Abstract
Objective
Angiogenesis requires tightly coordinated cross-talk between endothelial cells and stromal cells such as fibroblasts and smooth muscle cells. The specific molecular mechanisms moderating this process are still poorly understood.
Method and Results
Stromal cell-derived factors are essential for endothelial cell sprouting and lumen formation. We therefore compared the abilities of two primary fibroblast isolates and a primary smooth muscle cell isolate to promote in vitro angiogenesis and analyzed their secretomes using a combination of nanoLC-MS/MS, qPCR and ELISA. Each isolate exhibited a different level of angiogenic ability. Using quantitative MS, we then compared the secretomes of a fibroblast isolate exhibiting low angiogenic activity, a fibroblast isolate exhibiting high angiogenic activity and human umbilical vein endothelial cells. High angiogenic fibroblast supernatants exhibited an over-abundance of proteins associated with extracellular matrix constituents compared to low angiogenic fibroblasts or endothelial cells. Finally, siRNA technology and purified protein were used to confirm a role for stromal cell-derived hepatocyte growth factor and fibronectin in inducing endothelial cell sprouting.
Conclusion
Differences in stromal cell ability to induce angiogenesis are due to differences in the secreted proteomes of both extracellular matrix proteins and pro-angiogenic growth factors.
Keywords: Angiogenesis, endothelial cell, fibroblast, smooth muscle cell, extracellular matrix
Methods
Cell isolates and tissue culture
Primary human umbilical vein EC (HUVEC) were isolated from umbilical cords obtained from local hospitals under University of California, Irvine Institutional Review Board approval. HUVEC were routinely cultured in M199 (GIBCO) with 10% fetal bovine serum (FBS) and endothelial growth supplement (BD Biosciences). Primary, normal human lung fibroblasts (NHLF) were purchased from Lonza and routinely cultured in M199 with 10% FBS. These cells were originally from the same isolate, but since their original isolation the individual batches have shown phenotypic drift such that we now have sub-isolates that have developed difference in angiogenic capabilities. The fibroblasts are characterized by Lonza as being negative for von Willebrand Factor, cytokeratins 18 and 19, and alpha smooth muscle actin, indicating that they are not SMC. Normal human bronchial SMC were purchased from Lonza and routinely grown in DMEM (GIBCO) with 10% FBS. The SMC are characterized by Lonza as being negative for von Willebrand Factor and postive for alpha-smooth muscle actin.
Fibrin gel angiogenesis assay
The 3D in vitro model of angiogenesis was performed as described previously1. Briefly, p3 HUVEC were cultured on collagen-coated Cytodex™ micro carrier beads (Sigma-Aldrich Co., St Louis, MO). These beads were then embedded into a fibrin gel with or without stromal cells in 24-well plates. Gels were allowed to clot and 1 ml of EGM-2 (Lonza) with stromal cells was added to each well. Media was changed every-other day for the first 6 days. For studies involving protein addition, proteins were mixed at the indicated concentrations in EGM-2, which contains VEGF and other pro-angiogenic factors, and added to gels every other day until the assay was analyzed. For assays involving fibronectin addition, purified fibronectin (BD Biosciences, Bedford, MA) was added to fibrinogen before clotting at the indicated concentrations.
For quantification of sprouting, only sprouts with lengths greater than or equal to the diameter of the bead were counted and at least 30 beads per condition were counted. Lumen formation was quantified by counting the number of EC sprouts that had formed lumens and are expressed as a percentage (% lumenized).
Preparation of fibroblast, SMC and HUVEC CM
NHLF, SMC and HUVEC cultured in M199 containing 10% FBS were allowed to grow to 80% confluence. Medium was replaced with EGM-2 (Lonza) for one day and then replaced with serum-free EBM-2. CM was harvested and filtered through a 0.22µm filter 2 days later. Two volumes of acetonitrile containing 0.1% trifluoroacetic acid was added to the filtered supernatants and this solution was vortexed for two minutes before centrifuging at 14,000 g for five minutes.
In-solution Trypsin Digestion and Dimethyl labeling
Samples were suspended in 50mM of triethylammonium bicarbonate (TEAB, Sigma-Aldrich, St. Louis, MO) and 0.5% sodium deoxycholate (DOC) and subsequently treated with 5 mM tris 2-carboxyethyl phosphine (TCEP, 5 mM, Thermo Pierce, Rockford, IL) at room temperature (RT) for 20 min. Samples were then alkylated with iodoacetamide (10 mM final concentration, Sigma) in the dark at RT for 20 min. The reduced and alkylated samples were digested at a substrate-to enzyme ratio of 1:50 (w/w) with proteomics grade trypsin (Sigma) at 37 °C overnight. DOC were removed from the digest via phase transfer surfactant (PTS) method2. Samples were next vacuum-dried then labeled with stable isotope dimethyl labeling3. The resultant dimethyl labeled tryptic peptides were applied to C18-StageTips or fractionated into five vials using SCX-StageTips4.
NanoLC-MS System
NanoLC-MS/MS analyses were conducted using LTQ linear ion trap (Thermo Fisher Scientific, Rockford IL) coupled with Waters 600E HPLC pump (Waters, Milford, MA) with 1:1000 split and Famous autosampler (Dionex, Sunnyvale, CA). Halo C18 materials (MaC-MOD, Chadds Ford, PA) were packed into P-2000 (Sutter Instrument, Novato, CA) laser-pulled needle (15 cm × 75 µm i.d.) with nitrogen-pressuriezed Capillary Column Packing bomb (Western Analytical, Lake Elsinore, CA). Mobile phases consisted of 0.1% FA in water and 0.1% FA in acetonitrile, solvent A and B respectively, over 220 min at a flow rate of 200 nL/min with 2% to 35% gradient. The top 5 precursor ions were selected for MS/MS scans with normalized CID set to 35.
Database search and quantitation
Spectral peak processing used MASCOT Distiller, version 2.3.2.0 (Matrix Science, London, United Kingdom), with LCQ_plus_zoom.opt parameters and a precursor tolerance of 1.2 Da. Peptide searches were against SwissProt (57.1; human taxonomy) with (fixed mod) cysteine carbamiodomethylation and (variable mods) methionine oxidation, along with heavy, intermediated and light dimethyl label selected for both N-terminus and lysine3. The homology threshold (expected, 0.05) was applied for protein ID. Spectral quantitation was done using MASCOT Distiller with simple ratio selected and elution time shift set to 15 s. Protein abundance ratios were weighted by geometric means of values from individual peptides after filtering on the basis of goodness-of-fit and visual inspection5.
Transfection of siRNAs
siRNAs designed to HGF were obtained from Ambion, Inc. (Austin, TX). Transfection was performed using Lipofectamine™ (Invitrogen, St. Louis, MO) following the manufacturer’s recommended protocol.
Quantification RT-PCR, ELISA and Western Blots
RNA was isolated from NHLF and SMC using Trizol (Invitrogen, St. Louis, MO) and following the manufacturer’s recommended protocol. Three µg of RNA was used for cDNA synthesis using the iScript cDNA Synthesis kit (Bio-Rad, Madison, WI). Primers were synthesized by IDT and sequences can be found in Supplemental Table IV (please see http://atvb.ahajournals.org).
ELISA kits were purchased from R&D Systems (Minneapolis, MN).
Monoclonal anti-collagen, type I antibody, cat # C2456, was purchased from Sigma-Aldrich Co. Densitometry analysis was done using ImageJ (Adobe Systems Inc., San Jose, CA).
Microscopy/Imaging and statistical analysis
Visualization of fibrin gel bead assays was performed using brightfield or fluorescent images collected on an inverted microscope (Olympus IX70) with a SPOT Idea 3.0 megapixel color mosaic camera and SPOT software (SPOT Imaging Solutions, Sterling Heights, MI). Images were processed in ImageJ to adjust contrast and color balance. All images in a given experiment were treated identically.
Analyses of HUVEC sprouting and lumen formation in fibrin gel angiogenesis assays were performed by observers blinded to the experimental conditions at the indicated time points. Data shown are representative of three independent experiments unless otherwise indicated. Error bars represent standard error of the mean. For comparisons involving three cell isolates, a one-way analysis of variance (ANOVA) was performed to determine if the cell isolates affected the output. For comparisons between two isolates in the group of three the TukeyHSD p value was used to assess significance. The differences between experimental groups of equal variance when only two groups were being compared were analyzed using Student’s t-test.
