Abstract
Sensory rhodopsin II (SRII, also called pharaonis phoborhodopsin, ppR) is responsible for negative phototaxis in Natronomonas pharaonis. Photoisomerization of the retinal chromophore from all-trans to 13-cis initiates conformational changes in the protein, leading to activation of the cognate transducer protein (HtrII). We previously observed enhancement of the C14–D stretching vibration of the retinal chromophore at 2244 cm−1 upon formation of the K state and interpreted that a steric constraint occurs at the C14D group in SRIIK. Here, we identify the counterpart of the C14D group as Thr204, because the C14–D stretching signal disappeared in T204A, T204S, and T204C mutants as well as a C14–HOOP (hydrogen out-of-plane) vibration at 864 cm−1. Although the K state of the wild-type bacteriorhodopsin (BR), a light-driven proton pump, possesses neither 2244 nor 864 cm−1 bands, both signals appeared for the K state of a triple mutant of BR that functions as a light sensor (P200T/V210Y/A215T). We found a positive correlation between these vibrational amplitudes of the C14 atom at 77 K and the physiological phototaxis response. These observations strongly suggest that the steric constraint between the C14 group of retinal and Thr204 of the protein is a prerequisite for light-signal transduction by SRII.
Microbial rhodopsins convert light into chemiosmotic energy or cellular signals (1, 2). The primary reaction is all-trans to 13-cis photoisomerization of the retinal chromophore, and light energy is stored through altered chromophore–protein interaction (2, 3). Such energy is subsequently consumed by changing the protein structures, and ion pumping or transducer activation is achieved in light-energy or signal conversions, respectively. It is well-known that retinal isomerization in the protein is a selective and efficient reaction along the potential surface of the excited state in rhodopsins (2, 3). One of the characteristics of the photoisomerization is that the reaction occurs even at low temperatures. Since the protein motion is restricted at low temperatures, the structure of the retinal binding site is hardly altered. X-ray crystallography indeed showed little change in the retinal pocket in the primary K intermediate of bacteriorhodopsin (BR),1 a light-driven proton pump (4–6), and pharaonis sensory rhodopsin II (SRII, also called phoborhodopsin) (7), a light sensor for negative phototaxis, in measurements at 100 K. Prior to protein structural changes, the isomerized chromophore therefore has a distorted structure, by which light energy is stored.
How are such structural constraints observed experimentally? Current X-ray crystallographic structures of photointermediates are not sufficiently resolved for constructing the detailed structure of the retinal chromophore. In fact, the retinal structure is considerably different among three X-ray structures for the K intermediate of BR (4–6). The distorted retinal chromophore has been best investigated through vibrational analysis particularly on the hydrogen out-of-plane (HOOP) wagging vibrations in the 1000–800 cm−1 region (8). Previous resonance Raman spectroscopy of rhodopsins successfully detected enhanced HOOP vibrations in their primary intermediates, and they were interpreted in terms of the origin of the chromophore distortion (8). Similar enhanced HOOP modes were also observed for infrared bands. Thus, it is generally accepted that HOOP vibrations are a good probe of chromophore distortion in rhodopsins.
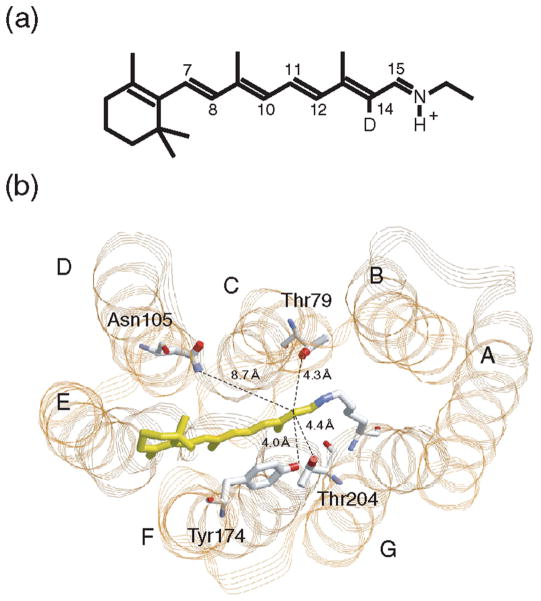
Recently, we reported a novel technique for monitoring steric constraint of the retinal chromophore in the binding pocket (9). In that study, all seven monodeuterated all-trans retinal analogues (positions 7, 8, 10–12, 14, and 15) were synthesized and incorporated into SRII (Figure 1a). The FTIR measurements at 77 K revealed that only the C14–D stretching vibration at 2244 cm−1 is significantly enhanced upon formation of the K state. We interpreted that the steric constraint occurs at the C14D group in SRIIK, where rotation of the C13=C14 bond probably leads to the movement of the C14D group (9). It is noted that SRIIK exhibits various HOOP vibrations, suggesting that the chromophore is widely distorted along the polyene chain (10). In contrast to the HOOP bands, the C–D stretching vibration was specific for the C14 group of SRIIK (9). This fact implies that the C–D stretch provides structural information about the direct contact of the C14 deuterium (hydrogen) with surroundings.
Figure 1.
(a) Chromophore molecule of SRII and BR. All-trans-retinal is bound to a lysine residue via a protonated Schiff base linkage. In this study, the C14 atom is deuterated. (b) X-ray crystallographic structure of SRII (PDB entry 1JGJ) in the extracellular-to-cytoplasmic view. Highlighted are the retinal chromophore (yellow) and residues that are mutated in this study. The distance from the C14 atom to the phenol oxygen of Tyr174 is 4.0 Å, to the hydroxyl oxygen of Thr204 is 4.4 Å, to the hydroxyl oxygen of Thr79 is 4.3 Å, and to the side chain nitrogen of Asn105 is 8.7 Å (7, 12). The OH group of Thr204 is within hydrogen bonding distance (3.2 Å) of the OH group of Tyr174.
The counterpart of the C14D group in SRIIK is of interest. According to the X-ray structure of SRII (11), the distance from the C14 atom to the phenol oxygen of Tyr174 is 4.0 Å, to the hydroxyl oxygen of Thr204 is 4.4 Å, and to the hydroxyl oxygen of Thr79 is 4.3 Å (Figure 1b). The structure of SRIIK shows that only the distance to Thr204 is significantly reduced, from 4.4 to 3.3 Å, by retinal photoisomerization (7, 11). This result suggested that Thr204 may be the counterpart of the C14D group (9). Interestingly, the functional importance of Thr204 has been emphasized by three articles after the report in 2005. Phototaxis function was lost in the mutants of Thr204 or Tyr174, suggesting the biological significance of the hydrogen bond between them (12). Sudo and Spudich successfully engineered BR into an SRII-like light sensor by only three residue replacements, threonine at SRII position 204 (Ala215 in BR) being the most critical (13). FTIR measurements of the triple mutant BR (BR-T) revealed a frequency shift only in the complex with the transducer protein, which can be ascribed to an O–H stretch of Thr215 which is the case for Thr204 of SRII (14). Thus, a crucial role of Thr204 is evident for light-signal transduction in SRII. Here we report that the C14D group indeed interacts with Thr204 in SRIIK by use of several mutant proteins. Such interaction was newly observed for the K state of the light-sensing BR-T, but not for the wild-type BR. Positive correlation between the vibrational amplitudes in the K state and phototaxis responses strongly suggests that the steric constraint between the C14 group of retinal and Thr204 of the protein is crucial for the activation of the transducer protein.
