characteristics of Disease: The Unique Disease Principle
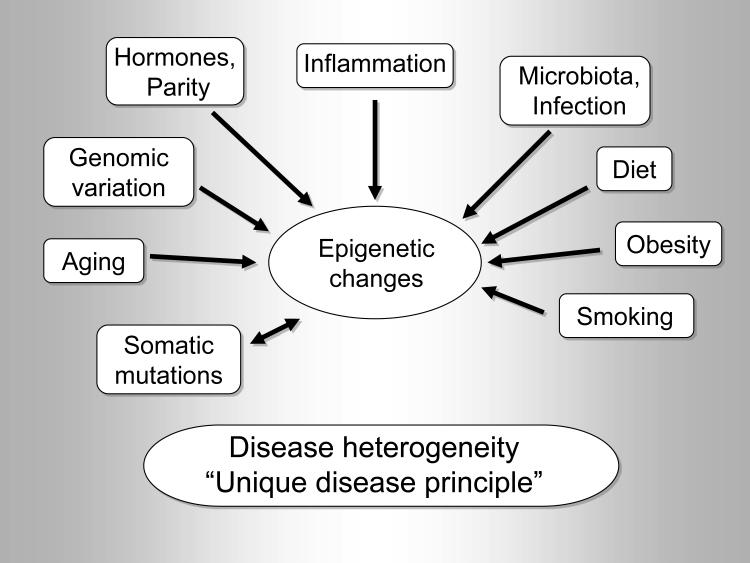
Human diseases are typically very complex processes (Figure 1), involving alterations in epigenomes, transcriptomes, proteomes, metabolomes, microbiomes and interactomes. Because each of us has a unique genome, and distinct combinations of exposome,38 epigenomes, transcriptomes, proteomes and metabolomes in specific cell-types, as well as unique microbiomes and interactomes in the tissue microenvironment, each disease process in each human must surely be unique, and distinct from what is nominally the same disease process in other individuals. This concept embodies the “unique disease principle”.
Figure 1.
A variety of endogenous and exogenous etiologic factors contribute to epigenetic changes leading to heterogeneity of disease processes, which is implicated by the “unique disease principle”. To simplify, only selected examples of those etiologic factors are demonstrated. There are numerous interactions between the factors, which are not depicted for simplicity.
To measure contribution of each of numerous molecular changes to disease pathogenesis will require enormous amounts of functional and correlative studies by both systems pathobiology and MPE approaches. Increasing roles of pathologists cannot be overemphasized in such efforts.
The “unique disease principle” poses a challenge to epidemiologic research, which is founded on the premise that we can predict disease occurrence and evolution, by inference from individuals with the disease with the same name. MPE, taking into account the unique disease principle, asserts that we can predict, to some extent, occurrence and evolution of a specific disease subtype by proper inference, which we will discuss further.
Molecular Pathological Epidemiology (MPE): Integrative Science
MPE has evolved through the integration of molecular pathology and epidemiology.21–23 The MPE paradigm has widely been utilized in a number of original and review articles,39–62 while further conceptual development of MPE remains active.63–67
MPE differs from conventional molecular epidemiology. MPE addresses the fundamental heterogeneity of disease processes, while conventional molecular epidemiology generally treats a given disease as a single entity.65,66 The conceptual framework of MPE resembles that of systems biology,68,69 and MPE integrates analyses of populations and the macroenvironment, with those of molecules and microenvironment. Importantly, the MPE paradigm encompasses all human diseases.
The MPE research approach allows investigators to examine the relationships between potential etiologic factors and disease subtypes based on molecular signatures. In addition, MPE permits the assessment of interactive effects of environmental influences and disease molecular signatures on disease progression.70–74 Thus, MPE research can provide insights into disease pathogenesis by demonstrating how specific etiologic factors influence mechanistic pathways in disease evolution and progression.
MPE possesses key advantages over traditional epidemiologic or pathologic research. Firstly, relationships can be uncovered between specific etiologic factors and molecular subtypes, supporting causality,21,22 and etiologic heterogeneity.67,75 Secondly, the risk of developing a specific disease subtype can be more accurately estimated.21,22 Thirdly, for individuals with susceptibility to a specific disease subtype or for patients with a specific disease subtype, personalized treatment and lifestyle modification strategies may be developed;21,22 examples include aspirin use for PIK3CA-mutant colorectal cancer patients,73 and physical activity recommendations for CTNNB1-negative colorectal cancer patients.72 These advantages are possible only with integrated MPE approach.
In the following sections we describe how epigenetics has contributed to MPE, with an emphasis on major disease epigenotypes.
Disease Epigenotypes
While the unique disease principal emphasizes the individuality of each disease process, molecular disease classification attempts to identify commonality in disease features, subgroup disease based on these shared characteristics, and predict disease evolution, progression, and therapeutic response.64,76 Epigenotyping can successfully classify cancers in various organs into distinct groups with different clinical, pathologic and molecular characteristics. Currently, clinical utility of epigenotyping remains limited, compared to conventional pathological assessment. However, accumulating evidence indicates distinct molecular signatures and phenotypes associated with specific epigenotypes, which cannot be discerned by pathological examination alone. Therefore, epigenotyping and pathological assessments should complement each other in the future.
The CpG Island Methylator Phenotype (CIMP)
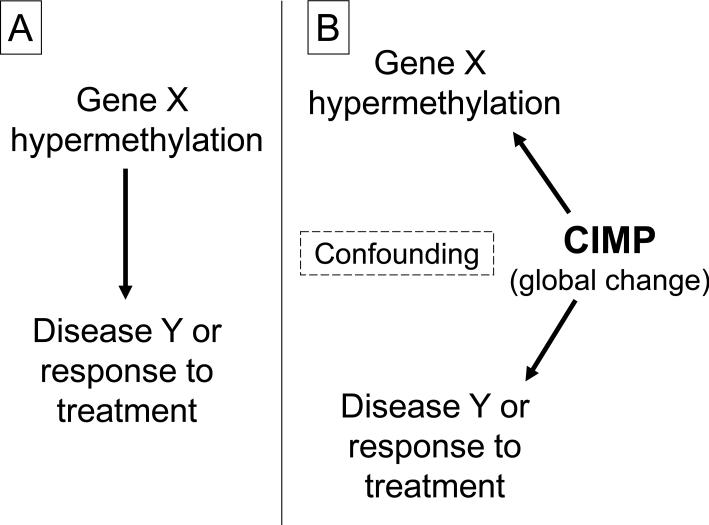
A specific tumor phenotype appears to exist that is characterized by the propensity of tumor cells to acquire widespread CpG island hypermethylation. This phenotype was named the CpG island methylator phenotype (CIMP), and was first described in colorectal cancer.77 The CIMP concept is important because it draws attention to the presence of an epigenome-wide driving force for CpG island hypermethylation. Therefore, when assessing CpG island methylation at a particular locus, CIMP must always be considered as a potential confounder (Figure 2).
Figure 2.
Why is examining global molecular phenomena so important? A. A typical study examining the relationship between gene X hypermethylation and disease Y, or response to treatment. Global molecular features, such as CIMP status, are not often considered. B. In reality, the significant relationship, if any, between gene X hypermethylation and disease, or treatment response, may reflect the association between CIMP and disease Y, or treatment response. CIMP status always needs to be considered as a potential confounder when examining locus-specific gene promoter hypermethylation and clinical outcome.
CIMP, CpG island methylator phenotype.
Normal differentiation state (including tissue of origin) likely influence epigenomic aberrations during neoplastic evolution.78 Although CIMP has been described in a variety of tumors,79–96 our discussion here focuses on colorectal cancer where CIMP has been most extensively characterized.39,44,76,97–99 The pathogenic basis of CIMP remains elusive, and CIMP may represent a multifactorial phenomenon.22 Although CIMP-high (high-level CIMP) is strongly associated with BRAF mutation in colorectal cancer,100–104 whether BRAF mutation causes CIMP remains uncertain.105,106 DNMT3B (DNA methyltransferase 3B) overexpression has been implicated in CIMP-high (high-level CIMP).107–112 CIMP-high colorectal cancer has been thought to arise from serrated precursor lesions such as sessile serrated polyp/adenoma.52,113–117 In addition to CIMP-high, a third CIMP category (“CIMP-low”) in colorectal cancers was found to be associated with KRAS mutations,118 which was confirmed by multiple studies.119–129 Features of CIMP-low colorectal cancers have been characterized.118–134 Prognostic and predictive roles of CIMP remains uncertain.39,44,76,132,135–139
CIMP-high is the cause of most colorectal cancers displaying high levels of microsatellite instability (MSI-high), which occur as a result of epigenetic inactivation of the mismatch repair gene MLH1.100–102,140 CIMP-high in colorectal cancer is associated with older age, female sex, proximal colonic location,100,103,141,142 poor tumor differentiation, mucinous and signet ring cell histology,143–146 immune and lymphocytic reactions,144,147–149 wild-type TP53,100,150 PTGS2 (cyclooxygenase-2) negativity,150 CDKN1A (p21) expression,151 loss of CDKN1B (p27) expression,152 CTNNB1 (β-catenin) membrane localization,153 wild-type APC,154 SIRT1 expression,155 PTGER2 expression,156 high-levels of LINE-1 methylation,103,157 loss of CDX2 expression,158,159 and low-level chromosomal instability.160–163 Tumor invasiveness and budding phenotype are inversely associated with MSI-high, rather than CIMP-high.164,165 Because BRAF mutation and MSI-high in colorectal cancer are associated with worse and better prognosis, respectively,121–123,132,166–182 it requires a large sample size and appropriate analyses to decipher prognostic significance of CIMP.132 CIMP-high is common in synchronous colorectal cancers (i.e., multiple separate primary cancers in a single patient),183–185 indicating that colorectal epithelial cells may be predisposed to CpG island methylation due to genetic and/or environmental factors.183,186
Despite the importance of CIMP phenomena, several caveats exist in CIMP research.44,76,187–190 Firstly, there has been a relative lack of consensus and validation in CIMP analysis methodologies.44,188 Validation of each component of analytical procedures is essential,191–194 as is replication of study findings in independent studies.102,103,195 Secondly, most previous studies on CIMP in human cancer specimens had convenience cohorts196 with small sample sizes, causing spurious findings, lack of robust statistics, and lack of generalizability.22,23,196,197
LINE-1 Methylation Epigenotypes
Owing to the relative simplicity of the assay, methylation levels in the long interspersed nucleotide element-1 (LINE-1; also called long interspersed nuclear element-1; long interspersed element-1; L1) have commonly been used as a surrogate measurement of cellular global DNA methylation level.198–200 In addition to its role as a surrogate marker of global DNA hypomethylation, LINE-1 hypomethylation by itself may have functional implications.201,202 Activation of LINE-1 retrotransposons may lead to the transcription of adjacent genes, gene disruptions, chromosomal instability, or the generation of transcripts that regulate gene expression.203–208
LINE-1 methylation level in colorectal cancer is widely distributed.157,209,210 LINE-1 hypomethylation has been associated with poor outcome in several cancer types,211–215 including colon cancer.216–218 LINE-1 hypomethylation is common in esophageal squamous cell carcinoma,45 metastatic prostate cancer,219 and metastatic pancreatic endocrine tumors.220 Average LINE-1 methylation levels in colorectal tumors decline as tumors progress from adenomas to invasive cancers, to highly invasive cancers.111,221 LINE-1 hypomethylation in colorectal cancer is associated with chromosomal instability,210,222 IGF2BP3 overexpression,223 and hypomethylation at the IGF2 differentially methylated region-0,224 which may be a somatic event in carcinogenesis.225 LINE-1 hypomethylated colorectal cancers are associated with family history of colorectal cancer and younger age of onset.210,226–228 Given that synchronous colorectal cancers show concordant LINE-1 hypomethylation patterns,183 there may be a predisposition to LINE-1 hypomethylation in colorectal epithelial cells.
Interplay of Genetic and Epigenetic Changes
When we consider epigenetic alterations, which may have importance in any stage of tumor development,229 we also need to consider genetic changes,230–234 which may be causes or consequences of epigenetic alterations. Genetic changes are increasingly studied on routine formalin-fixed paraffin-embedded archival tissue using next generation sequencing technologies,235 which is opening up opportunities for pathology research.
Epigenetic silencing of the DNA repair gene MGMT is implicated in somatic G>A mutations in various genes including KRAS, PIK3CA, APC, and TP53.120,236–242 Furthermore, CIMP-high in colorectal cancer is causally linked to MSI through epigenetic inactivation of mismatch repair gene MLH1;100–102,140 and then, MSI increases the rate of genome-wide mutational events,104,243–246 and influences cell cycle regulators.150–152,247–249 There are causally-uncertain relations between BRAF mutations and CIMP-high,100–103 and between KRAS mutations and CIMP-low.118–128
MPE of Etiologies and Epigenetics
Traditional epidemiology research has uncovered lifestyle, dietary, and environmental exposures that are positively or negatively associated with disease risk. In terms of cancer risk, these include smoking, some nutrients, alcohol consumption, energy metabolism status (energetics), aspirin use, hormone therapy, and some infectious / inflammatory conditions. However, generally, how these exposures influence disease pathogenesis remains not well understood. Lifestyle, dietary, and environmental factors likely influence the pathogenic process via altering the local tissue microenvironment, and epigenetics plays a key role in cellular response to microenvironmental change.
Various studies have adopted an MPE design to address the roles of potential etiologic factors. With the advent of technologies which enable analysis of genome-wide DNA methylation targets in archival tissue,250–252 we expect enormous opportunities to investigate environmental influences on somatic epigenetic alternations. Though this work is currently in its infancy, we highlight some examples, and the potential insights yielded, in the following sections.
