Abstract
Age-related macular degeneration (AMD) is associated with multiple genetic and cellular defects which lead to a common endpoint, retinal degeneration. Aging and oxidative stress, significant features in the pathogenesis of AMD, are associated with an increase in damaged intracellular organelles and defective autophagy flux in a range of age-related and neurodegenerative diseases. Autophagy is a key process in maintenance of cellular homeostasis that serves to remove dysfunctional organelles and proteins. Autophagy proteins are strongly expressed in the retina and there is now strong evidence that mitochondrial damage and defective autophagy are a feature of the aging retina and that this is further exacerbated in AMD. It is apparent that autophagy makes a significant contribution to lipofuscin accumulation in the RPE. Pharmacological manipulation of autophagy may offer an alternative therapeutic target in AMD.
Keywords: Autophagy, retinal pigment epithelium, age-related macular degeneration, retina, mitochondria, lipofuscin, oxidative damage
XX.1 Introduction
The significance of autophagy in health and disease has only become fully appreciated in the last decade. Under normal conditions, autophagy operates constitutively and serves as a housekeeping process through which cytoplasmic proteins and damaged cellular organelles, such as dysfunctional mitochondria, are removed (Marino et al. 2010). Of the three autophagic pathways (chaperone-mediated, micro-, and macroautophagy) that deliver cellular components of varying sizes to lysosomes, macroautophagy is the primary route for sequestration of organelles or large aggregates and their delivery to the lysosome (Cuervo 2008; Lieberthal 2008). It is evident that autophagy plays a key role in cellular homeostasis and that this process can be stimulated to cope with excessive organelle damage, aggregate removal and pathogen defense (Cuervo 2008).
XX.2 Molecular events in the autophagy process
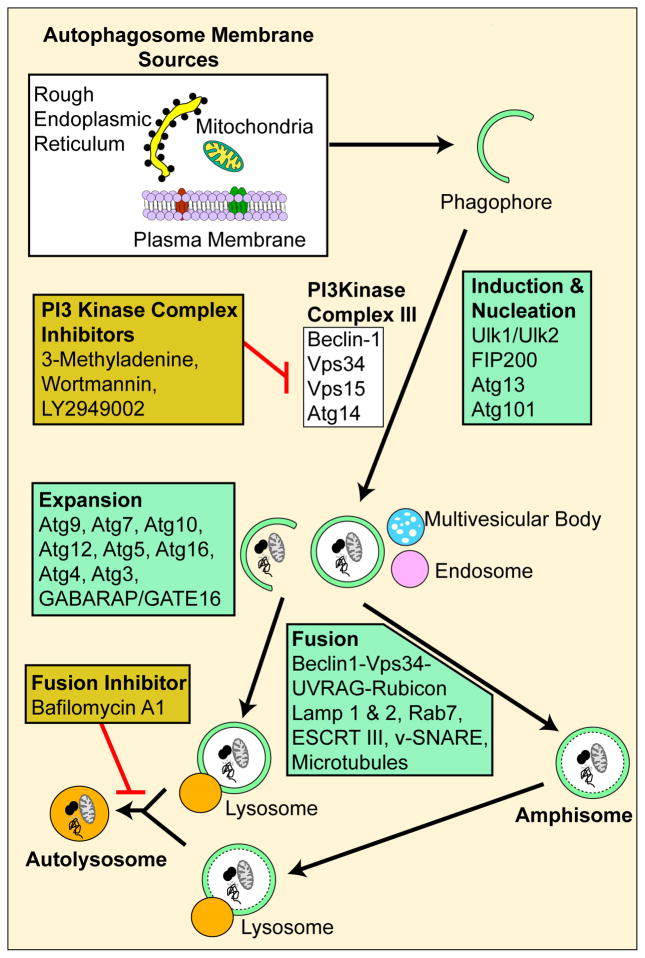
Since accumulation of proteins and damaged organelles are a general observation in the aging RPE as well as in AMD, it is postulated that a breakdown in the recycling capacity of autophagy may have a strong association. The process of autophagy is outlined in Figure 1 and involves over 30 autophagy-related proteins (Atg) which regulate different stages of the autophagic pathway. The autophagic process begins with the formation of an isolation membrane, also referred to as the phagophore (Yorimitsu and Klionsky 2005) that that is shown to originate primarily from the endoplasmic reticulum (Dunn 1990; Hamasaki and Yoshimori 2010). The phagophore gradually expands to engulf the cargo (e.g. mitochondria) to form a closed double-membrane structure termed an early autophagosome. Autophagosome maturation depend on two ubiquitin-like-conjugation systems namely the Atg12-Atg5-Atg16 complex and the LC3 conjugation system. Both the systems are regulated by the Atg7 molecule (Geng and Klionsky 2008). The mature autophagsome fuses with either the lysosome, late endosome or the multivesicular body to form the amphisome or late autophagosome which subsequently matures into the autolysosome. Lysosomes contain potent hydrolytic enzymes which then degrade the engulfed contents and the indigestible residual body, formed as an endpoint of lysosomal digestion, may subsequently be removed by exocytosis or may contribute to lipofuscin (Luzio et al. 2007; Settembre et al. 2008). A critical property of the lysosome that facilitates the fusion of the autophagosome to the lysosome and the digestive activity of the lysosomal enzymes is its pH which typically is acidic at around 4.5 (Kawai et al. 2007).
