Abstract
Purpose
Malignant astrocytomas (MAs) are aggressive central nervous system tumors with poor prognosis. Activating mutation of BRAF (BRAFV600E) has been reported in a subset of these tumors, especially in children. We have investigated the incidence of BRAFV600E in additional pediatric patient cohorts, and examined the effects of BRAF blockade in preclinical models of BRAFV600E and wild-type BRAF MA.
Experimental Design
BRAFV600E mutation status was examined in two pediatric MA patient cohorts. For functional studies, BRAFV600E MA cell lines were used to investigate the effects of BRAF shRNA knockdown in vitro, and to investigate BRAF pharmacologic inhibition, in vitro and in vivo.
Results
BRAFV600E mutations were identified in 11 and 10 percent of MAs from two distinct series of tumors (6 of 58 cases total). BRAF was expressed in all MA cell lines examined, among which BRAFV600E was identified in four instances. Using the BRAFV600E specific inhibitor PLX4720, pharmacologic blockade of BRAF revealed preferential anti-proliferative activity against BRAFV600E mutant cells in vitro, in contrast to the use of shRNA-mediated knockdown of BRAF, which inhibited cell growth of glioma cell lines regardless of BRAF mutation status. Using orthotopic MA xenografts, we demonstrate that PLX4720 treatment decreases tumor growth and increases overall survival in mice bearing BRAFV600E mutant xenografts, while being ineffective, and possibly tumor promoting, against xenografts with wild-type BRAF.
Conclusions
Our results indicate a 10% incidence of activating BRAFV600E among pediatric MAs. With regard to implications for therapy, our results support evaluation of BRAFV600E specific inhibitors for treating BRAFV600E MA patients.
Keywords: BRAF, V600E, in vivo, malignant astrocytoma, targeted therapy
Introduction
Malignant astrocytomas (MAs) are aggressive brain tumors that affect people of all ages. Current treatments are inadequate, with median survival being 14 months in adults (1). A recently published study involving the use of radiation and temozolomide in treating 90 pediatric patients with malignant astrocytoma revealed only 22% survival at 3 years (2). While the genomics of adult grade IV MA (i.e., glioblastoma: GBM) have been researched extensively, and in some cases comprehensively (3, 4), pediatric studies have been more limited. For the most part, our current understanding of the genetic etiology of pediatric MAs is based on the examination of selected genes most commonly altered in corresponding adult tumors. The results of such studies indicate that the incidence of two of the most common gene alterations in adult MAs, epidermal growth factor receptor (EGFR) amplification and PTEN inactivation, are significantly reduced in pediatric MAs (5,6). In contrast, other genetic alterations that have been linked with the pathogenesis of adult MA, such as those resulting in TP53 and CDKN2A inactivation, occur at significant frequencies in pediatric MAs as well (7–9).
The receptor tyrosine kinase (RTK)-RAS-RAF-MEK-ERK signaling pathway relays extracellular signals from cell membrane-based RTKs to the nucleus via a series of consecutive phosphorylation events (10,11). RTK-RAS-RAF-MEK-ERK signaling plays an important role in the pathogenesis of adult MAs (12), and increasing evidence supports the importance of this pathway in the development of pediatric MAs as well (13–15). Activation of the RTK-RAS-RAF-MEK-ERK signaling in adult MA is usually associated with abnormal signaling of upstream RTKs, such as EGFR and Platelet Derived Growth Factor Receptor (PDGFR) (3). Inactivation of the NF1 tumor suppressor gene, which encodes a RAS-GTPase, also leads to the activation of this pathway in adult MA (3, 16). Oncogenic mutation of other RTK-RAS-RAF-MEK-ERK signaling components, such as K-RAS, N-RAS or BRAF, which commonly occur in a wide variety of human cancers, is infrequent in adult MA (17).
Recent publications suggest that RAF gene alterations occur at a higher frequency in pediatric astrocytomas, including pilocytic astrocytomas, pleomorphic xanthoastrocytomas, and MAs (13, 18). There are three RAF family proteins: A-, B-, and CRAF (RAF-1). In rodent brain, ARAF is rarely expressed, whereas both BRAF and CRAF are expressed in normal CNS tissue (19). ARAF has the lowest intrinsic kinase activity, followed by CRAF, with BRAF having highest intrinsic activity (20). All three RAF isoforms share RAS as a common activator, and MEK as a common substrate (21). With respect to the genes encoding these proteins, BRAF T1799A (BRAFV600E) is the most common activating mutation, occurring in a broad spectrum of human tumors (22). BRAFV600E is constitutively active, requires neither interaction with RAS nor dimer formation in order to signal, and is refractory to negative feedback inhibition (23). A recent report from our group describes activation of MAP kinase signaling in nearly all pediatric MAs, some of which have associated BRAFV600E (13).