MATERIALS AND METHODS
Preparation of the SRII Samples
The wild-type and mutant proteins of SRII from Natronomonas pharaonis were prepared as described previously (15–17). Briefly, the SRII protein with a histidine tag at the C-terminus was expressed in Escherichia coli, solubilized with 1.5% n-dodecyl β-D-maltoside, and purified by Ni column chromatography. The SRII samples with retinal deuterated at C14 (Figure 1) were produced by adding all-trans-retinal (1 μM) labeled at C14 position to the E. coli culture instead of unlabeled all-trans-retinal (12). The purified SRII sample was then reconstituted into L-α-phosphatidylcholine (PC) liposomes by the removal of the detergent with Biobeads, where the molar ratio of the added PC to SRII was 50:1. The samples of SRII in PC liposomes were finally washed twice [2 mM phosphate buffer (pH 7.0)] for the FTIR measurements.
Preparation of the BR Samples
The wild-type and triple mutant [P200T/V210Y/A215T (BR-T)] proteins of BR were expressed in Halobacterium salinarum Pho81Wr− cells as described previously (13). The purified purple membrane suspension was used for the FTIR measurements of the unlabeled retinal (18). On the other hand, the C14D-labeled retinal was incorporated as follows. Two molar hydroxylamine (pH 7.0) was added to the purple membrane pellet. The sample was kept at room temperature and light through a cutoff filter (Y-52, >500 nm) focused onto the stirred sample. The suspension became colorless after 1.5 h, and the membrane was centrifuged and washed with 50 mM phosphate buffer (pH 7.0, 200 mM NaCl) five times to remove any residual traces of hydroxylamine that could interfere with the reconstitution reaction. The membrane was finally resuspended with 50 mM phosphate buffer (pH 7.0, 200 mM NaCl). C14D-labeled retinal in an ethanolic solution was added stepwise to the membrane suspension at room temperature. The reconstitution efficiency was >75% for both BR and BR-T. The BR and BR-T samples were finally washed twice [2 mM phosphate buffer (pH 7.0)] for the FTIR measurements.
FTIR Spectroscopy
FTIR spectroscopy was performed as described previously (19, 20). Ninety microliters of the SRII sample was dried on a BaF2 window with a diameter of 18 mm. The dry film was then rehydrated by putting an ~1 μL drop of water beside the film and sealing with another window and a silicon rubber O-ring (19, 20). After hydration, the sample was placed in a cell, which was mounted in an Oxford DN-1704 cryostat placed in the Bio-Rad FTS-40 spectrometer. All spectra were measured with 2 cm−1 resolution. The SRIIK minus SRII difference spectra were measured as follows (20). Illumination of the SRII film at pH 7 with 450 nm light at 77 K for 2 min converted SRII to SRIIK, and subsequent illumination with >560 nm light reverted SRIIK to SRII. On the other hand, BR and BR-T were illuminated at 500 nm (2 min) and >640 nm (60 s) for formation and reversion of the K state, respectively (14). The difference spectrum was calculated from spectra constructed with 128 interferograms collected before and after the illumination, and sixteen spectra obtained in this way were averaged for the difference spectra except for that of Y174F. In the case of Y174F, the spectrum for photoreversion of SRIIK to SRII was not a mirror image of the SRIIK minus SRII spectrum (i.e., the forward photoreaction). Since Tyr174 is located next to the retinal chromophore, the Y-to-F mutation must modify the local environment, leading to an unusual photoconvesion. While the detailed mechanism remains unclear, eight spectra independently obtained by illumination with 450 nm light were averaged to produce the SRIIK minus SRII spectrum of Y174F in this study.
Phototaxis Analysis
Phototaxis responses were measured as swimming reversal frequency changes of cell populations in response to photostimulus as described previously (12, 21). Swimming reversals were measured by stimulus-induced effects on the ratio of rate of change of direction (RCD) to speed (SPD) by computer-assisted motion analysis of individual cell tracks. The software used for the analysis system was Celltrack 1.2 Beta (Motion Analysis Corp., Santa Clara, CA); 200–2500 cells were assayed for each trace (Figure 4). Maximal stimuli from a 100 W Hg lamp through a heat filter and 40 nm band-pass interference filters delivered through microscope optics were as follows: 100 ms, 500 nm stimulus for wild-type SRII and SRII mutants; 100 ms, 580 nm stimulus for wild-type BR; and 100 ms, 550 nm stimulus for BR-T (13). The HtrII transducer protein that forms a complex with rhodopsin is from N. pharaonis in this study except for the measurements of BR and BR-T, where H. salinarum HtrII was used (13). The expression levels calculated from amplitudes of the pigments’ main absorption bands in the visible spectrum for SRII–HtrII, BR-T–HtrII, T204A–HtrII, T204S–HtrII, T204C–HtrII, Y174F–HtrII, T79A–HtrII, N105D–HtrII, and BR–HtrII complexes in transformants used in this study were all similar, namely, 2.0 × 104, 1.0 × 104, 2.7 × 104, 1.8 × 104, 1.7 × 104, 1.1 × 104, 1.8 × 104, 2.1 × 104, and 3.5 × 104 molecules per cell, respectively.
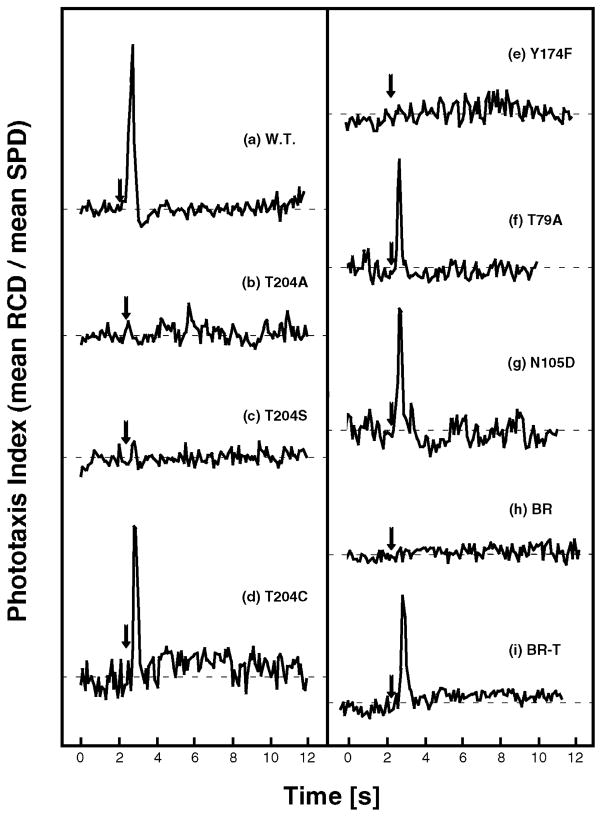
Figure 4.