INTRODUCTION
Angiogenesis is the growth of new blood vessels from the existing vasculature. Angiogenesis occurs during normal processes, such as development of the cardiovascular system and wound healing, but also during pathological events such as tumor growth, diabetic retinopathy and cardiovascular disease (CVD)6, 7. CVD is the leading cause of mortality in the Western World with over 16 million deaths per year and much of this is associated with atherosclerosis8, which is a major contributor to myocardial infarction (MI)9. Although there is debate as to whether angiogenesis is harmful or beneficial during atherosclerosis it is clear that this process occurs during disease progression6.
Endothelial cell (EC) tube formation requires coordinated crosstalk between EC and other cells present in the stromal compartment, such as fibroblasts and smooth muscle cells (SMC), that support blood vessel growth and maturation7, 10. During atherosclerosis, SMCs from the medial layer de-differentiate and migrate into the intima where they proliferate and synthesize extracellular matrix (ECM)11–13, proteases and angiogenic growth factors, all of which contribute to angiognenesis14–18. Fibroblasts are the predominant non-myocyte cell found in the heart and infiltrate to the infarcted area following MI, where they proliferate19 and promote angiogenesis20. A number of studies have indicated that fibroblasts, like SMC, promote angiogenesis through the production of ECM, proteases and growth factors21–24.
To date, there is no effective treatment to repair damaged heart tissue following MI and recent research has focused on tissue engineering approaches that replace damaged heart tissue25–27. One study showed that ECM scaffolds derived from cardiac tissue and injected into the hearts of Harlan Sprague-Dawley rats promoted vessel infiltration28. Kreutziger et al. found that stromal cells were required to prevascularize tissue grafts and grafts generated in the presence of high angiogenic stromal cells improved in vivo vessel formation in the infarct zone29. It is therefore important to carefully define, at the molecular level, the contributions of these cells to the angiogenic process so that optimal combinations and concentrations of proteins and cell types can be applied in these therapies.
In order to accomplish this, attention needs to be paid to the experimental systems used to identify and verify the function of these proteins. Studies from Bissel’s group, as well as others, have shown that cellular co-cultures in three-dimensional ECMs can accurately model the morphological changes that occur in vivo30, 31. In this study we make use of an in vitro model of angiogenesis where EC are co-cultured with stromal cells, such as fibroblasts or SMC in three-dimensional fibrin gels, to identify stromal cell-secreted proteins important for angiogenesis. Specifically, we compare secreted proteins from EC, fibroblasts and SMC. Additionally, we compare the levels of proteins secreted by a fibroblast isolate with high angiogenic activity to one that has low angiogenic activity and find that ECM proteins, including fibronectin, are more abundant in fibroblasts with high angiogenic activity. Finally, we confirm a role for hepatocyte growth factor (HGF) and fibronectin as stromal cell-secreted proteins important for vessel sprouting.
RESULTS
Stromal cells vary in their ability to support angiogenesis in vitro
In order to assess the ability of different stromal cell types to support angiogenesis we compared primary isolates of fibroblasts and SMC in a previously described in vitro angiogenesis assay1, 32. In this assay, human umbilical vein endothelial cells (HUVEC) are allowed to adhere to collagen-coated cytodex™ beads, which are then embedded in a 3D fibrin matrix. Stromal cells are plated within the gel or on top of the gel. The stromal cells secrete factors that, in combination with the growth factors present in the medium, induce EC sprouting and lumen formation. In the absence of the stromal cells there is minimal sprouting and no lumen formation.
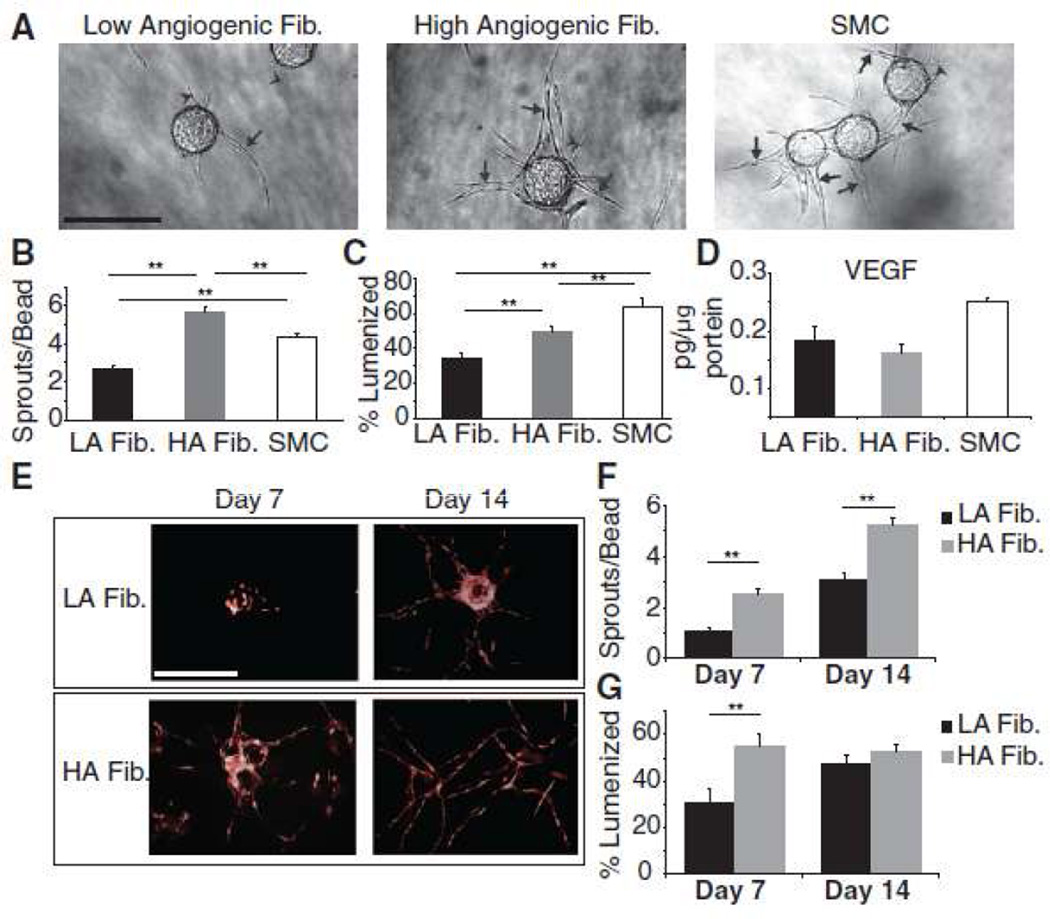
After 10 days of co-culture, with the stromal cells on top of the gel, we analyzed the number of EC sprouts and the percentage of sprouts that had formed lumens (Fig. 1). The stromal cells analyzed varied in their ability to support EC sprouting and lumen formation. One fibroblast isolate induced the highest number of EC sprouts (Fig. 1A,B) and will be referred to as high angiogenic fibroblasts (HA fibroblasts). A second fibroblast isolate induced the lowest level of EC sprouting and supported the lowest percentage of EC lumen formation, and this isolate is denoted low angiogenic (LA) fibroblasts. SMC supported a level of EC sprouting in between the fibroblast isolates, although a higher percentage of these sprouts formed lumens (Fig. 1C). The differences were found to be highly statistically significant by ANOVA (p < 0.01), confirming that stromal cells vary in their ability to support angiogenesis. We next assessed the level of vascular endothelial growth factor (VEGF) secreted by each cell isolate by harvesting cell culture supernatants and performing enzyme-linked immunosorbant assays (ELISA). Interestingly, there was only a slightly significant difference in the levels of VEGF produced by the three cell isolates (Fig. 1D, ANOVA p = 0.03), however, the amount of VEGF in the medium, which is on the order of ng/ml, is far greater than the amount of VEGF secreted by the stromal cells used in this study and, therefore, the differences in their abilities to support angiogenesis was not a result of differences in VEGF secretion.
Figure 1.