The discovery of activating BRAF mutation in pediatric MA’s provides a unique opportunity to improve treatment outcomes for a subset of patients with this devastating disease. Small molecule kinase inhibitors that specifically target BRAFV600E have recently been developed and shown remarkable efficacy against melanomas that harbor this mutation (24). A recent phase I study using BRAFV600E specific inhibitor PLX4032 showed a response rate of 81% in a group of 48 patients with BRAFV600E positive metastatic melanoma (25).
In the present study, we confirm the presence of BRAFV600E mutation in two additional cohorts of pediatric MA. To investigate the importance of BRAFV600E to MA growth, BRAF expression was suppressed in multiple MA cell lines by shRNA knockdown, with resultant determination that reduced levels of BRAF decreases ERK phosphorylation and results in decreased cell growth irrespective of tumor cell BRAFV600E status. In contrast, a BRAF pharmacologic inhibitor shows BRAFV600E dependency with regard to in vitro and in vivo MA anti-proliferative effects.
Materials and Methods
Cell lines, xenografts, and primary tumors
MA cell lines (Fig. 1) were obtained from the American Type Culture Collection, DSMZ – the German Resource Centre for Biological Material, and the Japan Health Sciences Foundation Health Science Research Resources bank. Normal human astrocytes (NHAs) were obtained from Clonetics and AllCells. All cell sources were authenticated through DNA fingerprinting using the Promega Powerplex platform.
Figure 1.
BRAF, CRAF, and downstream signaling mediator activation in MA cell lines. A. Cell lysates from 20 human MA cell lines were examined by Western Blot using antibodies against the indicated proteins. Cell lines harboring mutant BRAF are indicated by the dotted line. BRAF protein signals were normalized against corresponding α-TUBULIN signals, with ratios expressed in relation to a normal human astrocyte (Clonetics) value of 100. A second NHA cell source (AllCells) was determined as expressing nearly identical BRAF as the Clonetics NHAs that were used for establishing MA cell line BRAF expression levels.
Patient tissues from Royal Marsden Hospital, Sutton, and Newcastle Royal Infirmary, UK, were obtained after approval by Local and Multicenter Ethical Review Committees. Tumor DNAs were extracted from formalin fixed and paraffin embedded (FFPE) tissues and whole genome amplified, as described previously (15). For the St. Jude tumors, sections from FFPE tissue were reviewed by two neuropathologists for identification of specimens suitable for DNA extraction from corresponding snap-frozen specimens, as previously described (26).
BRAF sequencing
The BRAF V600E hotspot region was amplified using primers BRAF_Ex_15_FFPE_F (TTCATGAAGACCTCACAGTAAAAA) and BRAF_Ex_15_FFPE_R (CCACAAAATGGATCCAGACA), which are designed to yield correct target sequence product by touchdown PCR. DNA was sequenced in both directions using the di-deoxy chain termination method on an ABI 3730 DNA Sequencer. Results were examined using the Applied Biosystems Sequence Scanner Software v1.0.
Western Blot analysis
Cells and tissues were lysed in cell lysis buffer (Cell Signaling) supplemented with proteinase (Roche) and phosphatase (Sigma) inhibitor cocktails. Lysates were separated by SDS-PAGE and transferred to PVDF membranes. After probing with primary antibodies, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody, and visualized by ECL (Pierce). Antibodies specific for total ERK, p-ERK, total MEK1/2, p-MEK1/2, total AKT, p-AKT-Ser473, CYCLIN D1, CYCLIN D3, CDK4 and CDK6, and p27KIP1, were obtained from Cell Signaling Technologies. Antibody specific for BRAF was from Santa Cruz Biotechnology, CRAF from Upstate Biotechnology, and α-TUBULIN from Neomarkers (Fremont, CA).
Lentivirus mediated BRAF shRNA expression
Constructs for stable suppression of BRAF expression were obtained from The RNAi Consortium via Open Biosystems (Huntsville, AL). A total of 12 constructs were obtained and tested to identify those able to achieve efficient BRAF protein knockdown. Negative control constructs in the same vector system (vector alone and scrambled shRNA) were obtained from Addgene (Cambridge, MA). The lentiviral helper plasmids pHR'8.2 R and pCMV-VSV-G were also obtained from Addgene. The integrity of all plasmids was confirmed by restriction analysis, and the integrity of all shRNA inserts was confirmed by sequencing. Lentivirus preparation and infection of GBM cell lines have been described previously (27).
Cell growth assay
Stably infected GBM cells were plated into 96-well plates at a density of 100–500 cells/well and the medium was changed every three days. After selected incubation times, cell viabilities were determined with ‘CellTiter Glo Kit’ from Promega and/or ‘Cell Counting Kit-8’ from Dojindo Molecular Technologies. All cell growth analyses were conducted with 4–8 replicates.