Phototaxis responses measured as swimming reversal frequency changes to photostimulus (arrow): 100 ms, 500 nm stimulus for wild-type SRII and SRII mutants and 100 ms, 550 nm stimulus for wild-type BR and BR-T. The swimming reversal frequency in a cell population is monitored by the quotient of the rate of change of direction (RCD) and linear swimming speed (SPD), which increase and decrease, respectively, during a swimming reversal. Cells contain the wild-type SRII–HtrII (a), T204A–HtrII (b), T204S–HtrII (c), T204C–HtrII (d), Y174F–HtrII (e), T79A–HtrII (f), and N105D–HtrII (g) complexes and the wild-type BR–HtrII (h) and BR-T–HtrII (i) complexes.
RESULTS
C–D Stretching Vibrations of the Retinal in SRIIK of Mutants Show the Steric Constraint of the C14 Atom and Thr204
Figure 2A shows the SRIIK minus SRII difference spectra of the wild type (a) and several mutants (b–g) in the C–D stretching region. As reported previously (9), the spectra of wild-type SRII exhibit a strong positive peak at 2244 cm−1 for the C14D-labeled retinal (solid line in part a of Figure 2A), but not for the unlabeled retinal (dotted line in part a of Figure 2A). Such a C–D stretch was unique for the C14D group, and no peaks were observed for the C7D, C8D, C10D, C11D, C12D, and C15D groups (9). The lack of a corresponding negative peak suggests that the C14–D stretch for SRII is much smaller in amplitude than in SRIIK. Thus, it is likely that only the C14–D stretching vibration is greatly enhanced upon retinal photoisomerization of SRII, and we interpreted that the enhanced C–D stretch originates from the steric constraint with the surrounding protein moiety.
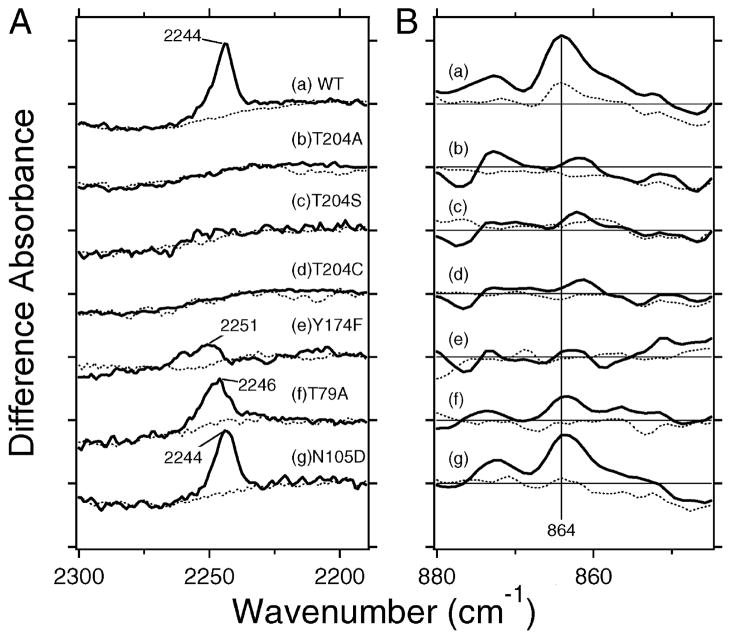
Figure 2.
(A) SRIIK minus SRII difference spectra of the wild type (a), T204A (b), T204S (c), T204C (d), Y174F (e), T79A (f), and N105D (g) in the 2330–2190 cm−1 region. The solid line represents the spectrum for the C14D-labeled retinal, while the dotted line corresponds to that for the unlabeled retinal. Vertical amplitudes of all spectra are normalized by the bands of Asn105 at 1704 (−)/1700 (+) cm−1 for spectra a–f. On the other hand, those in spectrum g were normalized to the wild type (a) by the C–C stretch at 1204 (−) cm−1 (unlabeled retinal) or the bands at 1743 (−)/1738 (+) cm−1 (C14D-labeled retinal) (see Figure S1 in detail). One division of the y-axis corresponds to 0.00018 absorbance unit. (B) SRIIK minus sRII difference spectra of the wild type (a), T204A (b), T204S (c), T204C (d), Y174F (e), T79A (f), and N105D (g) in the 880–845 cm−1 region. The solid line represents the spectrum for the unlabeled retinal, while the dotted line corresponds to that for the C14D-labeled retinal. One division of the y-axis corresponds to 0.0008 absorbance unit.
Then, we tested several mutant proteins. The SRIIK minus SRII difference spectra in the 1800–800 cm−1 region show that retinal photoisomerization takes place in all mutants as in the wild type (Figure S1 of the Supporting Information). Spectra b–d of Figure 2A show that the positive band completely disappears for the mutant of Thr204, implying that the counterpart of the steric interaction of the C14D group is Thr204. Spectrum e of Figure 2A shows that the positive band is significantly reduced in intensity also in the Y174F mutant. Since Tyr174 forms a hydrogen bond with Thr204 in SRII, the lack of a hydrogen bond in the Y174F mutant may influence the steric interaction between the C14 group and Thr204. In other words, these results strongly suggest that the counterpart of the C14D group is Thr204, not Tyr174. This is consistent with the X-ray structures of SRII and SRIIK, where the distance from the C14 atom changes from only 4.0 to 3.9 Å for the phenol oxygen of Tyr174, and a larger change from 4.4 Å to 3.3 Å occurs for the hydroxyl oxygen of Thr204 (7, 11). It also should be noted that the peak frequency is upshifted to 2251 cm−1 for Y174F, and the broadened band implies conformational heterogeneity for the C14D group (spectrum e of Figure 2A). The positive peak was preserved but considerably reduced in magnitude for T79A (spectrum f of Figure 2A), whose peak frequency was slightly upshifted (2246 cm−1). In contrast, the positive band looks identical for N105D (spectrum g of Figure 2A), although the peak intensity was slightly reduced.
Hydrogen Out-Of-Plane (HOOP) Vibrations of the Retinal in SRIIK of Mutants Show the Steric Constraint of the C14 Atom and Thr204
HOOP vibrations are also regarded as the probe of the distortion of the retinal chromophore (8). Therefore, if the steric constraint occurs at the C14 group, the C14–HOOP mode should also be influenced. We previously identified the positive band at 864 cm−1 as the C14–HOOP mode, because the 864 cm−1 band disappeared only in the C14D spectrum (10). Spectrum a of Figure 2B reproduces the previous result, where the positive band at 864 cm−1 (solid line) significantly reduces the intensity for the C14–D labeling (dotted line). The presence of the C14–HOOP band for SRIIK can be interpreted in terms of the chromophore distortion at this moiety. The positive peak at 864 cm−1 significantly reduces the intensity for T204A, T204S, T204C, and Y174F (spectra b–e of Figure 2B) which is the case for the C14–D stretching vibrations (spectra b–e of Figure 2A). The 864 cm−1 band was preserved in T79A, but the intensity became approximately half (spectrum f of Figure 2B). The positive band looks identical for N105D (spectrum g of Figure 2B), although the peak intensity was slightly reduced. These spectral features for T79A and N105D were also the same as for the C14–D stretching vibrations (spectra f and g of Figure 2A).
Since many HOOP and other vibrations exist in this frequency region, the spectra are not as clear as those for the C–D stretching vibrations. However, the spectral feature of the C14–HOOP band at 864 cm−1 is consistent with that of the C14–D stretching band at 2244 cm−1. It should be noted that the C14–HOOP mode is located at a frequency 33 cm−1 higher than that of the 13-cis-retinal (831 cm−1) (22) and is closer to that of the all-trans-retinal (876 cm−1) (23). This indicates that the chromophore of SRIIK is in an unrelaxed 13-cis configuration.