(A) Representative images (stromal cells on top of gel). Arrows indicate EC sprouts with mature lumens, arrow heads indicate EC sprouts without lumens. Quantitation of (B) EC sprouting and (C) lumen formation. (D) VEGF ELISA. (E) Representative images of EC sprouting and lumen formation (stromal cells in gel). Quantitation of (E) EC sprouting and (F) lumen formation. Data shown are the average of 3 independent experiments, each with n = 20. Error bars represent standard error of the mean. ** indicates TukeyHSD p < 0.01 for comparisons with three cell isolates and Student’s t test p < 0.01 for comparisons between two isolates. Scale bars: 50µm
During the course of our experiments we observed that HA fibroblasts invaded the fibrin gels after just two days of culture whereas LA fibroblasts did not exhibit significant gel invasiveness even after 10 days (data not shown). Previous work from our group showed that decreasing the distance between fibroblasts and EC in the fibrin gel angiogenesis assay correlates with increased vessel network formation22. We therefore reasoned that the higher angiogenic ability of HA fibroblasts could be due to the shorter distance between them and the EC as a result of them migrating into the fibrin gel. We tested this hypothesis by performing the angiogenesis assay with the fibroblasts embedded in the fibrin gel as opposed to on top, thus immediately placing them in closer proximity to the EC. HUVEC were transduced with retrovirus expressing mCherry to allow for distinction between them and the fibroblasts. Even in this orientation, the LA fibroblasts still failed to induce angiogenesis to the same extent as HA fibroblasts (Fig. 1E–G), thus ruling out the possibility that differences in fibroblast induction of angiogenesis were merely the result of increased invasiveness, and thereby closer apposition, of the HA fibroblasts.
Identification of fibroblast and smooth muscle cell secreted proteins
Based on the differences in the abilities of these stromal cells to induce angiogenesis, we decided to use these cells as a tool to identify secreted proteins that could regulate angiogenesis. We reasoned that differences in their ability to induce angiogenesis may have been the result of differences in the respective secretomes of LA fibroblasts, HA fibroblasts and SMC, that is, their ability to “condition” the local microenvironment, with either extracellular matrix proteins, growth factors or with both. In support of this hypothesis, we have previously shown that fibroblast conditioned medium supports EC sprouting and lumen formation in the absence of fibroblasts in our in vitro angiogenesis assay24. Therefore, we harvested supernatants from cell cultures grown in monolayer and used nanoLC-MS/MS to identify proteins that were secreted by each cell type. Table 1 lists a subset of proteins that were identified and the cell type(s) from which they were most prominently secreted. A complete list of the proteins identified can be found in Supplemental Table I (please see http://atvb.ahajournals.org). Of note, all cells expressed transforming growth factor beta-induced protein ig-h3 (βig-h3), collagen type 1, alpha-1 (Col1A1), insulin-like growth factor-binding protein 7 (IGFBP7), procollagen C-endopeptidase enhancer 1 (PCOLCE1) and secreted protein acidic and rich in cysteine (SPARC). These proteins have previously been identified by our laboratory as being necessary for EC lumen formation in the fibrin gel angiogenesis assay24. The fact that all cell types tested secreted these proteins is in agreement with the ability of each stromal cell type to support EC lumen formation, albeit to greater or lesser degrees.
Table 1. Abbreviated list of proteins found in LA fibroblasts, HA fibroblasts and SMC by MS.
Identification of proteins present in LA fibroblasts, HA fibroblasts and SMC conditioned media.
| Found In | |||||
|---|---|---|---|---|---|
| Symbol | Name | LA LF | HA LF | ||
| SMC | |||||
| A2MG | Alpha-2-macroglobulin precursor | yes | yes | yes | |
| ANG1 | Angiogenin | no | no | yes | |
| ANGL2 | Angiopoietin-related protein 2 | no | no | yes | |
| BGH3 | Transforming growth factor-beta-induced protein ig-h3 | yes | yes | yes | |
| BMP1 | Bone morphogenic protein 1 | no | no | yes | |
| CATB | Cathepsin B | yes | yes | yes | |
| CO1A1 | Collagen alpha-1(I) chain | yes | yes | yes | |
| CO1A2 | Collagen alpha-2(I) chain precursor | yes | yes | yes | |
| DKK3 | Dickkopf-related protein 3 | yes | yes | yes | |
| ENOA | Alpha-enolase | no | no | yes | |
| FINC | Fibronectin precursor | yes | yes | yes | |
| HGF | Hepatocyte growth factor | no | no | yes | |
| IBP2 | Insulin-like growth factor binding protein 2 precursor | yes | no | yes | |
| IBP3 | Insulin-like growth factor binding protein 3 precursor | no | no | yes | |
| IBP4 | Insulin-like growth factor binding protein 4 precursor | yes | yes | yes | |
| IBP5 | Insulin-like growth factor binding protein 5 precursor | yes | yes | yes | |
| IBP6 | Insulin-like growth factor binding protein 6 precursor | no | yes | yes | |
| IBP7 | Insulin-like growth factor binding protein 7 precursor | yes | yes | yes | |
| IC1 | Plasma protease C1 inhibitor precursor | yes | yes | yes | |
| IGF2 | Insulin-like growth factor II | no | no | yes | |
| LAMA2 | Laminin subunit alpha-2 | no | no | yes | |
| LAMA4 | Laminin subunit alpha-4 | no | no | yes | |
| LAMA5 | Laminin subunit alpha-5 | no | no | yes | |
| LAMB1 | Laminin subunit beta-1 | no | no | yes | |
| LAMB2 | Laminin subunit beta-2 | no | no | yes | |
| LAMC1 | Laminin subunit gama-1 precursor | yes | yes | yes | |
| LTBP1 | Latent-transforming growth factor beta-binding protein, isoform 1L | yes | yes | yes | |
| LTBP2 | Latent-transforming growth factor beta-binding protein 2 | yes | yes | yes | |
| LTBP4 | Latent-transforming growth factor beta-binding protein 4 | no | no | yes | |
| MMP1 | Interstitial collagenase precursor | no | yes | no | |
| MMP19 | Matrix metalloproteinase-19 | no | no | yes | |
| MMP2 | 72kDA type IV collagenase | yes | yes | yes | |
| PAI1 | Plaminogen activator inhibitor 1 | yes | yes | yes | |
| PCOC1 | Procollagen C-endopeptidase enhancer 1 precursor | yes | yes | yes | |
| PEDF | Pigment epithelium-derived factor precursor | no | yes | yes | |
| PGS2 | Decorin precursor | yes | yes | yes | |
| SDF1 | Stromal cell-derived factor 1 | no | no | yes | |
| SPRC | Secreted protein acidic and rich in cystein precursor (SPARC) | yes | yes | yes | |
| TETN | Tetranectin | yes | yes | yes | |
| TIMP1 | Metalloproteinase inhibitor 1 | yes | yes | yes | |