MPE of Cigarette Smoking and Epigenetics
Cigarette smoking has been associated with CIMP-high,253–256 MSI-high,253–260 and BRAF-mutant subtypes253–256,261 of colorectal cancer. Duration of smoking cessation is associated with a reduction of risk for CIMP-high colorectal cancer.256 Similar to colorectal cancer, smoking has been associated with hypermethylation of specific genes262,263 and CIMP81 in lung cancer.
MPE of One-Carbon Metabolism and Epigenetics
The methyl (CH3-) groups for DNA methylation are derived from one-carbon methyl donors, suggesting an intrinsic link between one-carbon nutrients and epigenetic alterations. However, the relationship between one-carbon nutrients and somatic molecular alterations appears complex.16 In most epidemiologic studies, low folate intake has been associated with an increased risk of colorectal cancer and adenomas.264 However, there have been concerns that supplementation with folic acid may have tumor-promoting effects.264–266 In mouse models, folate supplmentation may promote epigenomic and microbiomic changes, and intestinal tumor formation.267,268 Examining molecular changes in tumor cells in relation to folate intake may provide insights into the role of one-carbon metabolism in carcinogenesis.22
Altered levels of intracellular folate metabolites have been linked to aberrant DNA methylation patterns.269,270 The relationship between folate / alcohol intake and aberrant promoter hypermethylation in colorectal cancer remains unclear.124,271–277 The MTHFR rs1801131 polymorphism (codon 429) may (or may not124,278) be associated with CIMP-high cancer.279,280
With regard to LINE-1, or global DNA methylation level, experimental evidence suggests a link between folate deficiency and global DNA hypomethylation in the colonic epithelium.281 Folate supplementation increases global DNA methylation levels in glioma and colon cancer cells.282,283 Folate deficiency (or excess alcohol consumption) has been associated with increased risk of TP53-mutated,284 and LINE-1 hypomethylated colon cancers.285 Randomized trials suggest that folic acid supplementation may (or may not286) increase global DNA methylation levels in normal colonic mucosa.287 Collectively, there is suggestive evidence of a link between one-carbon nutrients and global DNA (LINE-1) hypomethylation, which may lead to carcinogenesis.
MPE of Energetics and Epigenetics
Energetics has been implicated in metabolic diseases and cancers.288–293 Nonetheless, analyses of potential links between energetics and epigenetic alterations in specific diseased cells are in their infancy.
Evidence suggests that caloric restriction in early life is associated with a lower risk of CIMP-high colorectal cancer.294 In contrast, obesity in adult has been associated with non-MSI colorectal cancer,49,295–297 and CIMP-low/negative colorectal cancer.274 A recent prospective study has shown that obesity is associated with an increased risk of fatty acid synthase (FASN)-negative colorectal cancer.298 FASN overexpression has been implicated in carcinogenesis,299,300 and associated with MSI-high colorectal cancer.301 The relationship between obesity and non-MSI (or CIMP-low/negative) cancer might be explained by the link between obesity and FASN-negative tumors.21
Energetic status has been shown to interact with tumor molecular signatures to modify the behavior of colorectal cancer. Such tumor markers include expression of CTNNB1 (β-catenin),72 FASN,70 PRKAA (AMP-activated protein kinase),302 STMN1,303 CDKN1A (p21),304 and CDKN1B (p27).305,306 This type of interaction analysis represents an emerging paradigm in MPE,21,22 and may inform the design of new clinical trials to assess lifestyle or pharmacologic interventions.
Endogenous and Exogenous Hormones, and Epigenetics
Hormone therapy has been linked to lower risk of certain cancers, but epigenetic and other molecular mechanisms remain poorly understood. In breast cancer, ESR1 (estrogen receptor 1) and PGR (progesterone receptor) expression status has been associated with methylome alteration patterns.307 Hormone therapy has been associated with ESR1 and PGR promoter methylation in colorectal cancer.308 However, recent prospective cohort studies failed to demonstrate a clear relationship between hormone therapy and CIMP status.47,48,309 Hormone therapy may be associated with a decreased risk of colorectal cancer lacking CDKN1A (p21) expression.309 There are possible interrelations between the vitamin D pathway, the RAS-PI3K-AKT pathway, and epigenetic modulations in colorectal cancer.310,311
Microbiota, Inflammation, Immunity, and Epigenetics
Microorganisms, such as viruses, bacteria, and parasites, have been increasingly implicated in human health and chronic disease.19,20,312 Recently, the interplay of microenvironment, microbiota, immunity, inflammation, cellular epigenetic alterations, and various chronic diseases has attracted increasing attention.15,17–20,23,59,313–316H. pylori infection has been associated with epigenetic changes in gastric epithelial cells,317,318 and Enterobacteriaceae and Tenericutes have been associated with epigenetic changes in head and neck squamous cell carcinoma.319
The role of viral infection in epigenetic changes has been extensively reviewed elsewhere.320 Evidence indicate roles of HBV, HPV and EBV in epigenetic alterations and carcinogenesis.320–323 In colorectal cancer, a role of JCV in epigenetic alterations has been controversial.324–326
Inflammation appears to play a crucial role in carcinogenesis,327–329 and has been linked to energetics,330,331 and epigenetics.332 Cellular epigenetic changes may be induced by inflammation and associated oxidative damage,333–335 while cellular epigenetic aberrations may cause inflammatory diseases.336 A study has demonstrated that the inflammatory mediator, prostaglandin E2, upregulates DNMT3B, resulting in promoter CpG island hypermethylation and promotion of intestinal tumorigenesis in mice.335 Regular use of anti-inflammatory drugs such as aspirin, an inhibitor of PTGS2 (cyclooxygenase 2), has been associated with a decrease in cancer incidence and mortality,57,329 Cancer-preventive effect of aspirin is apparent against PTGS2-positive colorectal cancer.71,337,338 Moreover, aspirin appears to be very effective to treat PIK3CA-mutated colorectal cancer, suggesting PIK3CA mutation as a predictive tumor biomarker for clinical use.73
The importance of tumor-host interactions, encompassing microbiota and inflammation, has been highlighted by the recent discovery of a continuum in the frequency of molecular features (including CIMP-high, MSI-high and BRAF mutation) in colorectal cancers along subsites in the proximal-distal axis of the bowel.142,339 Luminal microbial contents and immune infiltrates appear to change gradually along the bowel.142,339–341 Taken together, local host and environmental factors, such as luminal contents, microbiome, inflammation, and the innate immune response, likely contribute to the development of specific molecular subtypes of colorectal cancer.142
Implications of MPE in Disease Prevention and Therapy
Despite the “uniqueness” of each disease process, molecular disease classification exploits shared molecular features of disease processes in multiple patients, on the premise that disease evolution and progression can be, to some extent, generalized to other patients with the same molecular subtype, and that appropriate (“personalized” or “precise”) treatment measures can be initiated for the patients with the particular disease subtype.64,76 Essentially, pathology testing can guide treatment decision making. This is relevant to not only pharmacological and immunological interventions,23,73 but also lifestyle modifications.72,306
With regard to disease prevention, MPE research may identify risk factors for a specific disease subtype. For individuals who are susceptible to the specific disease subtype, appropriate preventive measures (such as avoiding the identified risk factors) can be taken, or early detection can be attempted. For example, genetic susceptibility and familial clustering have been suggested for colorectal cancer with LINE-1 hypomethylation,210,226–228, which is an aggressive subtype,216–218 but can be prevented by adequate folate intake and avoidance of alcohol.285
Analysis of Normal Tissue, Stool, Blood or Other Body Fluids in MPE Context
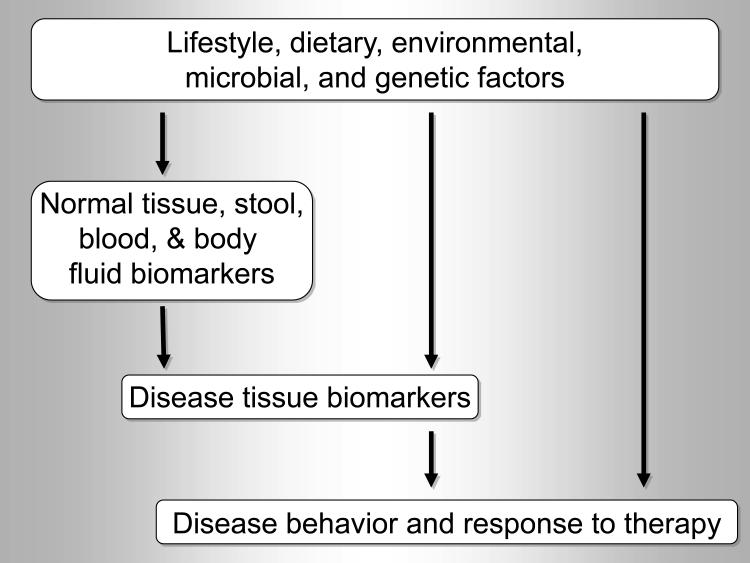
To date, most epigenetic studies on non-neoplastic diseases have relied on blood leukocytes as a surrogate for molecular processes in diseased cells,24–27,31–34 though there is very little evidence supporting the validity of this approach in non-hematologic diseases. Thus, the following discussion focuses on epigenetic or other molecular analyses of normal tissues or blood in the context of the MPE of cancer epigenetics. The ability to detect cancer or estimate cancer risk from normal tissue, stool, peripheral blood, or other body fluids (such as sputum and urine) has become the holy grail of biomarker discovery.342 Biomarkers in normal tissue, stool, or body fluids can represent: 1) a pathological outcome, analogous to established serum tumor markers; 2) a surrogate or shared indicator of etiologic exposure and disease predisposition; or 3) an intermediary in a causal pathway from etiology to downstream outcome (cancer incidence or behavior). Analysis of normal tissue, stool, and body fluids can expand the scope of, and add a novel dimension to MPE research (Figure 3).
Figure 3.
Biomarker analysis in normal tissue, stool, blood and body fluids adds new dimensions to molecular pathological epidemiology (MPE) research. Analyses of interactions among etiologic factors and biomarkers can be performed to test a specific research hypothesis.22,23,72,73
Bearing in mind the limitations of epigenetic and other molecular analyses on plasma or peripheral blood leukocytes, blood can conceivably carry a pathologic molecular signature, or diseased cells or cellular constituents, from any part of the body (e.g., bone marrow343). As a result of its abundance, ease of specimen collection, and practicality as a future clinical test, blood has been a common specimen type for studies on epigenetic changes in “normal” cells. Global DNA or LINE-1 methylation in leukocytes has attracted much interest as a potential cancer biomarker.344 Alterations in leukocyte DNA methylation have been associated with risk for a variety of solid tumors, including colon, bladder, stomach, and breast cancers.344–347 LINE-1 methylation level and other epigenetic changes in leukocytes appear to be influenced by a variety of exposures including smoking, early life or prenatal events.27,345–348 LINE-1 methylation in leukocytes and normal tissues is less variable compared to tumor, and may not correlate with tumor LINE-1 methylation levels.194,209,210,263,349
The study of interrelations between epidemiologic exposures, molecular changes in normal tissue or biological specimens, and cancer and other chronic diseases is an evolving field.350,351 Notably, several caveats must be applied to inferences drawn from study findings to date. Most studies have been relatively small and cross-sectional or retrospective in design, and replication in larger prospective studies is required. The type of assay employed and specific cell type analyzed appear to critically influence the determination of cellular DNA methylation levels.346–349,351–354 There is a relative lack of uniformity and robust validation data across laboratories in cell or nucleic acid isolation protocols, downstream processing, and assays. It remains a challenge to define exact biologic mechanisms to account for the associations between epigenetic alterations in normal blood and pathobiologic changes in specific cell types (e.g., breast duct epithelial cells which give rise to neoplasia). This has limited the potential for insights into disease pathogenesis and causality. Although blood biomarkers have the capability to reflect a disease process at a distant site in the body, epigenetic biomarkers, such as global DNA methylation in leukocytes, are often rather non-specific, and may be associated with a variety of different cancer types, as well as non-malignant conditions.27,345–347
Role of MPE in Post-GWAS Era
Over the past decade, genome-wide association studies (GWAS) have identified many germline genetic variants associated with numerous multifactorial diseases,355 and next steps such as fine locus mapping, gene-environment interaction analysis, family history analysis, and population structure analysis have been initiated.38,355–361 However, GWAS findings have made virtually no impact on clinical medicine and public health. Major shortcomings of existing GWAS approaches include insufficient consideration of disease heterogeneity,22 and relative lack of follow-up functional analyses of risk variants.362,363
Recently, the “GWAS-MPE approach” was proposed,22 to take disease heterogeneity into account following GWAS analyses. In a typical GWAS design, a disease of interest is regarded as a single entity without consideration of etiologic and biologic heterogeneity. Epigenetic analysis of diseased cells can provide ample opportunities to study biological significance of GWAS findings. By employing the MPE approach, molecular disease classification can help to identify a specific disease subtype that is more strongly associated with a given risk variant than other subtypes of the same disease. The “GWAS-MPE approach” has been applied to epidemiology research.364–366 The “GWAS-MPE approach”22 has advantages; 1) it may provide a possible causal link between the risk variant and molecular signatures in diseased cells; 2) it can more precisely refine risk estimates for each molecular subtype; and 3) it may identify new variant-subtype relationships, which may otherwise be obscured in conventional GWAS, dealing only with overall disease risk.