C14–D Stretching and C14–HOOP Vibrations in BR-TK
BR-T activates HtrII, and the BR mutant gains light signaling activity (13). How do the vibrations related to the C14 atom in BR-T differ from those of wild-type BR? In this study, we incorporated C14D-labeled retinal into wild-type BR and BR-T, and the isotope effect was examined by measuring the difference FTIR spectra between the K and unphotolyzed states. Unlike SRII, the wild-type BR containing C14D-labeled retinal exhibits no positive bands in the frequency region of C–D stretching (solid line in part a of Figure 3A), indicating that the 2244-cm−1 band is unique for SRII. In other words, formation of the K state does not entail steric constraint around the C14 atom in wild-type BR. In contrast, BR-T containing the C14-D labeled retinal shows an appearance of the band at 2250 cm−1 (solid line in part b of Figure 3A), which is absent for the unlabeled one (dotted line in part b of Figure 3A). The corresponding amino acid to Thr204 in SRII is Ala215 in BR, and Ala215 is replaced with threonine in BR-T (P200T/V210Y/A215T). Thus, the newly appearing C14–D stretch in part b of Figure 3A apparently originates from the steric constraint between the C14 atom and Thr215 in BR-T. It should be noted that we did not examine the single A215T mutant in this study. However, the spectrum of the K intermediate in the P200T/V210Y mutant BR is almost identical to that of wild-type BR (14), implying that the observed C14–D stretch arises from direct interaction with Thr215.
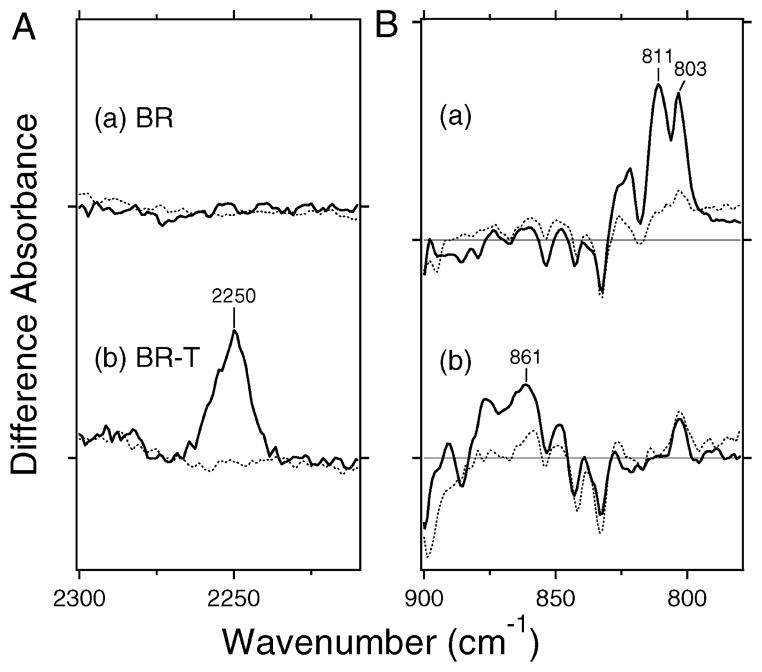
Figure 3.
(A) Difference spectra of BR (a) and BR-T (b) in the 2300–2210 cm−1 region. The solid line represents the spectrum for the C14D-labeled retinal, while the dotted line corresponds to that for the unlabeled retinal. Vertical amplitudes are normalized by the bands to wild-type SRII (Figure 2a) by the C–C stretch at 1204 (−) cm−1 (unlabeled retinal). Those for the C14D-labeled retinal are then normalized to those for the unlabeled retinal by the amide I bands at 1639 (−)/1623 (+) (a) and 1640 (−)/1623 (+) (b) cm−1 (see Figure S1 in detail). One division of the y-axis corresponds to 0.0003 absorbance unit. (B) Difference spectra of BR (a) and BR-T (b) in the 900–780 cm−1 region. The solid line represents the spectrum for the unlabeled retinal, while the dotted line corresponds to that for the C14D-labeled retinal. One division of the y-axis corresponds to 0.002 absorbance unit.
Parts a and b of Figure 3B show the HOOP region of wild-type BR and BR-T, respectively. We already reported the spectra for the unlabeled retinal (solid lines in parts a and b of Figure 3B) (14), while the spectra for the C14D-labeled retinal are newly measured (dotted lines). Two peaks at 811 and 803 cm−1 in part a of Figure 3B almost disappear upon deuteration of C14. Therefore, wild-type BR possesses the C14–HOOP mode at these frequencies, which is consistent with the previous resonance Raman study (24). In contrast, BR-T does not exhibit the bands at 811 and 803 cm−1. Instead, a new band appeared at 861 cm−1 (part b of Figure 3B) that is close in frequency to the C14–HOOP mode of wild-type SRII (part a of Figure 2B). In fact, the band significantly reduces the intensity for the C14–D labeling (dotted line), indicating that the band contains the C14–HOOP vibration.
Thus, both C–D stretching and HOOP vibrations in BR-T are fully consistent with the observations for SRII, indicating that the K state of BR-T exhibits a steric constraint at the C14 atom of retinal with Thr215. Therefore, BR-T was converted from the BR-type difference spectrum to the SRII-type difference spectrum by introduction of this steric constraint with the retinal chromophore (13). It should be noted that wild-type BR possesses the C14–HOOP mode at 811 and 803 cm−1 (part a of Figure 3B), but no C–D stretching vibrations (part a of Figure 3A), indicating that the C–D stretch is more specific to the structural change of the retinal chromophore.
Phototaxis Responses of the Cells Containing Mutant SRII Receptors
FTIR spectra show how the mutations affect early photocycle structural changes in SRII, BR, and BR-T. We measured the influence of each mutation on phototaxis responses by the motility behavior of cells containing the mutant proteins. Figure 4a shows that the SRII–HtrII complex mediated a phototaxis response evident as a transient increase in swimming reversal frequency. As we reported previously, the T204A–HtrII (Figure 4b), T204S–HtrII (Figure 4c), and Y174F–HtrII (Figure 4e) complexes did not mediate detectable responses, while the T204C–HtrII (Figure 4d) complex mediated a phototaxis response similar to that of the wild type (12). T79A and N105D mutants also exhibited phototaxis responses (Figure 4f,g).
Parts h and i of Figure 4 show the results for BR and BR-T, respectively. The former is a negative control, because BR does not form a complex with HtrII. On the other hand, BR-T forms a complex with HtrII and mediates negative phototaxis. Phototaxis responses from BR-T were found to be much greater with HtrII from H. salinarum than with HtrII from N. pharaonis, possibly because of tighter or more efficient coupling (13), and Figure 4i therefore shows the result with HtrII from H. salinarum.