| TIMP2 | Metalloproteinase inhibitor 2 precursor | yes | yes | yes | |
| TSP1 | Thrombospondin-1 precursor | no | yes | yes | |
| TSP2 | Thrombospondin-2 | no | no | yes | |
| VIME | Vimentin | yes | yes | yes | |
| VWA1 | von Willebrand factor A domain-containing protein 1 | no | no | yes | |
| WNT2BProtein | Wnt-2b | no | no | yes | |
| WNT5AProtein | Wnt-5a | no | no | yes | |
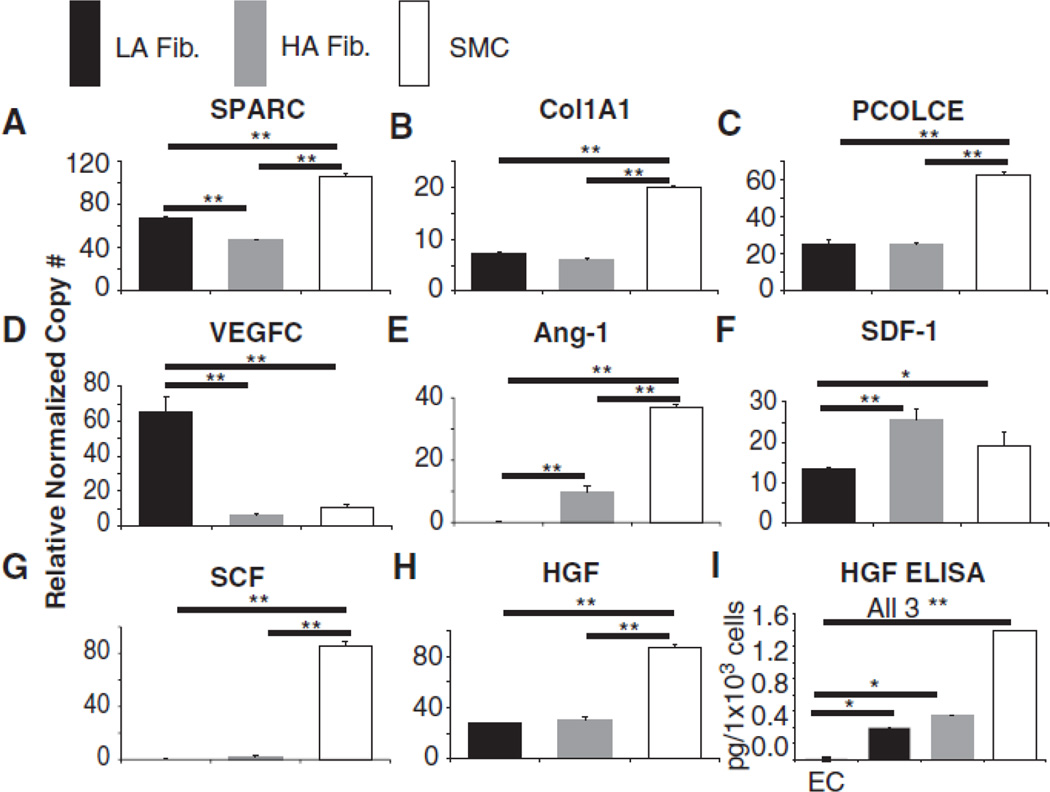
We designed primers to a subset of these genes and used quantitative real time-polymerase chain reaction (qRT-PCR) to confirm mRNA expression of the proteins previously identified by nano-LC-MS/MS. Figure 2 shows that SPARC, Col1A1 and PCOLCE mRNA was detected in all cell types, validating the results obtained using HPLC/MS. We also noted that several growth factors, including hepatocyte growth factor (HGF) and stromal cell derived factor (SDF), were only detected in the SMC sample during MS analysis (Table 1) while others, e.g. angiopoietin 1 (Ang-1), stem cell factor (SCF) and vascular endothelial growth factor C (VEGFC) were not detected in any cell type by MS. (Table 1). This could be due to the fact that many growth factors have KD values in the low nanomolar range and are therefore present in supernatants at much lower concentrations compared to the other proteins that were detected using HPLC/MS. For example, the ligand receptor pairs of SDF/CXCR4 and SCF/c-Kit have reported KD values of 5–10 nM and 2 nM respectively33–35 and the ligands are consequently secreted at similarly low levels. This observation prompted us to perform qRT-PCR on several of these growth factors and we found that, in contrast to the MS data, all of the genes tested were expressed at the mRNA level in all stromal cell types (Fig. 2D–H). HGF was also confirmed by ELISA to be present in cell culture supernatants of both fibroblast isolates and SMC, but not HUVEC. (Fig. 2I).
Figure 2.
Relative normalized copy number as measured by qRT-PCR of (A) SPARC, (B) Col1A1, (C) PCOLCE, (D) VEGFC, (E) Ang-1, (F) SDF-1α, (G) SCF and (H) HGF in LA fibroblasts, HA fibroblasts and SMC conditioned media. (I) HGF levels in LA fibroblasts, HA fibroblasts and SMC conditioned media as determined by ELISA. Data shown are representative of three independent experiments. * indicates TukeyHSD p < 0.05, ** indicates p < 0.01.
Assessment of protein levels in LA fibroblasts, HA fibroblasts and HUVEC secretomes
To gain a more quantitative understanding of protein expression by stromal cells in HA versus LA fibroblasts, we preformed nanoLC MS/MS on conditioned media from the isolates. Here, size-fractionated protein extracts of conditioned media from the three cell types were first digested with trypsin and the resulting peptides were then individually isotope labeled. The labeled samples were combined, and peptide fractionation was followed by nanoLC-MS/MS. Peptides were identified from fragmentation spectra, and their relative abundances in the three samples were quantitated from areas under the corresponding precursor level MS isotopologues. HUVEC served as a negative control as EC do not undergo angiogenic sprouting and lumen formation when cultured in the absence of stromal cells. Table 2 lists proteins identified that scored above the Mascot homology/identity threshold along with their corresponding abundance ratios in the three cell types analyzed (a more comprehensive list can be found in Supplemental Table II, please see http://atvb.ahajournals.org). Peptides from 687 unique accessions that passed quantitation quality filtering were identified in the three cell types tested. Of note, several proteins whose abundance ratios were skewed in favor of the angiogenic isolate have previously been shown to be pro-angiogenic, including matrix metalloproteinase 2 (MMP2)9, 36, fibronectin37 and clusterin38. These were found to be present at substantially higher levels in the HA fibroblast supernatants than in either the LA fibroblast or HUVEC supernatants (Table 2).
Table 2. Abbreviated list of protein abundance in LA fibroblasts, HA fibroblasts and HUVEC.
Quantitative mass spectrometry analysis of HA fibroblast, LA fibroblast and HUVEC conditioned media. Ratios show the relative abundance of the given protein for the given cell isolate comparison.
| Symbol | Name | HA:LA | |
|---|---|---|---|
| HA:HUVEC | |||
| CLUS | Clusterin | 13.9082* | 5.1733* |
| CO2A1 | Collagen alpha-1(II) chain | 13.5318* | 16.2867* |
| MMP3 | Stromelysin-1 | 5.6306* | 5.1600* |
| FINC | Fibronectin | 5.1414* | 5.3277* |
| FBN2 | Fibrillin-2 | 5.0633* | 6.9881* |
| LTBP2 | Latent-transforming growth factor beta-binding protein 2 | 4.8193* | 2.5569 |
| MMP2 | 72 kDa type IV collagenase | 4.3649* | 2.4096 |
| LAMB2 | Laminin subunit beta-2 | 3.2031 | 8.2372* |
| BGH3 | Transforming growth factor-beta-induced protein ig-h3 | 2.8145 | 5.4885* |
| FBLN3 | EGF-containing fibulin-like extracellular matrix protein 1 | 2.7655 | 0.2523 |
| TETN | Tetranectin | 2.4570 | 2.9735 |
| DKK3 | Dickkopf-related protein 3 | 2.3127 | 3.0102 |
| LAMB1 | Laminin subunit beta-1 | 2.2447 | 1.3296 |
| PGS2 | Decorin | 2.2109 | 5.0302* |
| CO1A2 | Collagen alpha-2(I) chain | 1.7458 | 3.4095 |
| CO1A1 | Collagen alpha-1(I) chain | 1.3132 | 2.0231 |
| SPRC | Secreted protein acidic and rich in cystein precursor (SPARC) | 1.1256 | 0.3640 |
| IBP7 | Insulin-like growth factor-binding protein 7 | 1.1131 | 0.4751 |
| PCOC1 | Procollagen C-endopeptidase enhancer 1 | 0.8750 | 2.2517 |
| EGLN | Endoglin | 0.4720† | 0.2116 |
indicates proteins that were found in the upper 10th percentile of the corresponding ratio
indicates proteins that were found in the lower 10th percentile of the corresponding ratio
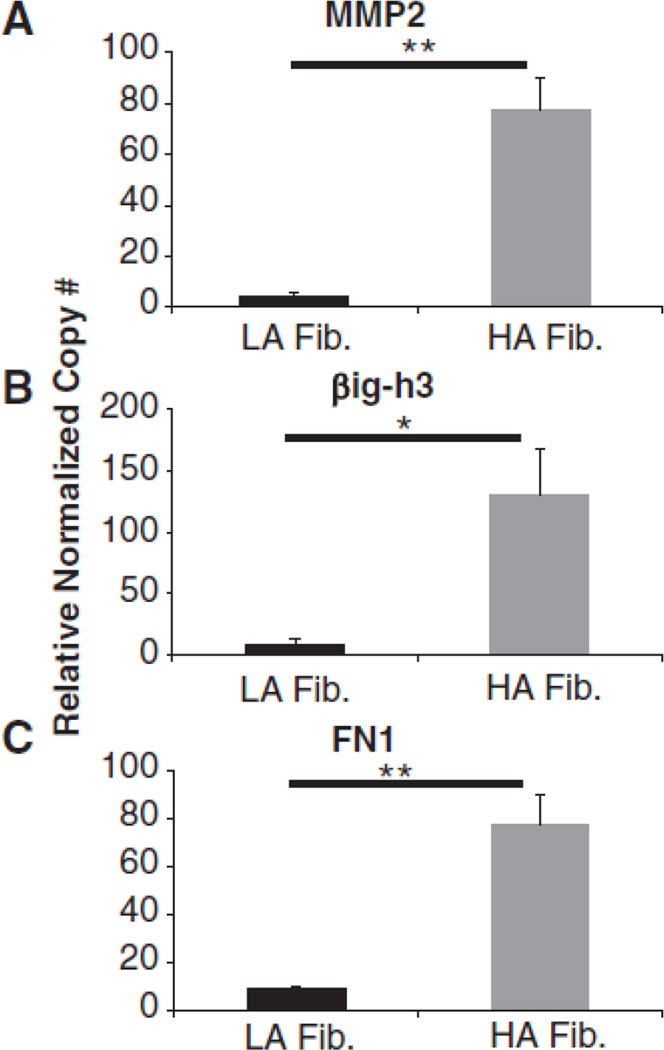
We addressed differentially abundant proteins at the mRNA level using qRT-PCR as well as western blot analysis. Figure 3 shows that MMP2, βig-h3 and fibronectin were all expressed at higher levels in HA fibroblasts than in LA fibroblasts. Interestingly, Col1A1 and PCOLCE had similar levels of mRNA in HA fibroblasts and LA fibroblasts (Fig. 2B–C), which is in agreement with data obtained via quantitative MS/HPLC. Supplemental Figure I (please see http://atvb.ahajournals.org) shows the level of various collagen 1 isoforms in LA and HA fibroblasts as well as SMC supernatants detected by western blotting. Here the high MW band corresponds to dimeric and trimeric collagen 1 and the low MW band corresponds to monomeric collagen 1. These cells secreted similar levels of collagen 1 which is in agreement with data obtained via quantitative MS.