Problems in Epigenome-wide Association Study
After flourishing GWAS research over the past decade, epigenome-wide association studies are attracting increasing attention.367 However, we must be aware of significant flaws and caveats associated with current epigenome-wide association study design.368 In essence, each human being possesses innumerable different epigenomes. Epigenomes differ between cell types even within a single organ (which consists of numerous different cell types). Even in a single cell, the epigenome changes over time in dynamic time-varying macro- and microenvironmental milieu. Despite the presence of numerous epigenomes in each individual, current epigenome-wide association study design assumes one representative epigenome for each individual. Furthermore, it is unlikely a totally valid approach to infer epigenomic variants in specific cells (non-leukocytes) from epigenomic analysis of leukocytes.
At this juncture, we must be prudent, and first develop the required technologies and biosensors capable of interrogating epigenomic variations and interactomes in different cell types (preferably, in vivo) in one human being. This is prerequisite for launching into very expensive, resource-intensive epigenome-wide association study consortia, which presently erroneously assume a single epigenome to be representative of all epigenomes in an individual.
Conclusions and Future Perspectives
Achieving the goal of personalized medicine and prevention requires the integration of molecular medicine and population health sciences, and the willingness to explore beyond conventional disease classification. Personalized medicine holds the promise of biomarkers that will help stratify patients and guide decisions on optimal disease treatment and prevention.369 However, there is an increasing gap between basic scientific discoveries and real impact on population health.370,371 Recently, “Molecular Pathological Epidemiology (MPE)” has emerged as the evolving transdisciplinary science that can help fill in this gap.21,22,65,66 MPE integrates molecular pathology and epidemiology, in an attempt to decipher disease at the molecular, cellular, organ, individual, and population levels. The application of molecular pathology is feasible in existing cohort studies with large amounts of accumulated multidimensional data on dietary, lifestyle and environmental exposures, and clinical outcomes. This represents a very cost-efficient research approach to advance our understanding of disease and improve medicine and public health.49,63,372–375 To advance this integrated science requires cooperation of all practicing pathologists, because it is necessary to gather tissue specimens (from various community and academic pathology laboratories, to minimize selection bias) within well-defined cohort populations for molecular analyses.
Most chronic diseases are complex, multifactorial, genetic and epigenetic diseases. Epigenetic research is promising because epigenetic mechanisms play critical roles in the regulation of cellular growth, differentiation, and behavior, and epigenetic changes are potential modifiable targets for therapy and chemoprevention. Analysis of normal tissue, and biological specimens, adds additional dimensions to MPE research, through we must keep in mind caveats of analysis of those specimens. Epigenetic analyses continue to contribute substantially to biomedical and population health sciences. The future widespread application of methylome and epigenome analyses250,376–378 to paraffin-embedded archival tissues represents a powerful investigative tool, capable of enhancing our understanding of disease heterogeneity and host-disease interactions. In the future, application of in-vivo real-time molecular pathology, encompassing genomic, epigenomic, transcriptomic, proteomic, metabolomic, microbiomic, and interactomic analyses, will further transform biomedical and population health sciences.
Ultimately, molecular disease classification should be the primary patho-phenotypic datum of entry in population registries and databases around the world; this will further advance integrated population health science. There has been and will be increasing contribution of modern pathology to broader public health sciences, which attests pivotal roles of pathologists in the integrated science towards our ultimate goal of personalized medicine and prevention.
Note added in proof
Spitz et al.379 have used the term “integrative epidemiology” to describe an integration of molecular analyses (on exposures and tumors) into epidemiology. Integrative epidemiology encompasses MPE and conventional molecular epidemiology. MPE differs from conventional epidemiology because MPE takes disease heterogeneity into analysis.
Acknowledgments
Funding: This work was supported by grants from USA National Institute of Health (NIH) [R01 CA151993 (to SO), R01 CA137178 (to ATC), R01 CA136950 (to EC), R01 CA149222 (to JAM), P50 CA127003 (to CSF), R01 CA124908 (to CSF), P01 CA87969 (to SE Hankinson), 1UM1 CA167552 (to WC Willett)]. PL is a Scottish Government Clinical Academic Fellow. ATC is a Damon Runyon Clinical Investigator. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH. Funding agencies did not have any role in the decision to submit the manuscript for publication, or the writing of the manuscript.
Footnotes
Conflicts of Interest: ATC was a consultant of Bayer Healthcare, Millennium Pharmaceuticals, and Pfizer Inc. This work was not funded by Bayer Healthcare, Millennium Pharmaceuticals, or Pfizer Inc. No other conflict of interest exists.
References
- 1.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 3.Park J, Xu K, Park T, et al. What are the determinants of gene expression levels and breadths in the human genome? Hum Mol Genet. 2012;21:46–56. doi: 10.1093/hmg/ddr436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 5.Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer discovery. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebert MP, Tanzer M, Balluff B, et al. TFAP2E-DKK4 and chemoresistance in colorectal cancer. N Engl J Med. 2012;366:44–53. doi: 10.1056/NEJMoa1009473. [DOI] [PubMed] [Google Scholar]
- 7.de Krijger I, Mekenkamp LJ, Punt CJ, et al. MicroRNAs in colorectal cancer metastasis. J Pathol. 2011;224:438–447. doi: 10.1002/path.2922. [DOI] [PubMed] [Google Scholar]
- 8.Wang F, Ma YL, Zhang P, et al. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J Pathol. 2012 doi: 10.1002/path.4078. [DOI] [PubMed] [Google Scholar]
- 9.Valeri N, Gasparini P, Fabbri M, et al. Modulation of mismatch repair and genomic stability by miR-155. Proc Natl Acad Sci U S A. 2010;107:6982–6987. doi: 10.1073/pnas.1002472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valeri N, Gasparini P, Braconi C, et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci U S A. 2010;107:21098–21103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartley AN, Yao H, Barkoh BA, et al. Complex patterns of altered MicroRNA expression during the adenoma-adenocarcinoma sequence for microsatellite-stable colorectal cancer. Clin Cancer Res. 2011;17:7283–7293. doi: 10.1158/1078-0432.CCR-11-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earle JS, Luthra R, Romans A, et al. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn. 2010;12:433–440. doi: 10.2353/jmoldx.2010.090154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alhopuro P, Sammalkorpi H, Niittymaki I, et al. Candidate driver genes in microsatellite-unstable colorectal cancer. Int J Cancer. 2012;130:1558–1566. doi: 10.1002/ijc.26167. [DOI] [PubMed] [Google Scholar]
- 14.Waldron L, Ogino S, Hoshida Y, et al. Expression profiling of archival tissues for long-term health studies. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-12-1915. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein RA. Epigenetics--the link between infectious diseases and cancer. JAMA. 2011;305:1484–1485. doi: 10.1001/jama.2011.446. [DOI] [PubMed] [Google Scholar]
- 16.Coppede F. Epigenetic biomarkers of colorectal cancer: focus on DNA methylation. Cancer Lett. 2012 doi: 10.1016/j.canlet.2011.12.030. in press (published online) [DOI] [PubMed] [Google Scholar]
- 17.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome research. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome research. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tjalsma H, Boleij A, Marchesi JR, et al. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 21.Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: The evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010;102:365–367. doi: 10.1093/jnci/djq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogino S, Chan AT, Fuchs CS, et al. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogino S, Galon J, Fuchs CS, et al. Cancer immunology-analysis of host and tumor factors for personalized medicine. Nature reviews Clinical oncology. 2011;8:711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baccarelli A, Wright R, Bollati V, et al. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21:819–828. doi: 10.1097/EDE.0b013e3181f20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bind M, Baccarelli A, Zanobetti A, et al. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23:332–340. doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang WY, Levin L, Talaska G, et al. Maternal Exposure to Polycyclic Aromatic Hydrocarbons and 5'-CpG Methylation of Interferon-gamma in Cord White Blood Cells. Environ Health Perspect. 2012;120:1195–1200. doi: 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flom JD, Ferris JS, Liao Y, et al. Prenatal smoke exposure and genomic DNA methylation in a multiethnic birth cohort. Cancer Epidemiol Biomarkers Prev. 2011;20:2518–2523. doi: 10.1158/1055-9965.EPI-11-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hernandez DG, Nalls MA, Gibbs JR, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ollikainen M, Smith KR, Joo EJ, et al. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet. 2010;19:4176–4188. doi: 10.1093/hmg/ddq336. [DOI] [PubMed] [Google Scholar]
- 30.Byun HM, Siegmund KD, Pan F, et al. Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet. 2009;18:4808–4817. doi: 10.1093/hmg/ddp445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan ES, Qiu W, Baccarelli A, et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum Mol Genet. 2012;21:3073–3082. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dempster EL, Pidsley R, Schalkwyk LC, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toperoff G, Aran D, Kark JD, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21:371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lambrou A, Baccarelli A, Wright RO, et al. Arsenic Exposure and DNA Methylation Among Elderly Men. Epidemiology. 2012;23:668–676. doi: 10.1097/EDE.0b013e31825afb0b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang WY, Su LJ, Hayes RB, et al. Prospective study of genomic hypomethylation of leukocyte DNA and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2012 doi: 10.1158/1055-9965.EPI-12-0700-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao Y, Killian K, Zhang H, et al. Leukocyte DNA methylation and colorectal cancer among male smokers. World journal of gastrointestinal oncology. 2012;4:193–201. doi: 10.4251/wjgo.v4.i8.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marusyk A, Almendro V, Polyak K. Intra-tumor heterogeneity: a looking glass for cancer. Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 38.Thomas DC, Lewinger JP, Murcray CE, et al. Invited commentary: GE-Whiz! Ratcheting gene-environment studies up to the whole genome and the whole exposome. Am J Epidemiol. 2012;175:203–207. doi: 10.1093/aje/kwr365. discussion 208–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Pathology Research International. 2011;2011:902674. doi: 10.4061/2011/902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes LA, Simons CC, van den Brandt PA, et al. Body size, physical activity and risk of colorectal cancer with or without the CpG island methylator phenotype (CIMP) PLoS One. 2011;6:e18571. doi: 10.1371/journal.pone.0018571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelley RK, Wang G, Venook AP. Biomarker use in colorectal cancer therapy. J Natl Compr Canc Netw. 2011;9:1293–1302. doi: 10.6004/jnccn.2011.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell PT, Newton CC, Dehal AN, et al. Impact of Body Mass Index on Survival After Colorectal Cancer Diagnosis: The Cancer Prevention Study-II Nutrition Cohort. J Clin Oncol. 2012;30:42–52. doi: 10.1200/JCO.2011.38.0287. [DOI] [PubMed] [Google Scholar]
- 43.Gehoff A, Basten O, Sprenger T, et al. Optimal Lymph Node Harvest in Rectal Cancer (UICC Stages II and III) after Preoperative 5-FU-based Radiochemotherapy. Acetone Compression is a New and Highly Efficient Method. The American journal of surgical pathology. 2012;36:202–213. doi: 10.1097/PAS.0b013e31823fa35b. [DOI] [PubMed] [Google Scholar]
- 44.Hughes LA, Khalid-de Bakker CA, Smits KM, et al. The CpG island methylator phenotype in colorectal cancer: Progress and problems. Biochim Biophys Acta. 2012;1825:77–85. doi: 10.1016/j.bbcan.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 45.Iwagami S, Baba Y, Watanabe M, et al. Pyrosequencing Assay to Measure LINE-1 Methylation Level in Esophageal Squamous Cell Carcinoma. Ann Surg Oncol. 2012;19:2726–2732. doi: 10.1245/s10434-011-2176-3. [DOI] [PubMed] [Google Scholar]
- 46.Esteban S, Moya P, Fernandez-Suarez A, et al. Diagnostic and prognostic utility of methylation and protein expression patterns of myopodin in colon cancer. Tumour Biol. 2012;33:337–346. doi: 10.1007/s13277-012-0320-8. [DOI] [PubMed] [Google Scholar]