Correlation between the Amplitudes of the Vibrations at the C14 Atom and Phototaxis Responses
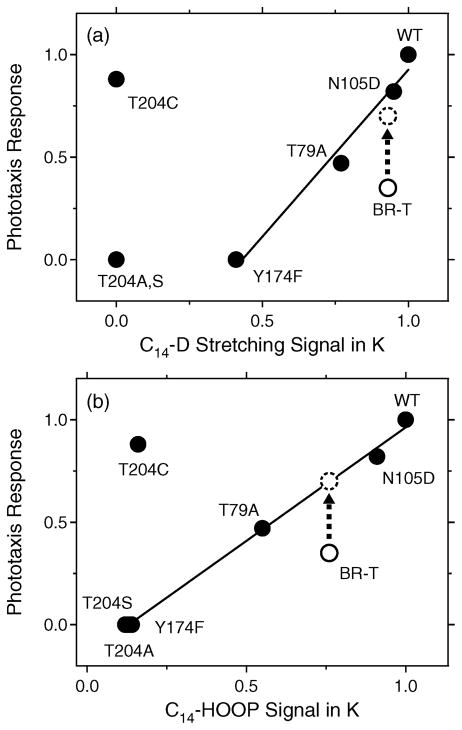
The C14–D stretching and C14–HOOP bands are observed for the K state, which appears on the picosecond (10−12) time scale. On the other hand, phototaxis responses appear on a subsecond (10−1) time scale (Figure 4). In spite of such different time scales, vibrational and phototaxis signals seem to be correlated for the mutants tested. The fraction of light-induced reversing of cells containing a mutant SRII–HtrII complex used in this study was plotted against photostimulus duration (2–500 ms) (13), and the Km values from Michaelis–Menten fits to the fluence–response curves were estimated as sensitivities of cells expressing BR-T–HtrII, T204A–HtrII, T204S–HtrII, T204C–HtrII, Y174F–HtrII, T79A–HtrII, and N105D–HtrII complexes (35, 0, 0, 88, 0, 47, and 82%, respectively, of that conferred by the wild-type SRII–HtrII complex). Figure 5a plots the Km values versus their C14–D stretching amplitudes, where the signal of the wild-type SRII is normalized to unity. The x-axis of Figure 5a represents the area of the difference spectra between C14D-labeled (solid lines) and unlabeled (dotted lines) retinals in Figure 2A. The straight line in Figure 5a implies a linear correlation between the C14–D stretching amplitude and phototaxis response except for the mutants of Thr204 and BR-T. Figure 5b similarly plots the Km values of SRII and BR-T versus their C14–HOOP signals, where the signal of the wild-type SRII is normalized to unity. Unlike the C14–D stretching vibration, deuteration of the C14 atom shows complex isotope effects, because there are multiple vibrations containing the C14–HOOP mode. Therefore, the x-axis of Figure 5b simply represents the subtracted amplitude of the C14D-labeled HOOP (dotted lines in Figures 2B and 3B) from unlabeled (solid lines) retinals at 864 cm−1. We observe a linear dependence except for T204C and BR-T. It should be noted that BR-T deviates from the straight line but BR-T possesses approximately half of the 13-cis form in the dark. If the low content of the all-trans form is corrected (dashed circles in Figure 5), positive correlations are also obtained for BR-T. These results strongly suggest that, if the steric constraint at the C14 position is weakened, receptor phototaxis signals become smaller.
Figure 5.
Correlation between the C14–D stretching (a) or C14–HOOP (b) signals in the K photointermediate and Km values of phototaxis responses. The Km values were calculated from Michaelis–Menten fits to the fluence–response curves as described in ref 14. (a) The abscissa corresponds to the normalized area of the positive bands at 2244 cm−1 in Figures 2A and 3A. The straight line is the best linear fit to the data excluding the T204 mutants and BR-T. (b) The abscissa corresponds to the subtracted amplitude of the C14D-labeled retinal HOOP (dotted line) from the unlabeled retinal HOOP (solid line) at 864 cm−1 in Figures 2B and 3B. The straight line is the best linear fit to the data except for T204C and BR-T. The values for the wild-type SRII are normalized to unity in panels a and b. Dashed circles represent the corrected data for the all-trans form of BR-T, where the phototaxis response is doubled because BR-T possesses approximately half of the 13-cis form.
Figure 5 also suggests some differences for the C14–D stretching (a) and C14–HOOP (b) vibrations. The Y174F mutant exhibits a small C14–D stretch (part e of Figure 2A), but no C14–HOOP vibration (part e of Figure 2B). As a consequence, the straight line in Figure 5a exhibits an offset at y = 0, while the straight line in Figure 5b approximately crosses the origin. Such a difference may originate from the differing structural information from the C–D stretch and the HOOP vibration, though both monitor steric constraint. As noted above, the counterpart of the C14D (H) group is probably the side chain of Thr204, and it is likely that the C–D vibration monitors more local events. Therefore, the mutants of Thr204 may have to be excluded from the linear correlation in the C–D stretch (Figure 5a), whereas the results for T204A and T204S are on the straight line for the HOOP vibration (Figure 5b).
T204C does not follow the correlation in Figure 5. There are neither C14–D stretching nor C14–HOOP vibrations for T204C (Figure 2d), but this mutant mediates normal phototaxis responses (Figure 4d). Apparently, the light-signal transduction in T204C does not need the steric constraint of the C14 atom of retinal. One possibility is that a similar steric constraint in T204C is already relaxed at 77 K, and lower temperatures would be required to observe C14–D stretching or C14–HOOP vibrations for T204C. Another possibility is that T204C, but not T204A or T204S, mimics the structure near position 204 found in the wild type after isomerization of the retinal and after the steric constraint is resolved. In other words, T204C may exhibit the local structure characteristic of the photoactivated state. However, the FTIR spectra at 77 K are otherwise similar between the wild type and T204C (Figure S1), suggesting the formation of the normal K state for T204C. Therefore, the intramolecular signal transduction mechanism of T204C is unclear at present.
In summary, the C14–D stretching and C14–HOOP bands are positively correlated with the phototaxis response in the presence of Thr204, the counterpart of the C14 group. Since their time scales are widely separated (10−12 s vs 10−1 s), we infer that the light-energy storage in the steric constraint at the retinal C14 group is a prerequisite for protein structural changes for the activation of HtrII, leading eventually to the function, phototaxis. Each mutation at Tyr174, Thr79, and Asn105 apparently weakens the chromophore–protein interaction, thereby reducing the phototaxis response. In other words, effects of mutation on the function may be explained in terms of light-energy storage in the steric constraint.
DISCUSSION
C14–D Stretching and C14–HOOP Vibrations in the K Intermediate
This study supports the notion that the enhanced C14–D stretching vibration at 2244 cm−1 in SRIIK originates from the interaction between the C14D group of the chromophore and Thr204 (Figure 2A). The enhanced C–D stretch must result from an enlarged dipole moment of the vibration, and we identify the local steric constraint between the C14D group and the side chain of Thr204 as the origin of the strong electric polarization. It should be noted that direct steric conflict may not occur, but rather a strong interaction between the C14D group and Thr204 may directly cause such electric polarization of the C14D group. While the mechanism of the enlarged dipole must be examined theoretically, the positive correlation between the C14–D stretching amplitude and phototaxis responses (Figure 5a) strongly suggests that the enhanced C14–D stretching vibration is functionally important.