Figure 3.
qRT-PCR validation of quantitative mass spectrometry data. Relative normalized copy number of (A) MMP2, (B) βig-h3 and (C) FN1 in LA and HA fibroblasts. Data shown are representative of three independent experiments. * indicates Student’s t test p < 0.05, ** p < 0.01
Numerous extracellular matrix proteins are over-abundantly expressed in HA fibroblast supernatants
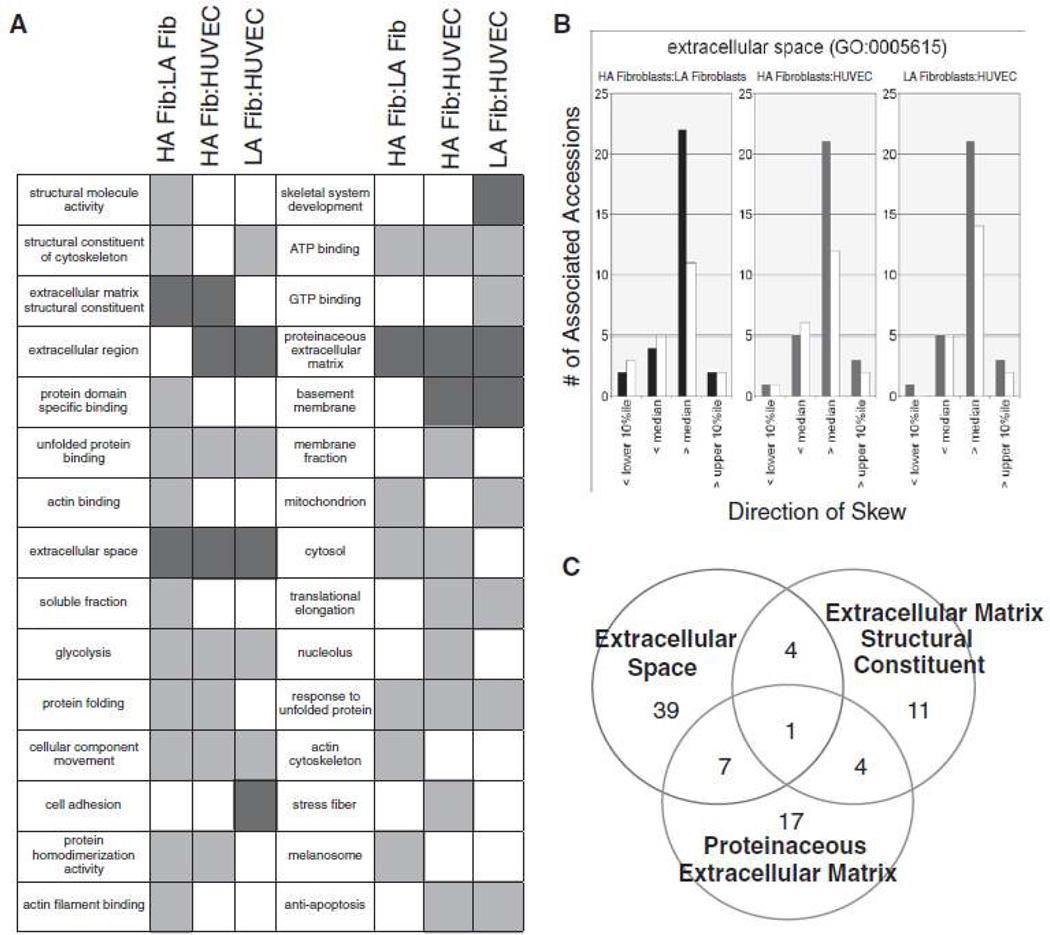
In order to ascertain whether specific functional classes of proteins were over-abundant in HA fibroblast secretomes versus those from LA fibroblasts or HUVEC, all nodes of the entire ~33,000 node Gene Ontology (GO) tree to which more than 9 of our 687 accession could be assigned, were screened for a “skew” in the distribution of all of the relative quant ratios (HA fibroblasts/LA fibroblasts, LA fibroblasts/HUVEC or HA fibroblasts/HUVEC). The criteria set for a “skew” was that the number of individual quant ratios lying on one side of the global median for all 687 unique accessions was at least 2.5× the number lying on the other side. Such a skew was taken to indicate that proteins of a particular functional class were substantially over- or under-abundant with respect to the 687-member secretome as a whole. Based on these criteria, 2,292 of the ~33,000 GO terms were assignable to accessions in our set and, of these, 28 exhibited the aforementioned “skew.” These are displayed in Figure 4A, with green boxes representing terms that were under-abundant for the indicated ratio and red boxes representing terms that were over-abundant for the given ratio.
Figure 4.
(A) List of GO terms exhibiting skew. Green boxes represent terms that were under-abundant, red boxes represent over-abundant. (B) For the GO term “Extracellular space”, histograms show numbers of associated accessions with quant ratios that exhibited the indicated direction of skew. Colored bars represent accessions quantitated on the basis of more than one peptide and white bars represent accessions that were quantitated on the basis of only one peptide. (C) Venn diagram of accessions associated with GO terms containing the string “extracellular space,” “proteinaceous extracellular matrix” and “extracellular matrix structural constituent” showing the minimal overlap of accessions matched to these terms.
Of the 28 GO terms exhibiting skew, three were found to be over-abundant in the HA fibroblasts compared to the LA fibroblasts (Fig. 4A). Interestingly, all three – extracellular matrix structural constituent, extracellular space, and proteinaceous extracellular matrix – relate to proteins associated with extracellular matrix deposition and assembly. Figure 4B shows histogram representations of proteins mapping to the “extracellular space” term that were over-abundant in HA fibroblast compared to LA fibroblasts and HUVEC. Since all three GO terms that exhibited skew between HA fibroblasts and LA fibroblasts were ECM terms and some accessions appeared in more than one GO term, we wanted to determine if this was due to the same accessions appearing in multiple GO terms. Therefore, we made a visual representation of the numbers of proteins that were identified in multiple ECM GO terms (Fig. 4C). As is evidenced by the Venn diagram, there is little overlap between the proteins identified as extracellular matrix structural constituent, exctracellular space, and proteinaceous extracellular matrix. Supplemental Table III (please see http://atvb.ahajournals.org) lists the specific proteins that were identified in each ECM GO term.