- 47.Limburg PJ, Limsui D, Vierkant RA, et al. Postmenopausal Hormone Therapy and Colorectal Cancer Risk in Relation to Somatic KRAS Mutation Status among Older Women. Cancer Epidemiol Biomarkers Prev. 2012;21:681–684. doi: 10.1158/1055-9965.EPI-11-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Limsui D, Vierkant RA, Tillmans LS, et al. Postmenopausal hormone therapy and colorectal cancer risk by molecularly defined subtypes among older women. Gut. 2012;61:1299–1305. doi: 10.1136/gutjnl-2011-300719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hughes LA, Williamson EJ, van Engeland M, et al. Body size and risk for colorectal cancers showing BRAF mutation or microsatellite instability: a pooled analysis. Int J Epidemiol. 2012;41:1060–1072. doi: 10.1093/ije/dys055. [DOI] [PubMed] [Google Scholar]
- 50.Ku CS, Cooper DN, Wu M, et al. Gene discovery in familial cancer syndromes by exome sequencing: prospects for the elucidation of familial colorectal cancer type X. Mod Pathol. 2012;25:1055–1068. doi: 10.1038/modpathol.2012.62. [DOI] [PubMed] [Google Scholar]
- 51.Kanthan R, Senger JL, Kanthan SC. Molecular Events in Primary and Metastatic Colorectal Carcinoma: A Review. Pathol Res Int. 2012;2012:597497. doi: 10.1155/2012/597497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rex DK, Ahnen DJ, Baron JA, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107:1315–1329. doi: 10.1038/ajg.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koshiol J, Lin SW. Can Tissue-Based Immune Markers be Used for Studying the Natural History of Cancer? Ann Epidemiol. 2012;22:520–530. doi: 10.1016/j.annepidem.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patai AV, Molnar B, Kalmar A, et al. Role of DNA methylation in colorectal carcinogenesis. Dig Dis. 2012;30:310–315. doi: 10.1159/000337004. [DOI] [PubMed] [Google Scholar]
- 55.Fini L, Grizzi F, Laghi L. Ettarh R, editor. Adaptive and Innate Immunity, Non Clonal Players in Colorectal Cancer Progression. Colorectal Cancer Biology - From Genes to Tumor: InTech. 2012:323–340. [Google Scholar]
- 56.Gay LJ, Mitrou PN, Keen J, et al. Dietary, lifestyle and clinico-pathological factors associated with APC mutations and promoter methylation in colorectal cancers from the EPIC-Norfolk Study. J Pathol. 2012;228:405–415. doi: 10.1002/path.4085. [DOI] [PubMed] [Google Scholar]
- 57.Chia WK, Ali R, Toh HC. Aspirin as adjuvant therapy for colorectal cancer-reinterpreting paradigms. Nature reviews Clinical oncology. 2012;9:561–570. doi: 10.1038/nrclinonc.2012.137. [DOI] [PubMed] [Google Scholar]
- 58.Dogan S, Shen R, Ang DC, et al. Molecular Epidemiology of EGFR and KRAS Mutations in 3026 Lung Adenocarcinomas: Higher Susceptibility of Women to Smoking-related KRAS-mutant Cancers. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galon J, Franck P, Marincola FM, et al. Cancer classification using the Immunoscore: a worldwide task force. Journal of translational medicine. 2012;10:205. doi: 10.1186/1479-5876-10-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haq S, Ali S, Mohammad R, et al. The complexities of epidemiology and prevention of gastrointestinal cancers. Int J Mol Sci. 2012;13:12556–12572. doi: 10.3390/ijms131012556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boyle T, Fritschi L, Heyworth J, et al. Long-term sedentary work and the risk of subsite-specific colorectal cancer. Am J Epidemiol. 2011;173:1183–1191. doi: 10.1093/aje/kwq513. [DOI] [PubMed] [Google Scholar]
- 62.Jacobs R, Voorneveld P, Kodach L, et al. Cholesterol metabolism and colorectal cancers. Current opinion in pharmacology. 2012 doi: 10.1016/j.coph.2012.07.010. [DOI] [PubMed] [Google Scholar]
- 63.Ogino S, Giovannucci E. Commentary: Lifestyle factors and colorectal cancer microsatellite instability - molecular pathological epidemiology science, based on unique tumour principle. In J Epidemiol. 2012;41:1072–1074. doi: 10.1093/ije/dys076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogino S, Fuchs CS, Giovannucci E. How many molecular subtypes? Implications of the unique tumor principle in personalized medicine. Expert Rev Mol Diagn. 2012;12:621–628. doi: 10.1586/erm.12.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ogino S, King EE, Beck AH, et al. Interdisciplinary education to integrate pathology and epidemiology: towards molecular and population-level health science. Am J Epidemiol. 2012;176:659–667. doi: 10.1093/aje/kws226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ogino S, Beck AH, King EE, et al. Ogino et al. respond to “The 21st century epidemiologist”. Am J Epidemiol. 2012;176:672–674. doi: 10.1093/aje/kws229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Begg CB, Zabor EC. Detecting and Exploiting Etiologic Heterogeneity in Epidemiologic Studies. Am J Epidemiol. 2012 doi: 10.1093/aje/kws128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ghosh S, Matsuoka Y, Asai Y, et al. Software for systems biology: from tools to integrated platforms. Nat Rev Genet. 2011;12:821–832. doi: 10.1038/nrg3096. [DOI] [PubMed] [Google Scholar]
- 69.Papp B, Notebaart RA, Pal C. Systems-biology approaches for predicting genomic evolution. Nat Rev Genet. 2011;12:591–602. doi: 10.1038/nrg3033. [DOI] [PubMed] [Google Scholar]
- 70.Ogino S, Nosho K, Meyerhardt JA, et al. Cohort study of fatty acid synthase expression and patient survival in colon cancer. J Clin Oncol. 2008;26:5713–5720. doi: 10.1200/JCO.2008.18.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan AT, Ogino S, Fuchs CS. Aspirin use and survival after diagnosis of colorectal cancer. JAMA. 2009;302:649–658. doi: 10.1001/jama.2009.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morikawa T, Kuchiba A, Yamauchi M, et al. Association of CTNNB1 (beta-catenin) alterations, body mass index, and physical activity with survival in patients with colorectal cancer. JAMA. 2011;305:1685–1694. doi: 10.1001/jama.2011.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao X, Lochhead P, Nishihara R, et al. Aspirin use, tumor PIK3CA mutation status, and colorectal cancer survival. N Eng1 J Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pasche B. Aspirin - from prevention to targeted therapy. N Engl J Med. 2012;367:1650–1651. doi: 10.1056/NEJMe1210322. [DOI] [PubMed] [Google Scholar]
- 75.Begg CB. A strategy for distinguishing optimal cancer subtypes. Int J Cancer. 2011;129:931–937. doi: 10.1002/ijc.25714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sproul D, Kitchen RR, Nestor CE, et al. Tissue of origin determines cancer-associated CpG island promoter hypermethylation patterns. Genome biology. 2012;13:R84. doi: 10.1186/gb-2012-13-10-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shinjo K, Okamoto Y, An B, et al. Integrated analysis of genetic and epigenetic alterations reveals CpG island methylator phenotype associated with distinct clinical characters of lung adenocarcinoma. Carcinogenesis. 2012;33:1277–1285. doi: 10.1093/carcin/bgs154. [DOI] [PubMed] [Google Scholar]
- 82.The Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kolbe DL, Deloia JA, Porter-Gill P, et al. Differential analysis of ovarian and endometrial cancers identifies a methylator phenotype. PLoS One. 2012;7:e32941. doi: 10.1371/journal.pone.0032941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ueki T, Toyota M, Sohn T, et al. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–1839. [PubMed] [Google Scholar]
- 85.Suzuki M, Shigematsu H, Iizasa T, et al. Exclusive mutation in epidermal growth factor receptor gene, HER-2, and KRAS, and synchronous methylation of nonsmall cell lung cancer. Cancer. 2006;106:2200–2207. doi: 10.1002/cncr.21853. [DOI] [PubMed] [Google Scholar]
- 86.Shaw RJ, Hall GL, Lowe D, et al. CpG island methylation phenotype (CIMP) in oral cancer: associated with a marked inflammatory response and less aggressive tumour biology. Oral Oncol. 2007;43:878–886. doi: 10.1016/j.oraloncology.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 87.Tanemura A, Terando AM, Sim MS, et al. CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res. 2009;15:1801–1807. doi: 10.1158/1078-0432.CCR-08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toyota M, Ahuja N, Suzuki H, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438–5442. [PubMed] [Google Scholar]
- 89.Fang F, Turcan S, Rimner A, et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011;3:75–25. doi: 10.1126/scitranslmed.3001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Warth A, Kloor M, Schirmacher P, et al. Genetics and epigenetics of small bowel adenocarcinoma: the interactions of CIN, MSI, and CIMP. Mod Pathol. 2011;24:564–570. doi: 10.1038/modpathol.2010.223. [DOI] [PubMed] [Google Scholar]
- 91.Abe M, Watanabe N, McDonell N, et al. Identification of genes targeted by CpG island methylator phenotype in neuroblastomas, and their possible integrative involvement in poor prognosis. Oncology. 2008;74:50–60. doi: 10.1159/000139124. [DOI] [PubMed] [Google Scholar]
- 92.Yagi K, Takahashi H, Akagi K, et al. Intermediate Methylation Epigenotype and Its Correlation to KRAS Mutation in Conventional Colorectal Adenoma. The American journal of pathology. 2012;180:616–625. doi: 10.1016/j.ajpath.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 93.Duncan CG, Barwick BG, Jin G, et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome research. 2012 doi: 10.1101/gr.132738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park SY, Kwon HJ, Choi Y, et al. Distinct patterns of promoter CpG island methylation of breast cancer subtypes are associated with stem cell phenotypes. Mod Pathol. 2012;25:185–196. doi: 10.1038/modpathol.2011.160. [DOI] [PubMed] [Google Scholar]
- 95.Zouridis H, Deng N, Ivanova T, et al. Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci Transl Med. 2012;4:156–140. doi: 10.1126/scitranslmed.3004504. [DOI] [PubMed] [Google Scholar]
- 96.Yamamoto E, Suzuki H, Yamano HO, et al. Molecular Dissection of Premalignant Colorectal Lesions Reveals Early Onset of the CpG Island Methylator Phenotype. The American journal of pathology. 2012;181:1847–1861. doi: 10.1016/j.ajpath.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 97.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686–700. doi: 10.1038/nrgastro.2011.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Greystoke A, Mullamitha SA. How many diseases is colorectal cancer? Gastroenterol Res Pract. 2012;2012:564741. doi: 10.1155/2012/564741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Beggs AD, Jones A, El-Bahwary M, et al. Whole-genome methylation analysis of benign and malignant colorectal tumours. J Pathol. 2013 doi: 10.1002/path.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Samowitz W, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 101.Ogino S, Cantor M, Kawasaki T, et al. CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut. 2006;55:1000–1006. doi: 10.1136/gut.2005.082933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 103.Nosho K, Irahara N, Shima K, et al. Comprehensive biostatistical analysis of CpG island methylator phenotype in colorectal cancer using a large population-based sample. PLoS ONE. 2008;3:e3698. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.The Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gazin C, Wajapeyee N, Gobeil S, et al. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hinoue T, Weisenberger DJ, Pan F, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin H, Yamada Y, Nguyen S, et al. Suppression of intestinal neoplasia by deletion of Dnmt3b. Mol Cell Biol. 2006;26:2976–2983. doi: 10.1128/MCB.26.8.2976-2983.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Linhart HG, Lin H, Yamada Y, et al. Dnmt3b promotes tumorigenesis in vivo by genespecific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Palakurthy RK, Wajapeyee N, Santra MK, et al. Epigenetic silencing of the RASSF1A tumor suppressor gene through HOXB3-mediated induction of DNMT3B expression. Mol Cell. 2009;36:219–230. doi: 10.1016/j.molcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nosho K, Shima K, Irahara N, et al. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clin Cancer Res. 2009;15:3663–3671. doi: 10.1158/1078-0432.CCR-08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ibrahim AE, Arends MJ, Silva AL, et al. Sequential DNA methylation changes are associated with DNMT3B overexpression in colorectal neoplastic progression. Gut. 2011;60:499–508. doi: 10.1136/gut.2010.223602. [DOI] [PubMed] [Google Scholar]
- 112.Steine EJ, Ehrich M, Bell GW, et al. Genes methylated by DNA methyltransferase 3b are similar in mouse intestine and human colon cancer. J Clin Invest. 2011;121:1748–1752. doi: 10.1172/JCI43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vaughn CP, Wilson AR, Samowitz WS. Quantitative evaluation of CpG island methylation in hyperplastic polyps. Mod Pathol. 2010;23:151–156. doi: 10.1038/modpathol.2009.150. [DOI] [PubMed] [Google Scholar]
- 114.Huang CS, Farraye FA, Yang S, et al. The Clinical Significance of Serrated Polyps. Am J Gastroenterol. 2011;106:229–240. doi: 10.1038/ajg.2010.429. [DOI] [PubMed] [Google Scholar]
- 115.Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol. 2011;42:1–10. doi: 10.1016/j.humpath.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 116.Patil DT, Shadrach BL, Rybicki LA, et al. Proximal colon cancers and the serrated pathway: a systematic analysis of precursor histology and BRAF mutation status. Mod Pathol. 2012;25:1423–1431. doi: 10.1038/modpathol.2012.98. [DOI] [PubMed] [Google Scholar]
- 117.Kriegl L, Neumann J, Vieth M, et al. Up and downregulation of p16(Ink4a) expression in BRAF-mutated polyps/adenomas indicates a senescence barrier in the serrated route to colon cancer. Mod Pathol. 2011;24:1015–1022. doi: 10.1038/modpathol.2011.43. [DOI] [PubMed] [Google Scholar]
- 118.Ogino S, Kawasaki T, Kirkner GJ, et al. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104:18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ogino S, kawasaki T, Kirkner GJ, et al. Molecular correlates with MGMT promoter methylation and silencing support CpG island methylator phenotype-low (CIMP-low) in colorectal cancer. Gut. 2007;56:1409–1416. doi: 10.1136/gut.2007.119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Barault L, Charon-Barra C, Jooste V, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 122.Kim JH, Shin SH, Kwon HJ, et al. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch. 2009;455:485–494. doi: 10.1007/s00428-009-0857-0. [DOI] [PubMed] [Google Scholar]