Steric constraint between the C14D group of the chromophore and Thr204 is also supported by the analysis of the C14–HOOP vibration at 864 cm−1. The C14–HOOP signals similarly diminished in the T204 mutants as well as Y174F (Figure 2B). We also observed a positive correlation of the C14–HOOP amplitude with the phototaxis response (Figure 5b). Thus, both C14–D stretch and C14–HOOP vibrations imply that the chromophore–protein interaction at the C14 position may be critical for protein function. Despite the common information gained from the vibrational analysis of the C14–D stretching and C14–HOOP vibrations, they are not necessarily identical. Previous study showed that the band at 864 cm−1 also contains a C12–HOOP vibration, because the amplitude is considerably reduced for C12D-labeled retinal (10). Thus, the band at 864 cm−1 originates from the coupled HOOP vibrations of C14 and C12 atoms, while the C14–HOOP vibration is predominant. On the other hand, the C–D stretching vibration is specific to the C14 group (9), implying that the C–D stretch provides structural information about the direct contact of deuterium (hydrogen) with surrounding atoms.
Mechanism of Light-Energy Storage and Signal Transduction
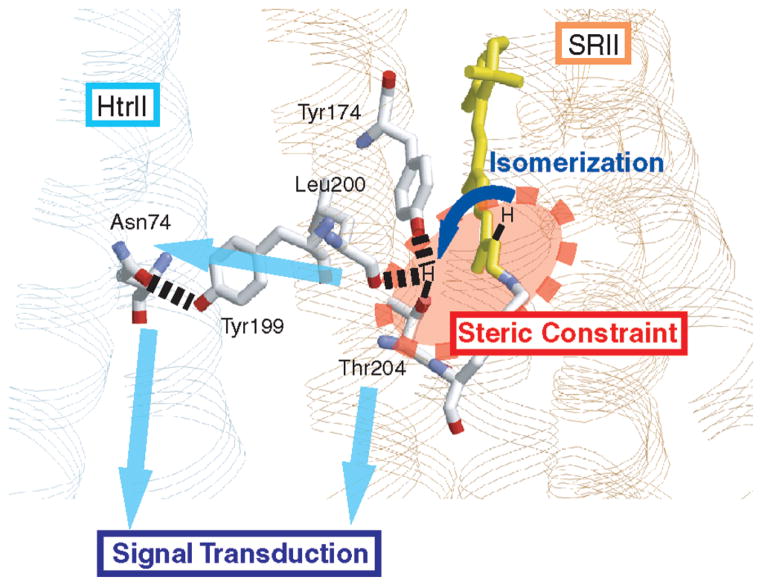
Our results argue that steric constraint at the retinal C14 group is likely a prerequisite for protein structural changes for the activation of HtrII, leading eventually to the function, phototaxis. Figure 6 schematically illustrates a possible intramolecular and intermolecular signal relay pathway based on the SRII–HtrII complex structure. In this model, retinal isomerization causes the steric conflict at the retinal C14 group with the side chain of Thr204, which strongly perturbs the hydrogen bonds of Thr204 with the phenol oxygen of Tyr174 and the peptide backbone of Leu200 (Figure 6). Such structural perturbation is relayed into the intermolecular hydrogen bond between Tyr199 of SRII and Asn74 of HtrII and/or nearby van der Waals contact regions (15, 25). The correlation in Figure 5 strongly suggests that the energy storage in the early intermediate is a prerequisite for the subsequent functional process.
Figure 6.
Schematic drawing of the primary step in the light-signal transduction in SRII (PDB entry 1H2S). Retinal photoisomerization causes the steric constraint between the C14H group and Thr204, by which the C14–D stretching vibration is enhanced. Light energy stored by the steric constraint is used for changing the protein structures, including the protein–protein interaction surface with HtrII.
With regard to the mechanism of light-energy storage, recent findings on the proton-pumping rhodopsins should be considered. Low-temperature FTIR studies showed a strong correlation between the presence of strongly hydrogen-bonded water molecule(s) and the proton pumping activity of rhodopsins (26). For example, strongly hydrogen-bonded water molecules are observed for BR (27), SRII (19), proteorhodopsin (28), Leptosphaeria rhodopsin (29), and azide-bound halorhodopsin (30), which all pump protons. Strongly hydrogen-bonded water molecules were not observed for salinarum halorhodopsin (31), pharaonis halorhodopsin (32), Anabaena sensory rhodopsin (33), Neurospora rhodopsin (34), or bovine rhodopsin (35), which have no proton pumping activity. Cl−-pumping BR_D85S has no strongly hydrogen-bonded water molecules (36), but proton-pumping BR_D212N(Cl−) has strongly hydrogen-bonded water molecules (37). These results were interpreted in terms of the proposal that the presence of a strong hydrogen bond of water is a prerequisite for proton pumping in rhodopsins (26). It is likely that destabilization of a water-containing hydrogen-bonding network plays an important role in light-energy storage in this case.
It should be noted that SRII (19) and the SRII–HtrII complex (38) both possess strongly hydrogen-bonded water molecules, whereas the latter does not pump protons but functions in light-signal transduction. Phylogenetic analysis suggests that rhodopsin photosensors like SRI and SRII evolved from light-driven proton pumps (39). Therefore, sensory rhodopsins can be considered pumps that have gained a mechanism for activating transducer proteins. This study indicates that photoisomerization-induced steric constraint between the C14H group of the retinal chromophore and Thr204 in SRII is one such mechanism.
The light-energy storage capacity of microbial rhodopsins is smaller than that in bovine rhodopsin (3). Low-temperature photocalorimetric studies reported the energy stored in the primary intermediates is 35 and 16 kcal/mol for bovine rhodopsin (40) and BR (41), respectively. This indicates that only 30% of the light energy is stored in the structure of BRK, which is approximately half of that in visual rhodopsin (60%). The loss of 70% of the light energy in the formation of BRK does not compromise BR function, since proton pumping by BR is a quantal process; i.e., BR pumps a single proton by use of one photon, and the free energy gain by pumping a proton is 6 kcal/mol (2, 3). The presence of strongly hydrogen-bonded water molecule in the proton-pumping rhodopsins suggests that hydrogen bonding stabilization of a water molecule and its destabilization in the K state is crucial for the pumping function. In fact, previous QM/MM calculations of BR and BRK estimated the hydrogen bonding destabilization to be 11 kcal/mol (70%) of the total energy storage of 16 kcal/mol (42, 43). On the other hand, optoacoustic spectroscopy revealed that the enthalpy change upon formation of SRIIK (21 kcal/mol) is larger than that for BRK (13 kcal/mol in this study) (44, 45). Thus, it is likely that the steric constraint at the C14 atom of retinal contributes to additional energy storage, acquired during the evolution of SRII from a BR-like pump, by the introduction of Thr204. This inference is supported by the fact that BR-T activates HtrII, demonstrating that BR and SRII share their fundamental mechanisms of phototransduction (13).
Supplementary Material
Footnotes
This work was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science, and Technology to H.K. (19370067, 20044012, and 20050015) and Y.F. (19045015) and National Institutes of Health Grant R37GM27750 and a Robert A. Welch Foundation grant to J.L.S.
Abbreviations: SRII, sensory rhodopsin II from Natronomonas pharaonis (also known as NpSRII or ppR); HtrII, halobacterial transducer protein II; BR, bacteriorhodopsin; BR-T, triple mutant of BR (P200T/V210Y/A215T) having photosensory signaling function; SRIIK, K intermediate of SRII; BRK, K intermediate of BR; FTIR, Fourier transform infrared; HOOP, hydrogen out-of-plane; PC, L-α-phosphatidylcholine; QM/MM, quantum mechanical/molecular mechanical.