Figure 1.
A schematic showing the basic steps of mammalian macroautophagy, the common autophagic molecules involved and pharmacological inhibitors used to block autophagy at different steps.
XX.3 Signaling mechanisms in autophagy
The mTOR kinase complexes have been widely studied as the central signaling molecules of autophagy and can sense regulating conditions such as nutrient abundance, energy state and growth factor levels (Ravikumar et al. 2004; Nobukuni et al. 2005). Interaction of Beclin1 with the antiapoptotic BH3 proteins such as Bcl-XL and Bcl-2 is also a critical aspect of autophagy regulation and could influence autophagy even independent of mTOR (Pattingre et al. 2005). However, it is likely that several other mTOR independent mechanisms of autophagy activation may also exist.
While starvation has been used as an inducer of autophagy in most studies, oxidative stress has also been acknowledged as a positive regulator of autophagy, at least in acute phases (Kiffin et al. 2006). However, it is now becoming evident that the pathways regulating baseline autophagy, starvation-induced autophagy and stress-induced autophagy have fundamental differences. It has been observed that autophagic deficient cells tend to accumulate p62 rich aggregates which in turn cause Nrf2 to be activated after separation from its interacting partner Keap1 which allows Nrf2 to mount an antioxidant response (Komatsu et al. 2010).
XX.4 Autophagy in the neural retina
We have demonstrated by immunohistochemistry that the autophagy proteins Atg9 and LC3 are strongly expressed in the ganglion cell layer, a subpopulation of cells in the inner nuclear layer, the outer nuclear layer, and the RPE in normal mouse retina (Figure 2). Interestingly, these represent cell layers with high metabolic demand and a propensity for mitochondrial damage (Jarrett et al. 2010).
Figure 2.
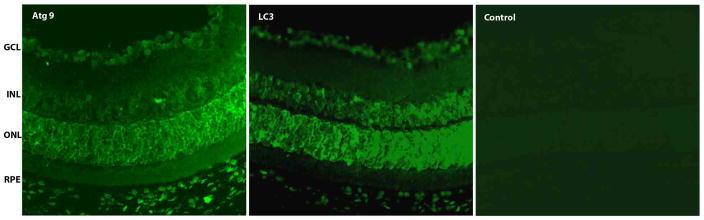
Immunolocalization of autophagy proteins (green) in normal mouse retina. Atg9 and LC3 were strongly expressed in the ganglion cell layer, retinal vessels, a subpopulation of the inner nuclear layer, the outer nuclear layer of rods and cones, and the RPE.
Autophagy in photoreceptor cells was first documented in 1977 (Reme and Young 1977). Autophagy in photoreceptor inner segments shows circadian rhythmicity (Reme and Wirz-Justice 1985) and is associated with lipofuscin accumulation in rod and cone photoreceptors (Iwasaki and Inomata 1988). Autophagic cell death in the retina has been reported to occur in a variety of retinal cells under oxidative stress (Kunchithapautham and Rohrer 2007) and autophagy occurs prior to programmed necrotic cell death of retinal neurons following ischemia (Rosenbaum et al. 2010). However, it has yet to be shown whether the autophagic response to ischemia is protective or detrimental to the neurons. Autophagy has been shown to induce axonal degeneration of retinal ganglion cells after crush lesion (Knoferle et al. 2010) while it can be protective to retinal ganglion cells following optic nerve transection (Kim et al. 2008). Stimulation of the insulin/mTOR pathway protects cone photoreceptors in a mouse model of retinitis pigmentosa (Punzo et al. 2009) and up-regulation of the autophagy protein Apg3 guards the retina from severe ischemic injury (Wu et al. 2006).