- 123.Dahlin AM, Palmqvist R, Henriksson ML, et al. The Role of the CpG Island Methylator Phenotype in Colorectal Cancer Prognosis Depends on Microsatellite Instability Screening Status. Clin Cancer Res. 2010;16:1845–1855. doi: 10.1158/1078-0432.CCR-09-2594. [DOI] [PubMed] [Google Scholar]
- 124.Van Guelpen B, Dahlin AM, Hultdin J, et al. One-carbon metabolism and CpG island methylator phenotype status in incident colorectal cancer: a nested case-referent study. Cancer Causes Control. 2010;21:557–566. doi: 10.1007/s10552-009-9484-y. [DOI] [PubMed] [Google Scholar]
- 125.Tanaka N, Huttenhower C, Nosho K, et al. Novel Application of Structural Equation Modeling to Correlation Structure Analysis of CpG Island Methylation in Colorectal Cancer. The American journal of pathology. 2010;177:2731–2740. doi: 10.2353/ajpath.2010.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Karpinski P, Szmida E, Misiak B, et al. Assessment of three epigenotypes in colorectal cancer by combined bisulfite restriction analysis. Molecular carcinogenesis. 2012;51:1003–1008. doi: 10.1002/mc.20871. [DOI] [PubMed] [Google Scholar]
- 127.Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome research. 2012;22:271–282. doi: 10.1101/gr.117523.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zlobec I, Bihl MP, Foerster A, et al. Stratification and Prognostic Relevance of Jass's Molecular Classification of Colorectal Cancer. Front Oncol. 2012;2:7. doi: 10.3389/fonc.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Imamura Y, Morikawa T, Liao X, et al. Specific Mutations in KRAS Codons 12 and 13, and Patient Prognosis in 1075 BRAF-wild-type Colorectal Cancers. Clin Cancer Res. 2012;18:4753–4763. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kawasaki T, Ohnishi M, Nosho K, et al. CpG island methylator phenotype-low (CIMP-low) colorectal cancer shows not only few methylated CIMP-high-specific CpG islands, but also low-level methylation at individual loci. Mod Pathol. 2008;21:245–255. doi: 10.1038/modpathol.3800982. [DOI] [PubMed] [Google Scholar]
- 131.Yagi K, Akagi K, Hayashi H, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 132.Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liao X, Morikawa T, Lochhead P, et al. Prognostic Role of PIK3CA Mutation in Colorectal Cancer: Cohort Study and Literature Review. Clin Cancer Res. 2012;18:2257–2268. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Whitehall VL, Rickman C, Bond CE, et al. Oncogenic PIK3CA mutations in colorectal cancers and polyps. Int J Cancer. 2012;131:813–820. doi: 10.1002/ijc.26440. [DOI] [PubMed] [Google Scholar]
- 135.Ogino S, Meyerhardt JA, Kawasaki T, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450:529–537. doi: 10.1007/s00428-007-0398-3. [DOI] [PubMed] [Google Scholar]
- 136.Teodoridis JM, Hardie C, Brown R. CpG island methylator phenotype (CIMP) in cancer: Causes and implications. Cancer Lett. 2008;268:177–186. doi: 10.1016/j.canlet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 137.Wu C, Bekaii-Saab T. CpG island methylation, microsatellite instability, and BRAF mutations and their clinical application in the treatment of colon cancer. Chemother Res Prac. 2012;2012:359041. doi: 10.1155/2012/359041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jover R, Nguyen TP, Perez-Carbonell L, et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140:1174–1181. doi: 10.1053/j.gastro.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Van Rijnsoever M, Elsaleh H, Joseph D, et al. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–2903. [PubMed] [Google Scholar]
- 140.Wong JJ, Hawkins NJ, Ward RL, et al. Methylation of the 3p22 region encompassing MLH1 is representative of the CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2011;24:396–411. doi: 10.1038/modpathol.2010.212. [DOI] [PubMed] [Google Scholar]
- 141.Sanchez JA, Krumroy L, Plummer S, et al. Genetic and epigenetic classifications define clinical phenotypes and determine patient outcomes in colorectal cancer. Br J Surg. 2009;96:1196–1204. doi: 10.1002/bjs.6683. [DOI] [PubMed] [Google Scholar]
- 142.Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chirieac LR, Shen L, Catalano PJ, et al. Phenotype of microsatellite-stable colorectal carcinomas with CpG island methylation. The American journal of surgical pathology. 2005;29:429–436. doi: 10.1097/01.pas.0000155144.53047.7d. [DOI] [PubMed] [Google Scholar]
- 144.Ogino S, Odze RD, kawasaki T, et al. Correlation of pathologic features with CpG island methylator phenotype (CIMP) by quantitative DNA methylation analysis in colorectal carcinoma. The American journal of surgical pathology. 2006;30:1175–1183. doi: 10.1097/01.pas.0000213266.84725.d0. [DOI] [PubMed] [Google Scholar]
- 145.Kawasaki T, Ohnishi M, Suemoto Y, et al. WRN promoter methylation possibly connects mucinous differentiation, microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2008;21:150–158. doi: 10.1038/modpathol.3800996. [DOI] [PubMed] [Google Scholar]
- 146.Kakar S, Deng G, Smyrk TC, et al. Loss of heterozygosity, aberrant methylation, BRAF mutation and KRAS mutation in colorectal signet ring cell carcinoma. Mod Pathol. 2012;25:1040–1047. doi: 10.1038/modpathol.2012.44. [DOI] [PubMed] [Google Scholar]
- 147.Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer and prognosis: cohort study and literature review. J Pathol. 2010;222:350–366. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dahlin AM, Henriksson ML, Van Guelpen B, et al. Colorectal cancer prognosis depends on T-cell infiltration and molecular characteristics of the tumor. Mod Pathol. 2011;24:671–682. doi: 10.1038/modpathol.2010.234. [DOI] [PubMed] [Google Scholar]
- 149.Ogino S, Nosho K, Irahara N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ogino S, Brahmandam M, kawasaki T, et al. Combined analysis of COX-2 and p53 expressions reveals synergistic inverse correlations with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Neoplasia. 2006;8:458–464. doi: 10.1593/neo.06247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ogino S, kawasaki T, Kirkner GJ, et al. Down-regulation of p21 (CDKN1A/CIP1) is inversely associated with microsatellite instability and CpG island methylator phenotype (CIMP) in colorectal cancer. J Pathol. 2006;210:147–154. doi: 10.1002/path.2030. [DOI] [PubMed] [Google Scholar]
- 152.Ogino S, kawasaki T, Kirkner GJ, et al. Loss of nuclear p27 (CDKN1B/KIP1) in colorectal cancer is correlated with microsatellite instability and CIMP. Mod Pathol. 2007;20:15–22. doi: 10.1038/modpathol.3800709. [DOI] [PubMed] [Google Scholar]
- 153.Kawasaki T, Nosho K, Ohnishi M, et al. Correlation of beta-catenin localization with cyclooxygenase-2 expression and CpG island methylator phenotype (CIMP) in colorectal cancer. Neoplasia. 2007;9:569–577. doi: 10.1593/neo.07334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Samowitz WS, Slattery ML, Sweeney C, et al. APC mutations and other genetic and epigenetic changes in colon cancer. Mol Cancer Res. 2007;5:165–170. doi: 10.1158/1541-7786.MCR-06-0398. [DOI] [PubMed] [Google Scholar]
- 155.Nosho K, Shima K, Irahara N, et al. SIRT1 histone deacetylase expression is associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2009;22:922–932. doi: 10.1038/modpathol.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Baba Y, Nosho K, Shima K, et al. PTGER2 overexpression in colorectal cancer is associated with microsatellite instability, independent of CpG island methylator phenotype. Cancer Epidemiol Biomarkers Prev. 2010;19:822–831. doi: 10.1158/1055-9965.EPI-09-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ogino S, Kawasaki T, Nosho K, et al. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Baba Y, Nosho K, Shima K, et al. Relationship of CDX2 loss with molecular features and prognosis in colorectal cancer. Clin Cancer Res. 2009;15:4665–4673. doi: 10.1158/1078-0432.CCR-09-0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zlobec I, Bihl M, Foerster A, et al. Comprehensive analysis of CpG Island Methylator Phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J Pathol. 2011;225:336–343. doi: 10.1002/path.2879. [DOI] [PubMed] [Google Scholar]
- 160.Ogino S, Kawasaki T, Kirkner GJ, et al. 18q loss of heterozygosity in microsatellite stable colorectal cancer is correlated with CpG island methylator phenotype-negative (CIMP-0) and inversely with CIMP-low and CIMP-high. BMC Cancer. 2007;7:72. doi: 10.1186/1471-2407-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Goel A, Nagasaka T, Arnold CN, et al. The CpG Island Methylator Phenotype and Chromosomal Instability Are Inversely Correlated in Sporadic Colorectal Cancer. Gastroenterology. 2007;132:127–138. doi: 10.1053/j.gastro.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 162.Cheng YW, Pincas H, Bacolod MD, et al. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res. 2008;14:6005–6013. doi: 10.1158/1078-0432.CCR-08-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kozlowska J, Karpinski P, Szmida E, et al. Assessment of chromosomal imbalances in CIMP-high and CIMP-low/CIMP-0 colorectal cancers. Tumour Biol. 2012;33:1015–1019. doi: 10.1007/s13277-012-0334-2. [DOI] [PubMed] [Google Scholar]
- 164.Morikawa T, Kuchiba A, Qian ZR, et al. Prognostic significance and molecular associations of tumor growth pattern in colorectal cancer. Ann Surg Oncol. 2012;19:1944–1953. doi: 10.1245/s10434-011-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zlobec I, Bihl MP, Foerster A, et al. The impact of CpG island methylator phenotype and microsatellite instability on tumour budding in colorectal cancer. Histopathology. 2012;61:777–787. doi: 10.1111/j.1365-2559.2012.04273.x. [DOI] [PubMed] [Google Scholar]
- 166.Ogino S, Shima K, Meyerhardt J, et al. Predictive and Prognostic Roles of BRAF Mutation in Stage III Colon Cancer: Results from Intergroup Trial CALGB 89803. Clin Cancer Res. 2012;18:890–900. doi: 10.1158/1078-0432.CCR-11-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.French AJ, Sargent DJ, Burgart LJ, et al. Prognostic Significance of Defective Mismatch Repair and BRAF V600E in Patients with Colon Cancer. Clin Cancer Res. 2008;14:3408–3415. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 169.Zlobec I, Bihl MP, Schwarb H, et al. Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer. 2010;127:367–380. doi: 10.1002/ijc.25042. [DOI] [PubMed] [Google Scholar]
- 170.Roth AD, Tejpar S, Delorenzi M, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 2010;28:466–474. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 171.Souglakos J, Philips J, Wang R, et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101:465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Farina-Sarasqueta A, van Lijnschoten G, Moerland E, et al. The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol. 2010;21:2396–2402. doi: 10.1093/annonc/mdq258. [DOI] [PubMed] [Google Scholar]
- 173.Ferracin M, Gafa R, Miotto E, et al. The methylator phenotype in microsatellite stable colorectal cancers is characterized by a distinct gene expression profile. J Pathol. 2008;214:594–602. doi: 10.1002/path.2318. [DOI] [PubMed] [Google Scholar]
- 174.Saridaki Z, Papadatos-Pastos D, Tzardi M, et al. BRAF mutations, microsatellite instability status and cyclin D1 expression predict metastatic colorectal patients' outcome. Br J Cancer. 2010;102:1762–1768. doi: 10.1038/sj.bjc.6605694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kalady MF, Sanchez JA, Manilich E, et al. Divergent oncogenic changes influence survival differences between colon and rectal adenocarcinomas. Dis Colon Rectum. 2009;52:1039–1045. doi: 10.1007/DCR.0b013e31819edbd4. [DOI] [PubMed] [Google Scholar]
- 176.Popat S, Hubner R, Houlston RS. Systematic Review of Microsatellite Instability and Colorectal Cancer Prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 177.Richman SD, Seymour MT, Chambers P, et al. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–5937. doi: 10.1200/JCO.2009.22.4295. [DOI] [PubMed] [Google Scholar]
- 178.Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 179.Price TJ, Hardingham JE, Lee CK, et al. Impact of KRAS and BRAF Gene Mutation Status on Outcomes From the Phase III AGITG MAX Trial of Capecitabine Alone or in Combination With Bevacizumab and Mitomycin in Advanced Colorectal Cancer. J Clin Oncol. 2011;29:2675–2682. doi: 10.1200/JCO.2010.34.5520. [DOI] [PubMed] [Google Scholar]
- 180.Phipps AI, Buchanan DD, Makar KW, et al. BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol Biomarkers Prev. 2012;21:1792–1798. doi: 10.1158/1055-9965.EPI-12-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gavin P, Colangelo LH, Fumagalli D, et al. Mutation Profiling and Microsatellite Instability in Stage II and III Colon Cancer: An Assessment of their Prognostic and Oxaliplatin Predictive Value. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-12-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Popovici V, Budinska E, Tejpar S, et al. Identification of a Poor-Prognosis BRAF-Mutant-Like Population of Patients With Colon Cancer. J Clin Oncol. 2012;30:1288–1295. doi: 10.1200/JCO.2011.39.5814. [DOI] [PubMed] [Google Scholar]