SUPPORTING INFORMATION AVAILABLE
SRIIK minus SRII difference FTIR spectra of the wild type, T204A, T204S, T204C, Y174F, T79A, and N105D and BRK minus BR difference FTIR spectra of the wild type and BR-T in the 1790-810 cm−1 region, where the chromophore molecule is the unlabeled and C14D-labeled retinal (Figure S1). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Spudich JL, Lanyi JK. Shuttling between two protein conformations: The common mechanism for sensory transduction and ion transport. Curr Opin Cell Biol. 1996;8:452–457. doi: 10.1016/s0955-0674(96)80020-2. [DOI] [PubMed] [Google Scholar]
- 2.Kandori H. In: Retinal Binding Proteins: From cis-trans Isomerization in Biochemistry. Dugave C, editor. Chapter 4. Wiley-VCH; Freiburg, Germany: 2006. p. 53. [Google Scholar]
- 3.Birge RR. Nature of the primary photochemical events in rhodopsin and bacteriorhodopsin. Biochim Biophys Acta. 1990;1016:293–327. doi: 10.1016/0005-2728(90)90163-x. [DOI] [PubMed] [Google Scholar]
- 4.Edman K, Nollert P, Royant A, Belrhali H, Pebay-Peyroula E, Hajdu J, Neutze R, Landau EM. High-resolution X-ray structure of an early intermediate in the bacteriorhodopsin photocycle. Nature. 1990;401:822–826. doi: 10.1038/44623. [DOI] [PubMed] [Google Scholar]
- 5.Schobert B, Cupp-Vickery J, Hornak V, Smith S, Lanyi J. Crystallographic Structure of the K Intermediate of Bacteriorhodopsin: Conservation of Free Energy after Phtoisomerization of the Retinal. J Mol Biol. 2002;321:715–726. doi: 10.1016/s0022-2836(02)00681-2. [DOI] [PubMed] [Google Scholar]
- 6.Matsui Y, Sakai K, Murakami M, Shiro Y, Adachi S, Okumura H, Kouyama T. Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary Photoreaction of Bacteriorhodopsin. J Mol Biol. 2002;324:469–481. doi: 10.1016/s0022-2836(02)01110-5. [DOI] [PubMed] [Google Scholar]
- 7.Edman K, Royant A, Nollert P, Maxwell CA, Pebay-Peyroula E, Navarro J, Neutze R, Landau EM. Early Structural Rearrangements in the Photocycle of an Integral Membrane Sensory Receptor. Structure. 2002;10:473–482. doi: 10.1016/s0969-2126(02)00736-0. [DOI] [PubMed] [Google Scholar]
- 8.Mathies RA, Smith SO, Palings I. In: Biological applications of Raman spectroscopy: Resonance Raman spectra of polyenes and aromatics. Spiro TG, editor. II. John Wiley and Sons; New York: 1987. pp. 59–108. [Google Scholar]
- 9.Sudo Y, Furutani Y, Wada A, Ito M, Kamo N, Kandori H. Steric Constraint in the Primary Photoproduct of an Archaeal Rhodopsin from Regiospecific Perturbation of C-D Stretching Vibration of the Retinyl Chromophore. J Am Chem Soc. 2005;127:16036–16037. doi: 10.1021/ja056203a. [DOI] [PubMed] [Google Scholar]
- 10.Furutani Y, Sudo Y, Wada A, Ito M, Shimono K, Kamo N, Kandori H. Assignment of the Hydrogen-Out-Of-Plane and -in-Plane Vibrations of the Retinal Choromophore in the K Intermediate of pharaonis Phoborhodopsin. Biochemistry. 2006;45:11836–11843. doi: 10.1021/bi0610597. [DOI] [PubMed] [Google Scholar]
- 11.Luecke H, Schobert B, Lanyi JK, Spudich EN, Spudich JL. Crystal Structure of Sensory Rhodopsin II at 2.4 Å: Insights into Color Tuning and Transducer Interaction. Science. 2001;293:1499–1503. doi: 10.1126/science.1062977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sudo Y, Furutani Y, Kandori H, Spudich JL. Functional Importance of the Interhelical Hydrogen Bond between Thr204 and Tyr174 of Sensory Rhodopsin II and Its Alteration during the Signaling Process. J Biol Chem. 2006;281:34239–34245. doi: 10.1074/jbc.M605907200. [DOI] [PubMed] [Google Scholar]
- 13.Sudo Y, Spudich JL. Three strategically placed hydrogen-bonding residues convert a proton pump into a sensory receptor. Proc Natl Acad Sci USA. 2006;103:16129–16134. doi: 10.1073/pnas.0607467103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudo Y, Furutani Y, Spudich JL, Kandori H. Early Photocycle Structural Changes in a Bacteriorhodopsin Mutant Engineered to Transmit Photosensory Signals. J Biol Chem. 2007;282:15550–15558. doi: 10.1074/jbc.M701271200. [DOI] [PubMed] [Google Scholar]
- 15.Sudo Y, Furutani Y, Shimono K, Kamo N, Kandori H. Hydrogen Bonding Alteration of Thr-204 in the Complex between pharaonis Phoborhodopsin and Its Transducer Protein. Biochemistry. 2003;42:14166–14172. doi: 10.1021/bi035678g. [DOI] [PubMed] [Google Scholar]
- 16.Iwamoto M, Sudo Y, Shimono K, Araiso T, Kamo N. Correlation of the O-Intermediate Rate with the pKa of Asp-75 in the Dark, the Counterion of the Schiff Base of Pharaonis Phoborhodopsin (Sensory Rhodopsin) Biophys J. 2005;88:1215–1223. doi: 10.1529/biophysj.104.045583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandori H, Shimono K, Shichida Y, Kamo N. Interaction of Asn 105 with the Retinal Chromophore during Photoisomerization of pharaonis phoborhodopsin. Biochemistry. 2002;41:4554–4559. doi: 10.1021/bi0120749. [DOI] [PubMed] [Google Scholar]
- 18.Tanimoto T, Furutani Y, Kandori H. Strucral Changes of Water in the Schiff Base Region of Bacteriorhodopsin: Proposal of Hydration Switch Model. Biochemistry. 2003;42:2300–2306. doi: 10.1021/bi026990d. [DOI] [PubMed] [Google Scholar]
- 19.Kandori H, Furutani Y, Shimono K, Shichida Y, Kamo N. Internal Water Molecules of pharaonis Phoborhodopsin Studied by Low-Temperature Infraed Spectroscopy. Biochemistry. 2001;40:15693–15698. doi: 10.1021/bi011621n. [DOI] [PubMed] [Google Scholar]
- 20.Kandori H, Shimono K, Sudo Y, Iwamoto M, Shichida Y, Kamo N. Structural Changes of pharaonis Phoborhodopsin upon Photoisomerization of the Retinal Chomophore: Infrared Spectral Comparison with Bacteriorhodopsin. Biochemistry. 2001;40:9238–9246. doi: 10.1021/bi0103819. [DOI] [PubMed] [Google Scholar]