HGF and fibronectin induce EC sprouting
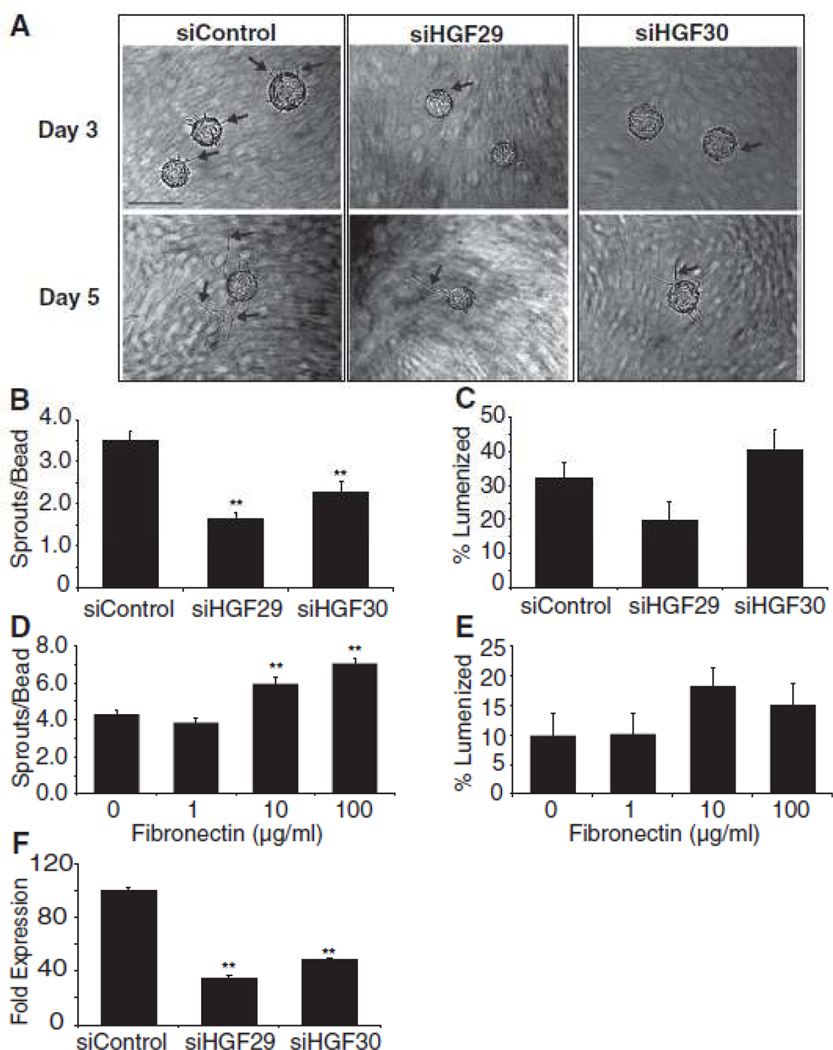
We have previously identified a combination of proteins, of which HGF is a component, that induces EC sprouting in the absence of fibroblasts24. In addition, we have shown here that fibroblasts and SMC make substantial amounts of HGF, while EC do not (Fig. 2I). In order to determine whether stromal cell-derived HGF was required for EC sprouting, we obtained two siRNAs targeted to different regions of the HGF gene and transfected HA fibroblasts to knock down HGF gene expression. These fibroblasts were then used in the fibrin gel angiogenesis assay. HGF knockdown was confirmed by qRT-PCR (Fig. 5F) and in order to ensure that HGF levels had not returned to basal levels, the fibrin gel angiogenesis assay was analyzed on Day 5. When fibroblasts were treated with either of the siRNAs designed to HGF, EC sprouting was significantly decreased (Fig. 5A,B) while lumen formation was unaffected (Fig. 5C). Similarly, knockdown of HGF in SMC also led to a decrease in EC sprouting and no change in EC lumen formation (Supplemental Fig. IIA, B, please see http://atvb.ahajournals.org). Thus, the ability of multiple stromal cell types to promote EC sprouting is critically dependent on secreted HGF.
Figure 5.
(A) Representative images of angiogenesis in the presence of HA fibroblasts treated with control or HGF siRNA. Quantitation of (B) EC sprouting and (C) lumen formation. Arrows indicate EC sprouts. Scale bar: 50µm (D) qRT-PCR confirmation of HGF knockdown. Quantitation of (E) EC sprouting and (F) lumen formation in the presence of the indicated amount of fibronectin. Data shown are representative of three independent experiments. ** p < 0.01.
Additionally, we examined the effect of HGF addition in the fibrin gel angiogenesis assay in the presence of LA fibroblasts in an attempt to increase EC sprouting. Indeed, addition of HGF increased EC sprouting and this increase was dependent on HGF concentration (Supplemental Fig. IIC, D, please see http://atvb.ahajournals.org). As the angiogenic response of LA fibroblasts is weaker and slower than the HA fibroblasts, these cultures were analyzed at Day 14.
We also attempted to increase EC sprouting in the presence of the LA fibroblasts by spiking ECM proteins into the fibrin gels. Fibronectin was chosen due to the fact that it was over-abundantly expressed by the HA fibroblasts compared to LA fibroblasts (Table 2). Addition of fibronectin induced a singnificant, dose-dependent increase in EC sprouting (Fig. 5D, E). These results demonstrate that fibroblasts and SMC contribute to angiogenic sprouting in multiple ways; by secreting HGF, which induces sprouting and also by secretion of a complex extracellular matrix, which contributes not only to sprouting, but also to lumen formation.
DISCUSSION
In this study, we demonstrate that different stromal cell populations vary in their ability to support in vitro vessel formation and that these differences can, at least in part, be attributed to differences in their secretomes. We took advantage of the differences in angiogenic activity in these stromal cell isolates to identify specific proteins that regulate this process. Interestingly, fibroblasts that exhibited high angiogenic activity expressed higher levels of proteins associated with the ECM compared to fibroblasts with low angiogenic activity. Importantly, addition of the ECM protein fibronectin, was shown to robustly restore angiogenic activity to the LA fibroblasts.
The fact that HA fibroblasts over-abundantly expressed ECM proteins is consistent with previous studies demonstrating a positive role for ECM proteins in regulating angiogenesis23, 24, 39. Fibronectin can bind other proteins present in the ECM such as heparin, collagen and fibrin; all of which are present in the fibrin gel angiogenesis assay40–42. With this in mind, it is plausible that ECM-fibronectin interactions could alter EC response to these matrices. Indeed, one study has shown that fibronectin regulates fibroblast migration into fibrin clots, suggesting a pivotal role for fibronectin – fibrin interactions in regulating cell migration43. ECs bind to ECM and respond to changes in ECM composition through integrins. A number of integrins have been shown to be important for EC migration including α1β1, α1β2 and αvβ344, 45. The fibronectin-binding integrin, α5β1, has been shown to regulate angiogenesis in vivo46. Also of interest to this study, Mitra et al. showed that blocking the α5β1 integrin with a function-blocking antibody decreased phosphorylation of the HGF receptor, cMET, in HUVEC plated on fibronectin47, and we show here a critical role for stromal cell-derived HGF in EC sprouting. Kreutziger et. al found that increased angiogenesis correlated with increased expression of the ECM component versican in co-cultures of EC and different mesenchymal stem cell lines29. Interestingly, we only detected versican expression in SMC in our MS analysis. Since no studies of versican knock down or inhibition were reported in the aforementioned study, it is difficult to determine whether versican indeed stimulates angiogenesis. Alternatively, different stromal cells may induce angiogenesis via different pathways.
HGF has previously been shown to regulate angiogenesis both in vivo and in vitro48–51. Our results further these findings by demonstrating that stromal cells, fibroblasts and SMCs in particular, are important sources of HGF during angiogenesis. Evidence for this is provided by ELISA data showing that LA fibroblasts, HA fibroblasts and SMCs secrete HGF (Fig. 2I), and that knocking down HGF expression in HA fibroblasts and SMC leads to decreased EC sprouting (Fig. 5A, B). It should be noted that HGF was not detected via MS analysis of HA or LA supernatents, but was detected via qPCR and ELISA. This is likely due to the fact that growth factors such as VEGF and HGF are secreted at much lower levels in comparison to extracellular matrix proteins such as those identified via MS. It would be interesting to identify the signaling proteins downstream of HGF-cMET signaling in EC that are responsible for mediating this increase in EC migration, and to determine to what extent crosstalk between HGF and fibronectin signaling through cMET is required.
The role that angiogenesis plays in CVD remains controversial,6 but it is clear that fibroblasts and SMC contribute to the process20 although the molecular mechanisms that regulate these activities remain poorly defined. The results presented here begin to unravel these molecular mechanisms, and have implications for the treatment of CVD by demonstrating that ECM proteins as well as HGF are important contributors to angiogenesis and that alterations in the levels of these proteins can either promote or inhibit blood vessel growth. This study also identifies a number of proteins that could be potentially useful in tissue engineering approaches to repair damaged heart tissue following MI.