- 183.Nosho K, Kure S, Irahara N, et al. A prospective cohort study shows unique epigenetic, genetic, and prognostic features of synchronous colorectal cancers. Gastroenterology. 2009;137:1609–1620. doi: 10.1053/j.gastro.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Konishi K, Shen L, Jelinek J, et al. Concordant DNA methylation in synchronous colorectal carcinomas. Cancer Prev Res (Phila Pa) 2009;2:814–822. doi: 10.1158/1940-6207.CAPR-09-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gonzalo V, Lozano JJ, Munoz J, et al. Aberrant gene promoter methylation associated with sporadic multiple colorectal cancer. PLoS One. 2010;5:e8777. doi: 10.1371/journal.pone.0008777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Leggett BA, Worthley DL. Synchronous colorectal cancer: not just bad luck? Gastroenterology. 2009;137:1559–1562. doi: 10.1053/j.gastro.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 187.Funkhouser WK, Lubin IM, Monzon FA, et al. Relevance, pathogenesis, and testing algorithm for mismatch repair-defective colorectal carcinomas: A report of the Association for Molecular Pathology. J Mol Diagn. 2012;14:91–103. doi: 10.1016/j.jmoldx.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 188.Tournier B, Chapusot C, Courcet E, et al. Why do results conflict regarding the prognostic value of the methylation status in colon cancers? The role of the preservation method. BMC Cancer. 2012;12:12. doi: 10.1186/1471-2407-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ang PW, Loh M, Liem N, et al. Comprehensive profiling of DNA methylation in colorectal cancer reveals subgroups with distinct clinicopathological and molecular features. BMC Cancer. 2010;10:227. doi: 10.1186/1471-2407-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rawson JB, Bapat B. Epigenetic biomarkers in colorectal cancer diagnostics. Expert Rev Mol Diagn. 2012;12:499–509. doi: 10.1586/erm.12.39. [DOI] [PubMed] [Google Scholar]
- 191.Morikawa T, Shima K, Kuchiba A, et al. No evidence for interference of h&e staining in DNA testing: usefulness of DNA extraction from h&e-stained archival tissue sections. Am J Clin Pathol. 2012;138:122–129. doi: 10.1309/AJCP28LAOOKSZSVW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ogino S, Kawasaki T, Brahmandam M, et al. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Ogino S, kawasaki T, Brahmandam M, et al. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Irahara N, Nosho K, Baba Y, et al. Precision of Pyrosequencing assay to measure LINE-1 methylation in colon cancer, normal colonic mucosa and peripheral blood cells. J Mol Diagn. 2010;12:177–183. doi: 10.2353/jmoldx.2010.090106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ogino S, Kawasaki T, Kirkner GJ, et al. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Rosenberg AR, Skapek SX, Hawkins DS. The Inconvenience of Convenience Cohorts: Rhabdomyosarcoma and the PAX-FOXO1 Biomarker. Cancer Epidemiol Biomarkers Prev. 2012;21:1012–1018. doi: 10.1158/1055-9965.EPI-12-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kitkumthorn N, Mutirangura A. Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications. Clin Epigenet. 2012;2:315–330. doi: 10.1007/s13148-011-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yang AS, Estecio MR, Doshi K, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Frigola J, Sole X, Paz MF, et al. Differential DNA hypermethylation and hypomethylation signatures in colorectal cancer. Hum Mol Genet. 2005;14:319–326. doi: 10.1093/hmg/ddi028. [DOI] [PubMed] [Google Scholar]
- 201.Aporntewan C, Phokaew C, Piriyapongsa J, et al. Hypomethylation of intragenic LINE-1 represses transcription in cancer cells through AGO2. PLoS One. 2011;6:e17934. doi: 10.1371/journal.pone.0017934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wissing S, Munoz-Lopez M, Macia A, et al. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum Mol Genet. 2012;21:208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Peaston AE, Evsikov AV, Graber JH, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 205.Faulkner GJ, Kimura Y, Daub CO, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 206.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 207.Yamada Y, Jackson-Grusby L, Linhart H, et al. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci U S A. 2005;102:13580–13585. doi: 10.1073/pnas.0506612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Howard G, Eiges R, Gaudet F, et al. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene. 2008;27:404–408. doi: 10.1038/sj.onc.1210631. [DOI] [PubMed] [Google Scholar]
- 209.Estecio MR, Gharibyan V, Shen L, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS ONE. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Baba Y, Huttenhower C, Nosho K, et al. Epigenomic diversity of colorectal cancer indicated by LINE-1 methylation in a database of 869 tumors. Mol Cancer. 2010;9:125. doi: 10.1186/1476-4598-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Bae JM, Shin S, Kwon H, et al. ALU and LINE-1 hypomethylations in multistep gastric carcinogenesis and their prognostic implications. Int J Cancer. 2012;131:1323–1331. doi: 10.1002/ijc.27369. [DOI] [PubMed] [Google Scholar]
- 212.Igarashi S, Suzuki H, Niinuma T, et al. A novel correlation between LINE-1 hypomethylation and the malignancy of gastrointestinal stromal tumors. Clin Cancer Res. 2010;16:5114–5123. doi: 10.1158/1078-0432.CCR-10-0581. [DOI] [PubMed] [Google Scholar]
- 213.Pattamadilok J, Huapai N, Rattanatanyong P, et al. LINE-1 hypomethylation level as a potential prognostic factor for epithelial ovarian cancer. Int J Gynecol Cancer. 2008;18:711–717. doi: 10.1111/j.1525-1438.2007.01117.x. [DOI] [PubMed] [Google Scholar]
- 214.Roman-Gomez J, Jimenez-Velasco A, Agirre X, et al. Promoter hypomethylation of the LINE-1 retrotransposable elements activates sense/antisense transcription and marks the progression of chronic myeloid leukemia. Oncogene. 2005;24:7213–7223. doi: 10.1038/sj.onc.1208866. [DOI] [PubMed] [Google Scholar]
- 215.Hoshimoto S, Kuo CT, Chong KK, et al. AIM1 and LINE-1 Epigenetic Aberrations in Tumor and Serum Relate to Melanoma Progression and Disease Outcome. J Invest Dermatol. 2012;132:1689–1697. doi: 10.1038/jid.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ogino S, Nosho K, Kirkner GJ, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–1738. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ahn JB, Chung WB, Maeda O, et al. DNA methylation predicts recurrence from resected stage III proximal colon cancer. Cancer. 2011;117:1847–1854. doi: 10.1002/cncr.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Rhee YY, Kim MJ, Bae JM, et al. Clinical Outcomes of Patients with Microsatellite-Unstable Colorectal Carcinomas Depend on L1 Methylation Level. Ann Surg Oncol. 2012;19:3441–3448. doi: 10.1245/s10434-012-2410-7. [DOI] [PubMed] [Google Scholar]
- 219.Yegnasubramanian S, Haffner MC, Zhang Y, et al. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res. 2008;68:8954–8967. doi: 10.1158/0008-5472.CAN-07-6088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Choi IS, Estecio MR, Nagano Y, et al. Hypomethylation of LINE-1 and Alu in well-differentiated neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors) Mod Pathol. 2007;20:802–810. doi: 10.1038/modpathol.3800825. [DOI] [PubMed] [Google Scholar]
- 221.Sunami E, de Maat M, Vu A, et al. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ogino S, Nosho K, Irahara N, et al. Prognostic significance and molecular associations of 18q loss of heterozygosity: a cohort study of microsatellite stable colorectal cancers. J Clin Oncol. 2009;26:5713–5720. doi: 10.1200/JCO.2009.22.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lochhead P, Imamura Y, Morikawa T, et al. Insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3) is a marker of unfavourable prognosis in colorectal cancer. Eur J Cancer. 2012 doi: 10.1016/j.ejca.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Baba Y, Nosho K, Shima K, et al. Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastroenterology. 2010;139:1855–1864. doi: 10.1053/j.gastro.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Ito Y, Koessler T, Ibrahim AE, et al. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum Mol Genet. 2008;17:2633–2643. doi: 10.1093/hmg/ddn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Antelo M, Balaguer F, Shia J, et al. A High Degree of LINE-1 Hypomethylation Is a Unique Feature of Early-Onset Colorectal Cancer. PLoS One. 2012;7:e45357. doi: 10.1371/journal.pone.0045357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Goel A, Xicola RM, Nguyen TP, et al. Aberrant DNA methylation in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. Gastroenterology. 2010;138:1854–1862. doi: 10.1053/j.gastro.2010.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ogino S, Nishihara R, Lochhead P, et al. Prospective study of family history and colorectal cancer risk by tumor LINE-1 methylation level. J Natl Cancer Inst. 2013 doi: 10.1093/jnci/djs482. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Carmona FJ, Villanueva A, Vidal A, et al. Epigenetic disruption of cadherin-11 in human cancer metastasis. J Pathol. 2012;228:230–240. doi: 10.1002/path.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ogino S, Meyerhardt JA, Irahara N, et al. KRAS mutation in stage III colon cancer and clinical outcome following intergroup trial CALGB 89803. Clin Cancer Res. 2009;15:7322–7329. doi: 10.1158/1078-0432.CCR-09-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lievre A, Blons H, Laurent-Puig P. Oncogenic mutations as predictive factors in colorectal cancer. Oncogene. 2010;29:3033–3043. doi: 10.1038/onc.2010.89. [DOI] [PubMed] [Google Scholar]
- 232.Myers MB, Wang Y, McKim KL, et al. Hotspot oncomutations: implications for personalized cancer treatment. Expert Rev Mol Diagn. 2012;12:603–620. doi: 10.1586/erm.12.51. [DOI] [PubMed] [Google Scholar]
- 233.Voutsina A, Tzardi M, Kalikaki A, et al. Combined analysis of KRAS and PIK3CA mutations, MET and PTEN expression in primary tumors and corresponding metastases in colorectal cancer. Mod Pathol. 2012 doi: 10.1038/modpathol.2012.150. [DOI] [PubMed] [Google Scholar]
- 234.Bond CE, Umapathy A, Buttenshaw RL, et al. Chromosomal Instability in BRAF Mutant, Microsatellite Stable Colorectal Cancers. PLoS One. 2012;7:e47483. doi: 10.1371/journal.pone.0047483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Adams MD, Veigl ML, Wang Z, et al. Global mutational profiling of formalin-fixed human colon cancers from a pathology archive. Mod Pathol. 2012 doi: 10.1038/modpathol.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Esteller M, Risques RA, Toyota M, et al. Promoter hypermethylation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res. 2001;61:4689–4692. [PubMed] [Google Scholar]
- 237.Esteller M, Toyota M, Sanchez-Cespedes M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–2371. [PubMed] [Google Scholar]
- 238.Halford S, Rowan A, Sawyer E, et al. O6-methylguanine methyltransferase in colorectal cancers: detection of mutations, loss of expression, and weak association with G:C>A:T transitions. Gut. 2005;54:797–802. doi: 10.1136/gut.2004.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Nosho K, Kawasaki T, Ohnishi M, et al. PIK3CA mutation in colorectal cancer: relationship with genetic and epigenetic alterations. Neoplasia. 2008;10:534–541. doi: 10.1593/neo.08336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Suehiro Y, Wong CW, Chirieac LR, et al. Epigenetic-Genetic Interactions in the APC/WNT, RAS/RAF, and P53 Pathways in Colorectal Carcinoma. Clin Cancer Res. 2008;14:2560–2569. doi: 10.1158/1078-0432.CCR-07-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Shima K, Morikawa T, Baba Y, et al. MGMT promoter methylation, loss of expression and prognosis in 855 colorectal cancers. Cancer Causes Control. 2011;22:301–309. doi: 10.1007/s10552-010-9698-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.de Vogel S, Weijenberg MP, Herman JG, et al. MGMT and MLH1 promoter methylation versus APC, KRAS and BRAF gene mutations in colorectal cancer: indications for distinct pathways and sequence of events. Ann Oncol. 2009;20:1216–1222. doi: 10.1093/annonc/mdn782. [DOI] [PubMed] [Google Scholar]
- 243.Timmermann B, Kerick M, Roehr C, et al. Somatic mutation profiles of MSI and MSS colorectal cancer identified by whole exome next generation sequencing and bioinformatics analysis. PLoS One. 2010;5:e15661. doi: 10.1371/journal.pone.0015661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Shah SN, Hile SE, Eckert KA. Defective mismatch repair, microsatellite mutation bias, and variability in clinical cancer phenotypes. Cancer Res. 2010;70:431–435. doi: 10.1158/0008-5472.CAN-09-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Dulak AM, Schumacher S, van Lieshout J, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012;72:4383–4393. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Donehower LA, Creighton CJ, Schultz N, et al. MLH1-Silenced and Non-Silenced Subgroups of Hypermutated Colorectal Carcinomas Have Distinct Mutational Landscapes. J Pathol. 2012 doi: 10.1002/path.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Nosho K, Kawasaki T, Chan AT, et al. Cyclin D1 is frequently overexpressed in microsatellite unstable colorectal cancer, independent of CpG island methylator phenotype. Histopathology. 2008;53:588–598. doi: 10.1111/j.1365-2559.2008.03161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Shima K, Nosho K, Baba Y, et al. Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: cohort study and literature review. Int J Cancer. 2011;128:1080–1094. doi: 10.1002/ijc.25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Belt EJ, Brosens RP, Delis-van Diemen PM, et al. Cell Cycle Proteins Predict Recurrence in Stage II and III Colon Cancer. Ann Surg Oncol. 2012;19:S682–692. doi: 10.1245/s10434-012-2216-7. [DOI] [PubMed] [Google Scholar]
- 250.Gu H, Bock C, Mikkelsen TS, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kibriya MG, Raza M, Jasmine F, et al. A genome-wide DNA methylation study in colorectal carcinoma. BMC Med Genomics. 2011;4:50. doi: 10.1186/1755-8794-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Jasmine F, Rahaman R, Roy S, et al. Interpretation of genome-wide infinium methylation data from ligated DNA in formalin-fixed, paraffin-embedded paired tumor and normal tissue. BMC Res Notes. 2012;5:117. doi: 10.1186/1756-0500-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Samowitz WS, Albertsen H, Sweeney C, et al. Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst. 2006;98:1731–1738. doi: 10.1093/jnci/djj468. [DOI] [PubMed] [Google Scholar]