- 21.Jung KH, Spudich JL. Protonatable residues at the cytoplasmic end of transmembrane helix-2 in the signal transducer HtrI control photochemistry and function of sensory rhodopsin I. Proc Natl Acad Sci USA. 1996;93:6557–6561. doi: 10.1073/pnas.93.13.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curry B, Palings I, Broek A, Pardoen JA, Mulder PPJ, Lugtenbrug J, Mathies R. Vibrational Analysis of 13-cis-Retinal. J Phys Chem. 1984;88:688–702. [Google Scholar]
- 23.Curry B, Broek A, Lugtenbrug J, Mathies R. Vibrational Analysis of all-trans-Retinal. J Am Chem Soc. 1982;104:5274–5286. [Google Scholar]
- 24.Palings I, van den Berg EM, Lugtenburg J, Mathies RA. Complete Assignment of the Hydrogen Out-of-Plane Wagging Vibrations of Bathorhodopsin: Chromophore Structure and Energy Storage in the Primary Photoproduct of Vision. Biochemistry. 1989;28:1498–1507. doi: 10.1021/bi00430a012. [DOI] [PubMed] [Google Scholar]
- 25.Furutani Y, Kamada K, Sudo Y, Shimono K, Kamo N, Kandori H. Structural Changes of the Complex between pharaonis Phoborhodopsin and Its Cognate Transducer upon Formation of the M Photointermediate. Biochemistry. 2005;44:2909–2915. doi: 10.1021/bi047893i. [DOI] [PubMed] [Google Scholar]
- 26.Furutani Y, Shibata M, Kandori H. Strongly hydrogen-bonded water molecules in the Schiff base region of rhodopsin. Photochem Photobiol Sci. 2005;4:661–666. doi: 10.1039/b416698a. [DOI] [PubMed] [Google Scholar]
- 27.Shibata M, Tanimoto T, Kandori H. Water Molecules in the Schiff Base Region of Bacteriorhodopisn. J Am Chem Soc. 2003;125:13312–13313. doi: 10.1021/ja037343s. [DOI] [PubMed] [Google Scholar]
- 28.Furutani Y, Ikeda D, Shibata M, Kandori H. Strongly hydrogen-bonded water molecules is observed only in the alkaline form of proteorhodopsin. Chem Phys. 2006;324:705–708. [Google Scholar]
- 29.Sumii M, Furutani Y, Waschuk SA, Brown LS, Kandori H. Strongly Hydrogen-Bonded Water Molecule Present near the Retinal Chromophore of Leptosphaeria Rhodopsin, the Bacteriorhodopsin-like Proton Pump from a Eukaryote. Biochemistry. 2005;44:15159–15166. doi: 10.1021/bi0513498. [DOI] [PubMed] [Google Scholar]
- 30.Muneda N, Shibata M, Demura M, Kandori H. Internal Water Molecules of the Proton-pumping Halorhodopsin in the Presence of Azide. J Am Chem Soc. 2006;128:6294–6295. doi: 10.1021/ja060253d. [DOI] [PubMed] [Google Scholar]
- 31.Shibata M, Muneda N, Ihara K, Sasaki T, Demura M, Kandori H. Internal water molecules of light-driven choride pump proteins. Chem Phys Lett. 2004;392:330–333. [Google Scholar]
- 32.Shibata M, Muneda N, Sasaki T, Shimono K, Kamo N, Demura M, Kandori H. Hydrogen-Bonding Alterations of the Protonated Schiff Base and Water Molecule in the Chloride Pump of Natronobacterium pharaonis. Biochemistry. 2005;44:12279–12286. doi: 10.1021/bi050726d. [DOI] [PubMed] [Google Scholar]
- 33.Furutani Y, Kawanabe A, Jung KH, Kandori H. FTIR Spectroscopy of the All-Trans Form of Anabaena Sensory Rhodopsin at 77 K: Hydrogen Bond of a Water between the Schiff Base and Asp75. Biochemistry. 2005;44:12287–12296. doi: 10.1021/bi050841o. [DOI] [PubMed] [Google Scholar]
- 34.Furutani Y, Bezerra AG, Jr, Waschuk S, Sumii M, Brown LS, Kandori H. FTIR Spectroscopy of the K Photointermediate of Neurospora Rhodopsin: Structural Changes of the Retinal, Protein, and Water Molecules after Photoisomerization. Biochemistry. 2004;43:9636–9646. doi: 10.1021/bi049158c. [DOI] [PubMed] [Google Scholar]
- 35.Furutani Y, Shichida Y, Kandori H. Structural Changes of Water Molecules during the Photoactivation Processes in Bovine Rhodopsin. Biochemistry. 2003;42:9619–9625. doi: 10.1021/bi034592k. [DOI] [PubMed] [Google Scholar]
- 36.Shibata M, Ihara K, Kandori H. Hydrogen-Bonding Interaction of the Protonated Schiff Base with Halides in a Chloride-Pumping Bacteriorhodopsin Mutant. Biochemistry. 2006;45:10633–10640. doi: 10.1021/bi060555s. [DOI] [PubMed] [Google Scholar]
- 37.Shibata M, Yoshitsugu M, Mizuide N, Ihara K, Kandori H. Halide Binding by the D212N Mutant of Bacteriorhodopsin Affects Hydrogen Bonding of Water in the Active Site. Biochemistry. 2007;46:7525–7535. doi: 10.1021/bi7004224. [DOI] [PubMed] [Google Scholar]
- 38.Furutani Y, Sudo Y, Kamo N, Kandori H. FTIR Spectroscopy of the Complex between pharaonis Phoborhodopsin and Its Transducer Protein. Biochemistry. 2003;42:4837–4842. doi: 10.1021/bi034317y. [DOI] [PubMed] [Google Scholar]
- 39.Sharma AK, Spudich JL, Doolittle WF. Microbial rhodopsins: Functional versatility and genetic mobility. Trends Microbiol. 2006;14:463–469. doi: 10.1016/j.tim.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Cooper A. Energy uptake in the first step of visual excitation. Nature. 1979;282:531–533. doi: 10.1038/282531a0. [DOI] [PubMed] [Google Scholar]
- 41.Birge RR, Cooper TM. Energy Storage in the Primary Step of the Photocycle of Bacteriorhodopsin. Biophys J. 1983;42:61–69. doi: 10.1016/S0006-3495(83)84369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi S, Tajkhorshid E, Schulten K. Structural Changes during the Formation of Early Intermediates in the Bacteriorhodopsin Photocycle. Biophys J. 2002;83:1281–1297. doi: 10.1016/S0006-3495(02)73900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayashi S, Tajkhorshid E, Kandori H, Schulten K. Role of Hydrogen-Bond Network in Energy Storage of Bacteriorhodopsin’s Light-Driven Proton Pump Revealed by ab Initio Normal-Mode Analysis. J Am Chem Soc. 2004;126:10516–10517. doi: 10.1021/ja047506s. [DOI] [PubMed] [Google Scholar]
- 44.Losi A, Wegener AA, Engelhard M, Gartner W, Braslavsky SE. Time-Resolved Absorption and Photo-thermal Measurements with Recombinant Sensory Rhodopsin II from Natronobacterium pharaonis. Biophys J. 1999;77:3277–3286. doi: 10.1016/S0006-3495(99)77158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang D, Mauzerall D. Volume and Enthalpy Changes in the Early Steps of Bacteriorhodopsin Photocycle Studied by Time-Resoleved Photoacoustics. Biophys J. 1996;71:381–388. doi: 10.1016/S0006-3495(96)79235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.