Tissue replacement strategies to repair damaged heart tissue are dependent on the ability of the grafted tissue to survive in the hostile ischemic microenvironment. One attractive strategy for improving graft tissue survival is to create prevascularized tissue constructs in the hopes that host vessels will rapidly anastomose with graft vessels. Indeed, prevascularized tissue constructs have been shown to improve tissue viability upon in vivo transplantation, with several studies showing that increased vascularization led to increased tissue survival29, 52–54. Knowledge of the cell types and signaling proteins that regulate and promote angiogenesis will help guide research into optimization of tissue constructs or injectable ECM gels. Our data indicate that tissue constructs containing stromal cells high in ECM protein production will improve graft viability by increasing angiogenesis.
Angiogenesis requires crosstalk between EC and other cells of the stromal compartment and a clear understanding of the molecules regulating this process will be critical for any strategies involving manipulation of angiogenesis.
Supplementary Material
ACKNOWLEDGEMENTS
None
SOURCES OF FUNDING
This work was supported by U.S. National Institutes of Health grant RO1HL60067. ACN received a pre-doctoral fellowship award from the Edward’s LifeSciences Center for Advanced Cardiovascular Technology. CCWH receives support from the Chao Family Comprehensive Cancer Center through the P30A062203 award from NCI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.C.N., W.C., K.M.W-R., S.A.P., D.P.T. and D.R.S. performed the experiments; A.H.F. and K.M.W-R helped prepare the manuscript; A.C.N., P.D.G. and C.C.W.H. designed experiments, analyzed the data and prepared the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Nakatsu MN, Hughes CC. An optimized three-dimensional in vitro model for the analysis of angiogenesis. Methods Enzymol. 2008;443:65–82. doi: 10.1016/S0076-6879(08)02004-1. [DOI] [PubMed] [Google Scholar]
- 2.Masuda T, Tomita M, Ishihama Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J Proteome Res. 2008;7:731–740. doi: 10.1021/pr700658q. [DOI] [PubMed] [Google Scholar]
- 3.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 4.Ishihama Y, Rappsilber J, Mann M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J Proteome Res. 2006;5:988–994. doi: 10.1021/pr050385q. [DOI] [PubMed] [Google Scholar]
- 5.Chou W, Ngo T, Gershon PD. An overview of the vaccinia virus infectome: a survey of the proteins of the poxvirus-infected cell. J Virol. 2012;86:1487–1499. doi: 10.1128/JVI.06084-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khurana R, Simons M, Martin JF, Zachary IC. Role of angiogenesis in cardiovascular disease: a critical appraisal. Circulation. 2005;112:1813–1824. doi: 10.1161/CIRCULATIONAHA.105.535294. [DOI] [PubMed] [Google Scholar]
- 7.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahlof B. Cardiovascular disease risk factors: epidemiology and risk assessment. Am J Cardiol. 2010;105:3A–9A. doi: 10.1016/j.amjcard.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Burke AP, Virmani R. Pathophysiology of acute myocardial infarction. Med Clin North Am. 2007;91:553–572. doi: 10.1016/j.mcna.2007.03.005. ix. [DOI] [PubMed] [Google Scholar]
- 10.Hughes CC. Endothelial-stromal interactions in angiogenesis. Curr Opin Hematol. 2008;15:204–209. doi: 10.1097/MOH.0b013e3282f97dbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newby AC, George SJ. Proliferation, migration, matrix turnover, and death of smooth muscle cells in native coronary and vein graft atherosclerosis. Curr Opin Cardiol. 1996;11:574–582. doi: 10.1097/00001573-199611000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Thyberg J, Blomgren K, Roy J, Tran PK, Hedin U. Phenotypic modulation of smooth muscle cells after arterial injury is associated with changes in the distribution of laminin and fibronectin. J Histochem Cytochem. 1997;45:837–846. doi: 10.1177/002215549704500608. [DOI] [PubMed] [Google Scholar]
- 13.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 14.Hughes SE, Crossman D, Hall PA. Expression of basic and acidic fibroblast growth factors and their receptor in normal and atherosclerotic human arteries. Cardiovasc Res. 1993;27:1214–1219. doi: 10.1093/cvr/27.7.1214. [DOI] [PubMed] [Google Scholar]
- 15.Barrett TB, Benditt EP. Platelet-derived growth factor gene expression in human atherosclerotic plaques and normal artery wall. Proc Natl Acad Sci U S A. 1988;85:2810–2814. doi: 10.1073/pnas.85.8.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bobik A, Agrotis A, Kanellakis P, Dilley R, Krushinsky A, Smirnov V, Tararak E, Condron M, Kostolias G. Distinct patterns of transforming growth factor-beta isoform and receptor expression in human atherosclerotic lesions. Colocalization implicates TGF-beta in fibrofatty lesion development. Circulation. 1999;99:2883–2891. doi: 10.1161/01.cir.99.22.2883. [DOI] [PubMed] [Google Scholar]
- 17.Nakata A, Miyagawa J, Yamashita S, Nishida M, Tamura R, Yamamori K, Nakamura T, Nozaki S, Kameda-Takemura K, Kawata S, Taniguchi N, Higashiyama S, Matsuzawa Y. Localization of heparin-binding epidermal growth factor-like growth factor in human coronary arteries. Possible roles of HB-EGF in the formation of coronary atherosclerosis. Circulation. 1996;94:2778–2786. doi: 10.1161/01.cir.94.11.2778. [DOI] [PubMed] [Google Scholar]
- 18.Lupu F, Heim DA, Bachmann F, Hurni M, Kakkar VV, Kruithof EK. Plasminogen activator expression in human atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 1995;15:1444–1455. doi: 10.1161/01.atv.15.9.1444. [DOI] [PubMed] [Google Scholar]
- 19.Dobaczewski M, Bujak M, Li N, Gonzalez-Quesada C, Mendoza LH, Wang XF, Frangogiannis NG. Smad3 signaling critically regulates fibroblast phenotype and function in healing myocardial infarction. Circ Res. 2012;107:418–428. doi: 10.1161/CIRCRESAHA.109.216101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao T, Zhao W, Chen Y, Ahokas RA, Sun Y. Acidic and basic fibroblast growth factors involved in cardiac angiogenesis following infarction. Int J Cardiol. 2011;152:307–313. doi: 10.1016/j.ijcard.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kellouche S, Mourah S, Bonnefoy A, Schoëvaert D, Podgorniak MP, Calvo F, Hoylaerts MF, Legrand C, Dosquet C. Platelets, thrombospondin-1 and human dermal fibroblasts cooperate for stimulation of endothelial cell tubulogenesis through VEGF and PAI-1 regulation. Exp Cell Res. 2007;313:486–499. doi: 10.1016/j.yexcr.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 22.Griffith CK, Miller C, Sainson RC, Calvert JW, Jeon NL, Hughes CC, George SC. Diffusion limits of an in vitro thick prevascularized tissue. Tissue Eng. 2005;11:257–266. doi: 10.1089/ten.2005.11.257. [DOI] [PubMed] [Google Scholar]
- 23.Berthod F, Germain L, Tremblay N, Auger FA. Extracellular matrix deposition by fibroblasts is necessary to promote capillary-like tube formation in vitro. J Cell Physiol. 2006;207:491–498. doi: 10.1002/jcp.20584. [DOI] [PubMed] [Google Scholar]
- 24.Newman AC, Nakatsu MN, Chou W, Gershon PD, Hughes CC. The requirement for fibroblasts in angiogenesis: fibroblast-derived matricellular proteins are essential for endothelial cell lumen formation. Molecular Biology of the Cell. 2011;20:3791–3800. doi: 10.1091/mbc.E11-05-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarig U, Machluf M. Engineering cell platforms for myocardial regeneration. Expert Opin Biol Ther. 2011;11:1055–1077. doi: 10.1517/14712598.2011.578574. [DOI] [PubMed] [Google Scholar]