- 254.Curtin K, Samowitz WS, Wolff RK, et al. Somatic alterations, metabolizing genes and smoking in rectal cancer. Int J Cancer. 2009;125:158–164. doi: 10.1002/ijc.24338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Limsui D, Vierkant RA, Tillmans LS, et al. Cigarette Smoking and Colorectal Cancer Risk by Molecularly Defined Subtypes. J Natl Cancer Inst. 2010;102:1012–1022. doi: 10.1093/jnci/djq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nishihara R, Morikawa T, Kuchiba A, et al. A prospective study of duration of smoking cessation and colorectal cancer risk by epigenetics-related tumor classification. Am J Epidemiol. 2013 doi: 10.1093/aje/kws431. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Poynter JN, Haile RW, Siegmund KD, et al. Associations between smoking, alcohol consumption, and colorectal cancer, overall and by tumor microsatellite instability status. Cancer Epidemiol Biomarkers Prev. 2009;18:2745–2750. doi: 10.1158/1055-9965.EPI-09-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lindor NM, Yang P, Evans I, et al. Alpha-1-antitrypsin deficiency and smoking as risk factors for mismatch repair deficient colorectal cancer: A study from the colon cancer family registry. Mol Genet Metab. 2010;99:157–159. doi: 10.1016/j.ymgme.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Eaton AM, Sandler R, Carethers JM, et al. 5,10-methylenetetrahydrofolate reductase 677 and 1298 polymorphisms, folate intake, and microsatellite instability in colon cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2023–2029. doi: 10.1158/1055-9965.EPI-05-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Chia VM, Newcomb PA, Bigler J, et al. Risk of microsatellite-unstable colorectal cancer is associated jointly with smoking and nonsteroidal anti-inflammatory drug use. Cancer Res. 2006;66:6877–6883. doi: 10.1158/0008-5472.CAN-06-1535. [DOI] [PubMed] [Google Scholar]
- 261.Rozek LS, Herron CM, Greenson JK, et al. Smoking, gender, and ethnicity predict somatic BRAF mutations in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:838–843. doi: 10.1158/1055-9965.EPI-09-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Russo AL, Thiagalingam A, Pan H, et al. Differential DNA hypermethylation of critical genes mediates the stage-specific tobacco smoke-induced neoplastic progression of lung cancer. Clin Cancer Res. 2005;11:2466–2470. doi: 10.1158/1078-0432.CCR-04-1962. [DOI] [PubMed] [Google Scholar]
- 263.Vaissiere T, Hung RJ, Zaridze D, et al. Quantitative analysis of DNA methylation profiles in lung cancer identifies aberrant DNA methylation of specific genes and its association with gender and cancer risk factors. Cancer Res. 2009;69:243–252. doi: 10.1158/0008-5472.CAN-08-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ulrich CM, Potter JD. Folate and cancer--timing is everything. JAMA. 2007;297:2408–2409. doi: 10.1001/jama.297.21.2408. [DOI] [PubMed] [Google Scholar]
- 265.Carroll C, Cooper K, Papaioannou D, et al. Meta-analysis: folic acid in the prevention of colorectal adenomas and the chemoprevention of colorectal cancer. Aliment Pharmacol Ther. 2010;31:708–718. doi: 10.1111/j.1365-2036.2010.04238.x. [DOI] [PubMed] [Google Scholar]
- 266.Cole BF, Baron JA, Sandler RS, et al. Folic acid for the prevention of colorectal adenomas: a randomized clinical trial. JAMA. 2007;297:2351–2359. doi: 10.1001/jama.297.21.2351. [DOI] [PubMed] [Google Scholar]
- 267.Kadaveru K, Protiva P, Greenspan EJ, et al. Dietary Methyl Donor Depletion Protects Against Intestinal Tumorigenesis in ApcMin/+ Mice. Cancer Prev Res (Phila) 2012;5:911–920. doi: 10.1158/1940-6207.CAPR-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Schaible TD, Harris RA, Dowd SE, et al. Maternal methyl-donor supplementation induces prolonged murine offspring colitis susceptibility in association with mucosal epigenetic and microbiomic changes. Hum Mol Genet. 2011;20:1687–1696. doi: 10.1093/hmg/ddr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Kim YI, Pogribny IP, Salomon RN, et al. Exon-specific DNA hypomethylation of the p53 gene of rat colon induced by dimethylhydrazine. Modulation by dietary folate. The American journal of pathology. 1996;149:1129–1137. [PMC free article] [PubMed] [Google Scholar]
- 270.Kawakami K, Ruszkiewicz A, Bennett G, et al. The folate pool in colorectal cancers is associated with DNA hypermethylation and with a polymorphism in methylenetetrahydrofolate reductase. Clin Cancer Res. 2003;9:5860–5865. [PubMed] [Google Scholar]
- 271.van Engeland M, Weijenberg MP, Roemen GM, et al. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res. 2003;63:3133–3137. [PubMed] [Google Scholar]
- 272.Schernhammer ES, Giovannucci E, Baba Y, et al. B vitamins, methionine and alcohol intake and risk of colon cancer in relation to BRAF mutation and CpG island methylator phenotype (CIMP) PLoS One. 2011;6:e21102. doi: 10.1371/journal.pone.0021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Razzak AA, Oxentenko AS, Vierkant RA, et al. Alcohol intake and colorectal cancer risk by molecularly defined subtypes in a prospective study of older women. Cancer Prev Res (Phila) 2011;4:2035–2043. doi: 10.1158/1940-6207.CAPR-11-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Slattery ML, Curtin K, Sweeney C, et al. Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer. 2007;120:656–663. doi: 10.1002/ijc.22342. [DOI] [PubMed] [Google Scholar]
- 275.Curtin K, Samowitz WS, Ulrich CM, et al. Nutrients in Folate-Mediated, One-Carbon Metabolism and the Risk of Rectal Tumors in Men and Women. Nutrition and cancer. 2011;63:357–366. doi: 10.1080/01635581.2011.535965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Curtin K, Ulrich CM, Samowitz WS, et al. Candidate pathway polymorphisms in one-carbon metabolism and risk of rectal tumor mutations. Int J Mol Epidemiol Genet. 2011;2:1–8. [PMC free article] [PubMed] [Google Scholar]
- 277.Razzak AA, Oxentenko AS, Vierkant RA, et al. Associations Between Intake of Folate and Related Micronutrients with Molecularly Defined Colorectal Cancer Risks in the Iowa Women's Health Study. Nutrition and cancer. 2012;64:899–910. doi: 10.1080/01635581.2012.714833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.de Vogel S, Wouters KA, Gottschalk RW, et al. Genetic variants of methyl metabolizing enzymes and epigenetic regulators: associations with promoter CpG island hypermethylation in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:3086–3096. doi: 10.1158/1055-9965.EPI-09-0289. [DOI] [PubMed] [Google Scholar]
- 279.Curtin K, Slattery ML, Ulrich CM, et al. Genetic polymorphisms in one-carbon metabolism: associations with CpG island methylator phenotype (CIMP) in colon cancer and the modifying effects of diet. Carcinogenesis. 2007;28:1672–1679. doi: 10.1093/carcin/bgm089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Hazra A, Fuchs CS, Kawasaki T, et al. Germline polymorphisms in the one-carbon metabolism pathway and DNA methylation in colorectal cancer. Cancer Causes Control. 2010;21:331–345. doi: 10.1007/s10552-009-9464-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Linhart HG, Troen A, Bell GW, et al. Folate deficiency induces genomic uracil misincorporation and hypomethylation but does not increase DNA point mutations. Gastroenterology. 2009;136:227–235. e223. doi: 10.1053/j.gastro.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Hervouet E, Debien E, Campion L, et al. Folate supplementation limits the aggressiveness of glioma via the remethylation of DNA repeats element and genes governing apoptosis and proliferation. Clin Cancer Res. 2009;15:3519–3529. doi: 10.1158/1078-0432.CCR-08-2062. [DOI] [PubMed] [Google Scholar]
- 283.Wasson GR, McGlynn AP, McNulty H, et al. Global DNA and p53 region-specific hypomethylation in human colonic cells is induced by folate depletion and reversed by folate supplementation. J Nutr. 2006;136:2748–2753. doi: 10.1093/jn/136.11.2748. [DOI] [PubMed] [Google Scholar]
- 284.Schernhammer ES, Ogino S, Fuchs CS. Folate intake and risk of colon cancer in relation to p53 status. Gastroenterology. 2008;135:770–780. doi: 10.1053/j.gastro.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Schernhammer ES, Giovannucci E, Kawasaki T, et al. Dietary folate, alcohol, and B vitamins in relation to LINE-1 hypomethylation in colon cancer. Gut. 2010;59:794–799. doi: 10.1136/gut.2009.183707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Figueiredo JC, Grau MV, Wallace K, et al. Global DNA hypomethylation (LINE-1) in the normal colon and lifestyle characteristics and dietary and genetic factors. Cancer Epidemiol Biomarkers Prev. 2009;18:1041–1049. doi: 10.1158/1055-9965.EPI-08-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Pufulete M, Al-Ghnaniem R, Khushal A, et al. Effect of folic acid supplementation on genomic DNA methylation in patients with colorectal adenoma. Gut. 2005;54:648–653. doi: 10.1136/gut.2004.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Vrieling A, Kampman E. The role of body mass index, physical activity, and diet in colorectal cancer recurrence and survival: a review of the literature. Am J Clin Nutr. 2010;92:471–490. doi: 10.3945/ajcn.2010.29005. [DOI] [PubMed] [Google Scholar]
- 289.Ballard-Barbash R, Friedenreich CM, Courneya KS, et al. Physical Activity, Biomarkers, and Disease Outcomes in Cancer Survivors: A Systematic Review. J Natl Cancer Inst. 2012;104:815–840. doi: 10.1093/jnci/djs207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Jeon JY, Meyerhardt JA. Energy in and energy out: what matters for survivors of colorectal cancer? J Clin Oncol. 2012;30:7–10. doi: 10.1200/JCO.2011.39.6374. [DOI] [PubMed] [Google Scholar]
- 291.Giovannucci EL. Physical activity as a standard cancer treatment. J Natl Cancer Inst. 2012;104:797–799. doi: 10.1093/jnci/djs229. [DOI] [PubMed] [Google Scholar]
- 292.Clague J, Bernstein L. Physical Activity and Cancer. Current oncology reports. 2012 doi: 10.1007/s11912-012-0265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Betof AS, Dewhirst MW, Jones LW. Effects and potential mechanisms of exercise training on cancer progression: A translational perspective. Brain, behavior, and immunity. 2012 doi: 10.1016/j.bbi.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Hughes LA, van den Brandt PA, de Bruine AP, et al. Early life exposure to famine and colorectal cancer risk: a role for epigenetic mechanisms. PLoS One. 2009;4:e7951. doi: 10.1371/journal.pone.0007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Slattery ML, Curtin K, Anderson K, et al. Associations between cigarette smoking, lifestyle factors, and microsatellite instability in colon tumors. J Natl Cancer Inst. 2000;92:1831–1836. doi: 10.1093/jnci/92.22.1831. [DOI] [PubMed] [Google Scholar]
- 296.Satia JA, Keku T, Galanko JA, et al. Diet, lifestyle, and genomic instability in the north Carolina colon cancer study. Cancer Epidemiol Biomarkers Prev. 2005;14:429–436. doi: 10.1158/1055-9965.EPI-04-0486. [DOI] [PubMed] [Google Scholar]
- 297.Campbell PT, Jacobs ET, Ulrich CM, et al. Case-control study of overweight, obesity, and colorectal cancer risk, overall and by tumor microsatellite instability status. J Natl Cancer Inst. 2010;102:391–400. doi: 10.1093/jnci/djq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kuchiba A, Morikawa T, Yamauchi M, et al. Body mass index and risk of colorectal cancer according to fatty acid synthase expression in the nurses' health study. J Natl Cancer Inst. 2012;104:415–420. doi: 10.1093/jnci/djr542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Uddin S, Hussain AR, Ahmed M, et al. High Prevalence of Fatty Acid Synthase Expression in Colorectal Cancers in Middle Eastern Patients and Its Potential Role as a Therapeutic Target. Am J Gastroenterol. 2009;104:1790–1801. doi: 10.1038/ajg.2009.230. [DOI] [PubMed] [Google Scholar]
- 300.Migita T, Ruiz S, Fornari A, et al. Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. J Natl Cancer Inst. 2009;101:519–532. doi: 10.1093/jnci/djp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Ogino S, kawasaki T, Ogawa A, et al. Fatty acid synthase overexpression in colorectal cancer is associated with microsatellite instability, independent of CpG island methylator phenotype. Hum Pathol. 2007;38:842–849. doi: 10.1016/j.humpath.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 302.Baba Y, Nosho K, Shima K, et al. Prognostic significance of AMP-activated protein kinase expression and modifying effect of MAPK3/1 in colorectal cancer. Br J Cancer. 2010;103:1025–1033. doi: 10.1038/sj.bjc.6605846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ogino S, Nosho K, Baba Y, et al. A cohort study of STMN1 expression in colorectal cancer: body mass index and prognosis. Am J Gastroenterol. 2009;104:2047–2056. doi: 10.1038/ajg.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Ogino S, Nosho K, Shima K, et al. p21 expression in colon cancer and modifying effects of patient age and body mass index on prognosis. Cancer Epidemiol Biomarkers Prev. 2009;18:2513–2521. doi: 10.1158/1055-9965.EPI-09-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Ogino S, Shima K, Nosho K, et al. A cohort study of p27 localization in colon cancer, body mass index, and patient survival. Cancer Epidemiol Biomarkers Prev. 2009;18:1849–1858. doi: 10.1158/1055-9965.EPI-09-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Meyerhardt JA, Ogino S, Kirkner GJ, et al. Interaction of molecular markers and physical activity on mortality in patients with colon cancer. Clin Cancer Res. 2009;15:5931–5936. doi: 10.1158/1078-0432.CCR-09-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Li L, Lee KM, Han W, et al. Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum Mol Genet. 2010;19:4273–4277. doi: 10.1093/hmg/ddq351. [DOI] [PubMed] [Google Scholar]