XX.5 Autophagy in the RPE
There is now considerable evidence that the RPE, like most other cells maintains a basal autophagy for cellular homeostasis and that this changes with both age and disease. We and others have shown that autophagy proteins are strongly expressed in the RPE (Figure 2) (Wang et al. 2009b; Wang et al. 2009a; Krohne et al. 2010; Viiri et al. 2010). Furthermore, RPE cells can accumulate lipofuscin even in the absence of a photoreceptor substrate strongly suggestive that autophagy is involved (Boulton et al. 1989; Burke and Skumatz 1998; Kurz et al. 2009). The autophagy and phagocytic pathways are interdependent and both culminate in lysosomal degradation of the substrate (Figure 3). Thus autophagy flux in the RPE is likely to be highly susceptible to changes in lysosomal pH or the accumulation of lipofuscin as both will impede the fusion of autophagosomes with lysosomes. Despite the paucity of corroborative data it does appear that autophagy proteins and flux show an age-related increase within mouse and human RPE (Wang et al. 2009c).
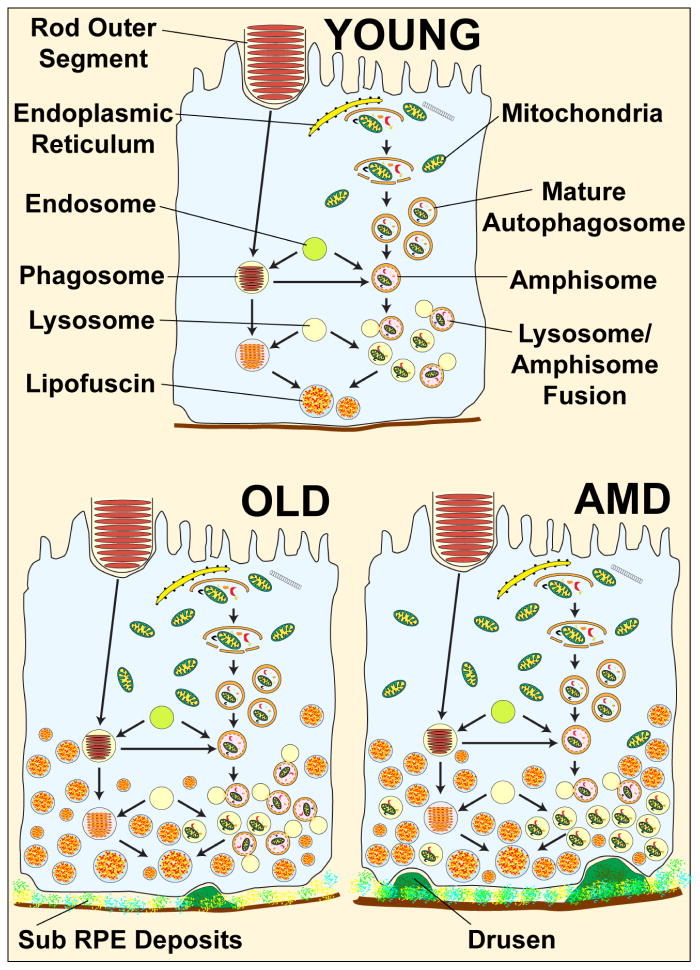
Figure 3.
Hypothetical schematic showing possible outcomes for aging and AMD on autophagy in RPE cells. Young: Autophagy will occur at a basal level and remove dysfunctional organelles and aggregates. Formation of autophagosomes, fusion of these with lysosomes and clearance are all balanced. Lipofuscin in residual bodies is minimally present. Sub-RPE drusen is absent. Old: Autophagy is increased in the old RPE cells compared to young, in line with the increased accumulation of aggregates and damaged organelles with age that need to be cleared. Levels of lipofuscin in residual bodies are increased and drusen are present. AMD: Autophagy is dysfunctional in the RPE of AMD patients. Although autophagosome formation may be equivalent to an old RPE cell, fusion with the lysosomes and degradation of the engulfed material may be greatly impaired leading to an accumulation of autophagosomes. The cell is laden with secondary lysosomes containing partially degraded material, there are high levels of lipofuscin and sub RPE drusen are prominent.