- 26.Ye KY, Black LD. 3rd Strategies for tissue engineering cardiac constructs to affect functional repair following myocardial infarction. J Cardiovasc Transl Res. 2011;4:575–591. doi: 10.1007/s12265-011-9303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beitnes JO, Lunde K, Brinchmann JE, Aakhus S. Stem cells for cardiac repair in acute myocardial infarction. Expert Rev Cardiovasc Ther. 2011;9:1015–1025. doi: 10.1586/erc.11.108. [DOI] [PubMed] [Google Scholar]
- 28.Seif-Naraghi SB, Salvatore MA, Schup-Magoffin PJ, Hu DP, Christman KL. Design and characterization of an injectable pericardial matrix gel: a potentially autologous scaffold for cardiac tissue engineering. Tissue Eng Part A. 2010;16:2017–2027. doi: 10.1089/ten.tea.2009.0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreutziger KL, Muskheli V, Johnson P, Braun K, Wight TN, Murry CE. Developing vasculature and stroma in engineered human myocardium. Tissue Eng Part A. 2011;17:1219–1228. doi: 10.1089/ten.tea.2010.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inman JL, Bissell MJ. Apical polarity in three-dimensional culture systems: where to now? J Biol. 2010;9:2. doi: 10.1186/jbiol213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo AT, Mori H, Mott J, Bissell MJ. Constructing Three-Dimensional Models to Study Mammary Gland Branching Morphogenesis and Functional Differentiation. J Mammary Gland Biol Neoplasia. 2012;2:103–110. doi: 10.1007/s10911-012-9251-7. [DOI] [PubMed] [Google Scholar]
- 32.Nakatsu MN, Sainson RC, Aoto JN, Taylor KL, Aitkenhead M, Perez-del-Pulgar S, Carpenter PM, Hughes CC. Angiogenic sprouting and capillary lumen formation modeled by human umbilical vein endothelial cells (HUVEC) in fibrin gels: the role of fibroblasts and Angiopoietin-1. Microvasc Res. 2003;66:102–112. doi: 10.1016/s0026-2862(03)00045-1. [DOI] [PubMed] [Google Scholar]
- 33.Hesselgesser J, Liang M, Hoxie J, Greenberg M, Brass LF, Orsini MJ, Taub D, Horuk R. Identification and characterization of the CXCR4 chemokine receptor in human T cell lines: ligand binding, biological activity, and HIV-1 infectivity. J Immunol. 1998;160:877–883. [PubMed] [Google Scholar]
- 34.Lev S, Blechman J, Nishikawa S, Givol D, Yarden Y. Interspecies molecular chimeras of kit help define the binding site of the stem cell factor. Mol Cell Biol. 1993;13:2224–2234. doi: 10.1128/mcb.13.4.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lev S, Yarden Y, Givol D. Dimerization and activation of the kit receptor by monovalent and bivalent binding of the stem cell factor. J Biol Chem. 1992;267:15970–15977. [PubMed] [Google Scholar]
- 36.Rojiani MV, Alidina J, Esposito N, Rojiani AM. Expression of MMP-2 correlates with increased angiogenesis in CNS metastasis of lung carcinoma. Int J Clin Exp Pathol. 2010;3:775–781. [PMC free article] [PubMed] [Google Scholar]
- 37.Stenzel D, Lundkvist A, Sauvaget D, et al. Integrin-dependent and -independent functions of astrocytic fibronectin in retinal angiogenesis. Development. 2011;138:4451–4463. doi: 10.1242/dev.071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson JK, Gleave ME, Gleave J, Burt HM. The inhibition of angiogenesis by antisense oligonucleotides to clusterin. Angiogenesis. 2005;8:229–238. doi: 10.1007/s10456-005-9018-5. [DOI] [PubMed] [Google Scholar]
- 39.Davis GE, Senger DR. Extracellular matrix mediates a molecular balance between vascular morphogenesis and regression. Curr Opin Hematol. 2008;15:197–203. doi: 10.1097/MOH.0b013e3282fcc321. [DOI] [PubMed] [Google Scholar]
- 40.Busby TF, Argraves WS, Brew SA, Pechik I, Gilliland GL, Ingham KC. Heparin binding by fibronectin module III-13 involves six discontinuous basic residues brought together to form a cationic cradle. J Biol Chem. 1995;270:18558–18562. doi: 10.1074/jbc.270.31.18558. [DOI] [PubMed] [Google Scholar]
- 41.Balian G, Click EM, Crouch E, Davidson JM, Bornstein P. Isolation of a collagen-binding fragment from fibronectin and cold-insoluble globulin. J Biol Chem. 1979;254:1429–1432. [PubMed] [Google Scholar]
- 42.Makogonenko E, Tsurupa G, Ingham K, Medved L. Interaction of fibrin(ogen) with fibronectin: further characterization and localization of the fibronectin-binding site. Biochemistry. 2002;41:7907–7913. doi: 10.1021/bi025770x. [DOI] [PubMed] [Google Scholar]
- 43.Greiling D, Clark RA. Fibronectin provides a conduit for fibroblast transmigration from collagenous stroma into fibrin clot provisional matrix. J Cell Sci. 1997;110(Pt 7):861–870. doi: 10.1242/jcs.110.7.861. [DOI] [PubMed] [Google Scholar]
- 44.Senger DR, Ledbetter SR, Claffey KP, Papadopoulos-Sergiou A, Peruzzi CA, Detmar M. Stimulation of endothelial cell migration by vascular permeability factor/vascular endothelial growth factor through cooperative mechanisms involving the alphavbeta3 integrin, osteopontin, and thrombin. Am J Pathol. 1996;149:293–305. [PMC free article] [PubMed] [Google Scholar]
- 45.Senger DR, Peruzzi CA, Streit M, Koteliansky VE, de Fougerolles AR Detmar M. The alpha(1)beta(1) and alpha(2)beta(1) integrins provide critical support for vascular endothelial growth factor signaling, endothelial cell migration, and tumor angiogenesis. Am J Pathol. 2002;160:195–204. doi: 10.1016/s0002-9440(10)64363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim S, Bell K, Mousa SA, Varner JA. Regulation of angiogenesis in vivo by ligation of integrin alpha5beta1 with the central cell-binding domain of fibronectin. Am J Pathol. 2000;156:1345–1362. doi: 10.1016/s0002-9440(10)65005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitra AK, Sawada K, Tiwari P, Mui K, Gwin K, Lengyel E. Ligand-independent activation of c-Met by fibronectin and alpha(5)beta(1)-integrin regulates ovarian cancer invasion and metastasis. Oncogene. 2011;30:1566–1576. doi: 10.1038/onc.2010.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bussolino F, Di Renzo MF, Ziche M, Bocchietto E, Olivero M, Naldini L, Gaudino G, Tamagone L, Coffer A, Comogolio PM. Hepatocyte growth factor is a potent angiogenic factor which stimulates endothelial cell motility and growth. J Cell Biol. 1992;119:629–641. doi: 10.1083/jcb.119.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghajar CM, Kachgal S, Kniazeva E, Mori H, Costes SV, George SC, Putnam AJ. Mesenchymal cells stimulate capillary morphogenesis via distinct proteolytic mechanisms. Exp Cell Res. 2010;316:813–825. doi: 10.1016/j.yexcr.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grant DS, Kleinman HK, Goldberg ID, Bhargava MM, Nickoloff BJ, Kinsella JL, Polverini P, Rosen EM. Scatter factor induces blood vessel formation in vivo. Proc Natl Acad Sci U S A. 1993;90:1937–1941. doi: 10.1073/pnas.90.5.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sengupta S, Gherardi E, Sellers LA, Wood JM, Sasisekharan R, Fan TP. Hepatocyte growth factor/scatter factor can induce angiogenesis independently of vascular endothelial growth factor. Arterioscler Thromb Vasc Biol. 2003;23:69–75. doi: 10.1161/01.atv.0000048701.86621.d0. [DOI] [PubMed] [Google Scholar]
- 52.Caspi O, Lesman A, Basevitch Y, Gepstein A, Arbel G, Habib IH, Gepstein L, Levenberg S. Tissue engineering of vascularized cardiac muscle from human embryonic stem cells. Circ Res. 2007;100:263–272. doi: 10.1161/01.RES.0000257776.05673.ff. [DOI] [PubMed] [Google Scholar]
- 53.Stevens KR, Kreutziger KL, Dupras SK, Korte FS, Regnier M, Muskheli V, Nourse MB, Bendixen K, Reinecke H, Murry CE. Physiological function and transplantation of scaffold-free and vascularized human cardiac muscle tissue. Proc Natl Acad Sci U S A. 2009;106:16568–16573. doi: 10.1073/pnas.0908381106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen X, Aledia AS, Ghajar CM, Griffith CK, Putnam AJ, Hughes CC, George SC. Prevascularization of a fibrin-based tissue construct accelerates the formation of functional anastomosis with host vasculature. Tissue Eng Part A. 2009;15:1363–1371. doi: 10.1089/ten.tea.2008.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.