- 308.Wu AH, Siegmund KD, Long TI, et al. Hormone therapy, DNA methlyation and colon cancer. Carcinogenesis. 2010;31:1060–1067. doi: 10.1093/carcin/bgq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Lin JH, Morikawa T, Chan AT, et al. Postmenopausal hormone therapy is associated with a reduced risk of colorectal cancer lacking CDKN1A expression. Cancer Res. 2012;72:3020–3028. doi: 10.1158/0008-5472.CAN-11-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Kure S, Nosho K, Baba Y, et al. Vitamin D receptor expression is associated with PIK3CA and KRAS mutations in colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2765–2772. doi: 10.1158/1055-9965.EPI-09-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Pereira F, Barbachano A, Silva J, et al. KDM6B/JMJD3 histone demethylase is induced by vitamin D and modulates its effects in colon cancer cells. Hum Mol Genet. 2011;20:4655–4665. doi: 10.1093/hmg/ddr399. [DOI] [PubMed] [Google Scholar]
- 312.Blumberg R, Powrie F. Microbiota, disease, and back to health: a metastable journey. Sci Transl Med. 2012;4:137–137. doi: 10.1126/scitranslmed.3004184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Larsson E, Tremaroli V, Lee YS, et al. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Ly NP, Litonjua A, Gold DR, et al. Gut microbiota, probiotics, and vitamin D: interrelated exposures influencing allergy, asthma, and obesity? J Allergy Clin Immunol. 2011;127:1087–1094. doi: 10.1016/j.jaci.2011.02.015. quiz 1095–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Allen M, Louise Jones J. Jekyll and Hyde: the role of the microenvironment on the progression of cancer. J Pathol. 2011;223:162–176. doi: 10.1002/path.2803. [DOI] [PubMed] [Google Scholar]
- 316.Fridman WH, Pages F, Sautes-Fridman C, et al. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 317.Ding SZ, Goldberg JB, Hatakeyama M. Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol. 2010;6:851–862. doi: 10.2217/fon.10.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Peterson AJ, Menheniott TR, O'Connor L, et al. Helicobacter pylori infection promotes methylation and silencing of trefoil factor 2, leading to gastric tumor development in mice and humans. Gastroenterology. 2010;139:2005–2017. doi: 10.1053/j.gastro.2010.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Bebek G, Bennett KL, Funchain P, et al. Microbiomic subprofiles and MDR1 promoter methylation in head and neck squamous cell carcinoma. Hum Mol Genet. 2012;21:1557–1565. doi: 10.1093/hmg/ddr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Fernandez AF, Esteller M. Viral epigenomes in human tumorigenesis. Oncogene. 2010;29:1405–1420. doi: 10.1038/onc.2009.517. [DOI] [PubMed] [Google Scholar]
- 321.Fernandez AF, Rosales C, Lopez-Nieva P, et al. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome research. 2009;19:438–451. doi: 10.1101/gr.083550.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.De-Castro Arce J, Gockel-Krzikalla E, Rosl F. Silencing of multi-copy HPV16 by viral self-methylation and chromatin occlusion: a model for epigenetic virus-host interaction. Hum Mol Genet. 2012;21:1693–1705. doi: 10.1093/hmg/ddr604. [DOI] [PubMed] [Google Scholar]
- 323.Hino R, Uozaki H, Murakami N, et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766–2774. doi: 10.1158/0008-5472.CAN-08-3070. [DOI] [PubMed] [Google Scholar]
- 324.Goel A, Li MS, Nagasaka T, et al. Association of JC Virus T-Antigen Expression With the Methylator Phenotype in Sporadic Colorectal Cancers. Gastroenterology. 2006;130:1950–1961. doi: 10.1053/j.gastro.2006.02.061. [DOI] [PubMed] [Google Scholar]
- 325.Nosho K, Shima K, Kure S, et al. JC virus T-antigen in colorectal cancer is associated with p53 expression and chromosomal instability, independent of CpG island methylator phenotype. Neoplasia. 2009;11:87–95. doi: 10.1593/neo.81188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Karpinski P, Myszka A, Ramsey D, et al. Detection of viral DNA sequences in sporadic colorectal cancers in relation to CpG island methylation and methylator phenotype. Tumour Biol. 2011;32:653–659. doi: 10.1007/s13277-011-0165-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Wang D, Dubois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29:781–788. doi: 10.1038/onc.2009.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Wang D, Xia D, DuBois RN. The crosstalk of PTGS2 and EGF signaling pathways in colorectal cancer. Cancers. 2011;3:3894–3908. doi: 10.3390/cancers3043894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Wang D, Dubois RN. Associations Between Obesity and Cancer: The Role of Fatty Acid Synthase. J Natl Cancer Inst. 2012;104:343–345. doi: 10.1093/jnci/djs010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Girnun G. PPARG: a new independent marker for colorectal cancer survival. Gastroenterology. 2009;136:1157–1160. doi: 10.1053/j.gastro.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 332.Kanai Y, Hirohashi S. Alterations of DNA methylation associated with abnormalities of DNA methyltransferases in human cancers during transition from a precancerous to a malignant state. Carcinogenesis. 2007;28:2434–2442. doi: 10.1093/carcin/bgm206. [DOI] [PubMed] [Google Scholar]
- 333.O'Hagan HM, Wang W, Sen S, et al. Oxidative Damage Targets Complexes Containing DNA Methyltransferases, SIRT1, and Polycomb Members to Promoter CpG Islands. Cancer Cell. 2011;20:606–619. doi: 10.1016/j.ccr.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Brigati C, Banelli B, di Vinci A, et al. Inflammation, HIF-1, and the epigenetics that follows. Mediators Inflamm. 2010;2010:263914. doi: 10.1155/2010/263914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Xia D, Wang D, Kim SH, et al. Prostaglandin E(2) promotes intestinal tumor growth via DNA methylation. Nat Med. 2012;18:224–226. doi: 10.1038/nm.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Kellermayer R, Balasa A, Zhang W, et al. Epigenetic maturation in colonic mucosa continues beyond infancy in mice. Hum Mol Genet. 2010;19:2168–2176. doi: 10.1093/hmg/ddq095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Chan AT, Ogino S, Fuchs CS. Aspirin and the Risk of Colorectal Cancer in Relation to the Expression of COX-2. N Engl J Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 338.Ogino S, Kirkner GJ, Nosho K, et al. Cyclooxygenase-2 expression is an independent predictor of poor prognosis in colon cancer. Clin Cancer Res. 2008;14:8221–8227. doi: 10.1158/1078-0432.CCR-08-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Yamauchi M, Lochhead P, Morikawa T, et al. Colorectal cancer: a tale of two sides or a continuum? Gut. 2012;61:794–797. doi: 10.1136/gutjnl-2012-302014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Li X, Leblanc J, Truong A, et al. A metaproteomic approach to study human-microbial ecosystems at the mucosal luminal interface. PLoS One. 2011;6:e26542. doi: 10.1371/journal.pone.0026542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Edin S, Wikberg ML, Dahlin AM, et al. The distribution of macrophages with a m1 or m2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS One. 2012;7:e47045. doi: 10.1371/journal.pone.0047045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Cortese R, Kwan A, Lalonde E, et al. Epigenetic markers of prostate cancer in plasma circulating DNA. Hum Mol Genet. 2012;21:3619–3631. doi: 10.1093/hmg/dds192. [DOI] [PubMed] [Google Scholar]
- 343.Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 344.Terry MB, Delgado-Cruzata L, Vin-Raviv N, et al. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–837. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Mirabello L, Savage SA, Korde L, et al. LINE-1 methylation is inherited in familial testicular cancer kindreds. BMC Med Genet. 2010;11:77. doi: 10.1186/1471-2350-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Wu HC, Delgado-Cruzata L, Flom JD, et al. Global methylation profiles in DNA from different blood cell types. Epigenetics. 2011;6:76–85. doi: 10.4161/epi.6.1.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Wu HC, John EM, Ferris JS, et al. Global DNA methylation levels in girls with and without a family history of breast cancer. Epigenetics. 2011;6:29–33. doi: 10.4161/epi.6.1.13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Breitling LP, Yang R, Korn B, et al. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.van Bemmel D, Lenz P, Liao LM, et al. Correlation of LINE-1 Methylation Levels in Patient-Matched Buffy Coat, Serum, Buccal Cell, and Bladder Tumor Tissue DNA Samples. Cancer Epidemiol Biomarkers Prev. 2012;21:1143–1148. doi: 10.1158/1055-9965.EPI-11-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Breton CV, Byun HM, Wenten M, et al. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Breton CV, Salam MT, Wang X, et al. Particulate Matter, DNA Methylation in Nitric Oxide Synthase, and Childhood Respiratory Disease. Environ Health Perspect. 2012;120:1320–1326. doi: 10.1289/ehp.1104439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Iacopetta B, Grieu F, Phillips M, et al. Methylation levels of LINE-1 repeats and CpG island loci are inversely related in normal colonic mucosa. Cancer Sci. 2007;98:1454–1460. doi: 10.1111/j.1349-7006.2007.00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Liu J, Hesson LB, Meagher AP, et al. Relative Distribution of Folate Species Is Associated with Global DNA Methylation in Human Colorectal Mucosa. Cancer Prev Res (Phila) 2012;5:921–929. doi: 10.1158/1940-6207.CAPR-11-0577. [DOI] [PubMed] [Google Scholar]
- 354.Buda A, De Bona M, Dotti I, et al. Prevalence of Different Subtypes of Serrated Polyps and Risk of Synchronous Advanced Colorectal Neoplasia in Average-Risk Population Undergoing First-Time Colonoscopy. Clin Transl Gasntroenterol. 2012;3:e6. doi: 10.1038/ctg.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Hunter DJ. Lessons from genome-wide association studies for epidemiology. Epidemiology. 2012;23:363–367. doi: 10.1097/EDE.0b013e31824da7cc. [DOI] [PubMed] [Google Scholar]
- 356.Spain SL, Carvajal-Carmona LG, Howarth KM, et al. Refinement of the associations between risk of colorectal cancer and polymorphisms on chromosomes 1q41 and 12q13.13. Hum Mol Genet. 2012;21:934–946. doi: 10.1093/hmg/ddr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Price AL, Zaitlen NA, Reich D, et al. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Thomas D. Gene--environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Hutter CM, Chang-Claude J, Slattery ML, et al. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 2012;72:2036–2044. doi: 10.1158/0008-5472.CAN-11-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.On beyond GWAS. Nat Genet. 2010;42:551. doi: 10.1038/ng0710-551. [DOI] [PubMed] [Google Scholar]
- 361.Ghosh A, Hartge P, Purdue MP, et al. Assessing Disease Risk in Genome-wide Association Studies Using Family History. Epidemiology. 2012;23:616–622. doi: 10.1097/EDE.0b013e31825583a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Freedman ML, Monteiro AN, Gayther SA, et al. Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet. 2011;43:513–518. doi: 10.1038/ng.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Tuupanen S, Turunen M, Lehtonen R, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 364.Gruber SB, Moreno V, Rozek LS, et al. Genetic variation in 8q24 associated with risk of colorectal cancer. Cancer Biol Ther. 2007;6:1143–1147. doi: 10.4161/cbt.6.7.4704. [DOI] [PubMed] [Google Scholar]
- 365.Slattery ML, Herrick J, Curtin K, et al. Increased Risk of Colon Cancer Associated with a Genetic Polymorphism of SMAD7. Cancer Res. 2010;70:1479–1485. doi: 10.1158/0008-5472.CAN-08-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Garcia-Albeniz X, Nan H, Valeri L, et al. Phenotypic and tumor molecular characterization of colorectal cancer in relation to a susceptibility SMAD7 variant associated with survival. Carcinogenesis. 2013 doi: 10.1093/carcin/bgs335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Rakyan VK, Down TA, Balding DJ, et al. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Heijmans BT, Mill J. Commentary: The seven plagues of epigenetic epidemiology. Int J Epidemiol. 2012;41:74–78. doi: 10.1093/ije/dyr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw. 2011;9(Suppl 5):S1–32. doi: 10.6004/jnccn.2011.0137. quiz S33. [DOI] [PubMed] [Google Scholar]
- 370.Khoury MJ, Gwinn M, Ioannidis JP. The emergence of translational epidemiology: from scientific discovery to population health impact. Am J Epidemiol. 2010;172:517–524. doi: 10.1093/aje/kwq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Butler D. Translational research: crossing the valley of death. Nature. 2008;453:840–842. doi: 10.1038/453840a. [DOI] [PubMed] [Google Scholar]
- 372.Colditz GA, Winn DM. Criteria for the evaluation of large cohort studies: an application to the nurses' health study. J Natl Cancer Inst. 2008;100:918–925. doi: 10.1093/jnci/djn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Colditz GA, Hankinson SE. The Nurses' Health Study: lifestyle and health among women. Nat Rev Cancer. 2005;5:388–396. doi: 10.1038/nrc1608. [DOI] [PubMed] [Google Scholar]
- 374.Robison LL, Armstrong GT, Boice JD, et al. The Childhood Cancer Survivor Study: a National Cancer Institute-supported resource for outcome and intervention research. J Clin Oncol. 2009;27:2308–2318. doi: 10.1200/JCO.2009.22.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Colditz GA. Ensuring long-term sustainability of existing cohorts remains the highest priority to inform cancer prevention and control. Cancer Causes Control. 2010;21:649–656. doi: 10.1007/s10552-009-9498-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Hansen KD, Timp W, Bravo HC, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Xu Y, Hu B, Choi AJ, et al. Unique DNA methylome profiles in CpG island methylator phenotype colon cancers. Genome research. 2012;22:283–291. doi: 10.1101/gr.122788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Spitz MR, Caporaso NE, Sellers TA. Integrative cancer epidemiology — the next generation. Cancer Discovery. 2012;2:1087–1090. doi: 10.1158/2159-8290.CD-12-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]