XX.6 The association between autophagy and AMD
Many of the important pathogenic features of the RPE in AMD, e.g. lipofuscin accumulation, susceptibility to oxidative stress, mitochondrial damage and lysosomal dysregulation have an association with autophagy. However, it remains to be determined whether changes in autophagy flux are a cause or consequence of disease and whether autophagy changes reflect alterations in the formation or elimination of autophagosomes. A recent study by Wang et al suggest that drusen, a feature of early AMD, may reflect an increase in both mitochondrial damage and autophagy (Wang et al. 2009a). Using cultured RPE cells and ex vivo AMD donor tissue they observed that a) under conditions of increased mtDNA damage, autophagy markers and exosome markers were upregulated and b) that drusen in AMD donor eyes contain markers for autophagy and exosomes. They speculated that increased autophagy and the release of intracellular proteins via exosomes by the aged RPE may contribute to the formation of drusen. The occupation of lysosomal volume by lipofuscin may alone be sufficient to impair autophagosome-lysosome fusion, while, alternatively an increase in lysosomal pH may be responsible. A2E, a component lipofuscin, (Boulton 2009) has been reported to inhibit the lysosomal ATP-driven proton pump resulting in an increase of the lysosomal pH (Bergmann et al. 2004). Furthermore, lipid peroxidation products reduce autophagy flux and increase lipofuscin accumulation in the RPE (Krohne et al. 2010). Interestingly, the effect of A2E on lysosomal pH can be reversed through reacidification (Liu et al. 2008).
There is substantial cross-talk between autophagy and proteasomal degradation pathways (Kaarniranta et al. 2009; Ryhanen et al. 2009; Kaarniranta 2010). HSP70, proteasomes, and macroautophagy combine to regulate protein turnover in human RPE cells (Ryhanen et al. 2009). The p62/sequestosome 1 links the proteasomal and lysosomal clearance systems (Korolchuk et al. 2009) and, via association with LC3, directs aggregates to the lysosome for autophagic degradation (Seibenhener et al. 2007; Kirkin et al. 2009). Proteasome inhibition in RPE cells evoked the accumulation of perinuclear aggregates that strongly colocalized with p62 and HSP70 and the silencing of p62, rather than HSP70, evoked suppression of autophagy (Viiri et al. 2010).
XX.7 Conclusion
It is evident that autophagy plays a significant role as a house keeping pathway in the retina and that autophagy flux is less effective with aging. Figure 3 provides a hypothetical schematic showing possible outcomes for aging and AMD on autophagy in RPE cells. Furthermore, the interactive role between autophagy, phagocytosis, lysosomal function and the proteosomal system, all of which are dysregulated in AMD, require further study. Autophagy modulation may offer a treatment regime for various ocular diseases including AMD.
Acknowledgments
This work was funded by NIH grant EY019688 and AHAF grant M2009024.
XX.9 References
- Bergmann M, Schutt F, Holz FG, et al. Inhibition of the ATP-driven proton pump in RPE lysosomes by the major lipofuscin fluorophore A2-E may contribute to the pathogenesis of age-related macular degeneration. FASEB J. 2004;18:562–564. doi: 10.1096/fj.03-0289fje. [DOI] [PubMed] [Google Scholar]
- Boulton M, McKechnie NM, Breda J, et al. The formation of autofluorescent granules in cultured human RPE. Invest Ophthalmol Vis Sci. 1989;30:82–89. [PubMed] [Google Scholar]
- Boulton ME. Lipofuscin of the RPE. In: Lois M, Forrester J, editors. Fundus Autofluorescence. Philadelphia: Lipincott Williams and Wilkins; 2009. pp. 14–26. [Google Scholar]
- Burke JM, Skumatz CM. Autofluorescent inclusions in long-term postconfluent cultures of retinal pigment epithelium. Invest Ophthalmol Vis Sci. 1998;39:1478–1486. [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Yoshimori T. Where do they come from? Insights into autophagosome formation. FEBS Lett. 2010;584:1296–1301. doi: 10.1016/j.febslet.2010.02.061. [DOI] [PubMed] [Google Scholar]
- Iwasaki M, Inomata H. Lipofuscin granules in human photoreceptor cells. Invest Ophthalmol Vis Sci. 1988;29:671–679. [PubMed] [Google Scholar]
- Jarrett SG, Lewin AS, Boulton ME. The importance of mitochondria in age-related and inherited eye disorders. Ophthalmic Res. 2010;44:179–190. doi: 10.1159/000316480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaarniranta K. Autophagy--hot topic in AMD. Acta Ophthalmol. 2010;88:387–388. doi: 10.1111/j.1755-3768.2009.01840.x. [DOI] [PubMed] [Google Scholar]
- Kaarniranta K, Salminen A, Eskelinen EL, et al. Heat shock proteins as gatekeepers of proteolytic pathways-Implications for age-related macular degeneration (AMD) Ageing Res Rev. 2009;8:128–139. doi: 10.1016/j.arr.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Kawai A, Uchiyama H, Takano S, et al. Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy. 2007;3:154–157. doi: 10.4161/auto.3634. [DOI] [PubMed] [Google Scholar]
- Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8:152–162. doi: 10.1089/ars.2006.8.152. [DOI] [PubMed] [Google Scholar]
- Kim SH, Munemasa Y, Kwong JM, et al. Activation of autophagy in retinal ganglion cells. J Neurosci Res. 2008;86:2943–2951. doi: 10.1002/jnr.21738. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, et al. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Knoferle J, Koch JC, Ostendorf T, et al. Mechanisms of acute axonal degeneration in the optic nerve in vivo. Proc Natl Acad Sci U S A. 2010;107:6064–6069. doi: 10.1073/pnas.0909794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, Menzies FM, et al. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohne TU, Stratmann NK, Kopitz J, et al. Effects of lipid peroxidation products on lipofuscinogenesis and autophagy in human retinal pigment epithelial cells. Exp Eye Res. 2010;90:465–471. doi: 10.1016/j.exer.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Kunchithapautham K, Rohrer B. Apoptosis and autophagy in photoreceptors exposed to oxidative stress. Autophagy. 2007;3:433–441. doi: 10.4161/auto.4294. [DOI] [PubMed] [Google Scholar]
- Kurz T, Karlsson M, Brunk UT, et al. ARPE-19 retinal pigment epithelial cells are highly resistant to oxidative stress and exercise strict control over their lysosomal redox-active iron. Autophagy. 2009;5:494–501. doi: 10.4161/auto.5.4.7961. [DOI] [PubMed] [Google Scholar]
- Lieberthal W. Macroautophagy: a mechanism for mediating cell death or for promoting cell survival? Kidney Int. 2008;74:555–557. doi: 10.1038/ki.2008.325. [DOI] [PubMed] [Google Scholar]
- Liu J, Lu W, Reigada D, et al. Restoration of lysosomal pH in RPE cells from cultured human and ABCA4(−/−) mice: pharmacologic approaches and functional recovery. Invest Ophthalmol Vis Sci. 2008;49:772–780. doi: 10.1167/iovs.07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- Marino G, Madeo F, Kroemer G. Autophagy for tissue homeostasis and neuroprotection. Curr Opin Cell Biol. 2010 doi: 10.1016/j.ceb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Punzo C, Kornacker K, Cepko CL. Stimulation of the insulin/mTOR pathway delays cone death in a mouse model of retinitis pigmentosa. Nat Neurosci. 2009;12:44–52. doi: 10.1038/nn.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Reme C, Wirz-Justice A. Circadian rhythm, the retina and light. Klin Monbl Augenheilkd. 1985;186:175–179. [PubMed] [Google Scholar]
- Reme CE, Young RW. The effects of hibernation on cone visual cells in the ground squirrel. Invest Ophthalmol Vis Sci. 1977;16:815–840. [PubMed] [Google Scholar]
- Rosenbaum DM, Degterev A, David J, et al. Necroptosis, a novel form of caspase-independent cell death, contributes to neuronal damage in a retinal ischemia-reperfusion injury model. J Neurosci Res. 2010;88:1569–1576. doi: 10.1002/jnr.22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryhanen T, Hyttinen JM, Kopitz J, et al. Crosstalk between Hsp70 molecular chaperone, lysosomes and proteasomes in autophagy-mediated proteolysis in human retinal pigment epithelial cells. J Cell Mol Med. 2009;13:3616–3631. doi: 10.1111/j.1582-4934.2008.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener ML, Geetha T, Wooten MW. Sequestosome 1/p62--more than just a scaffold. FEBS Lett. 2007;581:175–179. doi: 10.1016/j.febslet.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Fraldi A, Jahreiss L, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- Viiri J, Hyttinen JM, Ryhanen T, et al. p62/sequestosome 1 as a regulator of proteasome inhibitor-induced autophagy in human retinal pigment epithelial cells. Mol Vis. 2010;16:1399–1414. [PMC free article] [PubMed] [Google Scholar]
- Wang AL, Lukas TJ, Yuan M, et al. Autophagy and exosomes in the aged retinal pigment epithelium: possible relevance to drusen formation and age-related macular degeneration. PLoS One. 2009a;4:e4160. doi: 10.1371/journal.pone.0004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AL, Boulton ME, Dunn WA, Jr, et al. Using LC3 to monitor autophagy flux in the retinal pigment epithelium. Autophagy. 2009b;5:1190–1193. doi: 10.4161/auto.5.8.10087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Lao U, Edgar BA. TOR-mediated autophagy regulates cell death in Drosophila neurodegenerative disease. J Cell Biol. 2009c;186:703–711. doi: 10.1083/jcb.200904090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BX, Darden AG, Laser M, et al. The rat Apg3p/Aut1p homolog is upregulated by ischemic preconditioning in the retina. Mol Vis. 2006;12:1292–1302. [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]