Cell cycle analysis
Cell cycle distributions were determined by flow cytometric analysis following staining with propidium iodide. In brief, exponentially growing cells were harvested and fixed in 70% ethanol, then stained in PBS-buffered saline containing 0.1% Triton X-100, 50 µg/ml RNase, and 50 µg/ml propidium iodide, for 60 min at room temperature. The fluorescence was measured on a FACSort flow cytometer (Becton Dickinson, Franklin Lakes, NJ), and data were analyzed using ModFit software (Verity Software House, Topsham, ME). Small molecule inhibitors used in association with cell cycle analysis were PLX4720 (Plexxikon Inc., Berkeley, CA), Sorafenib, and GDC-0941, with the latter two obtained from Selleck Chemicals (Houston, TX).
Modification of tumor cells with firefly luciferase reporter
Lentiviral vectors containing firefly luciferase (Fluc) were generated as previously described (28). AM-38 human MA cells were transduced with Fluc lentivirus as previously described for U87 cells (29). Cells were screened in vitro for transduction efficiency by treatment with luciferin (D-luciferin potassium salt, 150 mg/kg, Gold Biotechnology, St Louis, MO), and analysis for luminescence using a Xenogen IVIS Lumina System (Xenogen Corp., Alameda, CA).
In Vivo Experiments
Five-week-old female athymic mice (nu/nu genotype, BALB/c background) were purchased from Simonsen Laboratories (Gilroy, CA). Animals were housed under aseptic conditions. The UCSF Institutional Animal Care and Use Committee approved all animal protocols. Tumor cells were implanted into the brains of athymic mice as previously described (30). In vivo bioluminescence imaging (BLI) was performed using the Xenogen IVIS Lumina System coupled to LivingImage data-acquisition software (Xenogen Corp.). Mice were anesthetized with 100 mg/kg of ketamine and 10 mg/kg of xylazine and imaged 10 minutes after intraperitoneal injection of luciferin. Signal intensity was quantified using LivingImage software.
To evaluate the therapeutic response of intracranial glioma xenografts, mice implanted with luciferase-modified MA cells were randomized to vehicle control (DMSO) or PLX4720 treatment groups. Mice receiving PLX4720 treatments were administered a daily dose of 20 mg/kg by intraperitoneal injection for 14 consecutive days. Treatment was initiated at day 7 post implantation of tumor cells. All mice were monitored every day for the development of symptoms related to tumor burden, and 1–2× weekly by BLI. Mice were euthanized when they exhibited symptoms indicative of significant compromise to neurologic function. The Kaplan–Meier estimator was used to generate survival curves, and differences between survival curves were calculated using a log-rank test. In addition to the mice used for the survival analysis, two pre-symptomatic mice within each cohort were sacrificed two hours after their seventh treatment, with their brains resected and either placed in formalin and prepared for immunohistochemical analysis, or dissected from surrounding normal brain and snap frozen for immunoblot analysis. IHC was performed as previously described (30), using Ki-67 antibody from Ventana Inc.
Results
BRAFV600E Mutation in Malignant Astrocytomas
In follow-up to our previous finding of BRAFV600E mutation in pediatric MAs (13), BRAFV600E status was determined in two additional tumor cohorts. As shown in Table 1, BRAFV600E mutation was found in three out of 28 tumors in the ICR cohort, and three out of thirty tumors in the cohort from St Jude. Interestingly, all BRAFV600E mutations in these series were identified in MAs of grade IV malignancy (i.e., GBM). Our previously published study (13) combined with results in the current report indicate a BRAFV600E mutation frequency of 14% (11 of 78) in grade III + grade IV pediatric MAs. As in our previously report (13), BRAFV600E mutations and PDGFRA amplifications were found as mutually exclusive in the additional cohorts being reported here (supplemental Tables 1s, 2s). For the combined 78 MA’s in 3 series of pediatric tumors, 9 instances of PDGFRA amplification were determined (12%: Tables 1s–3s).
Table 1. BRAFV600E and PDGFRA Status of Pediatric MAs.
Thirty patients from St Jude Children Research Hospital and 28 patients from the Institute of Cancer Research were screened for the presence of BRAFV600E and PDGFRA amplifications. Diagnosis, age at diagnosis, and PDGFRA amplification status are also shown.
| St Jude's Cohort (30 patients) |
BRAFV600E | Wildtype BRAF | All |
|---|---|---|---|
| Diagnosis- AA | 0 | 8 | 8 |
| Diagnosis-GBM | 3 | 19 | 22 |
| Median Age at Diagnosis (months) |
165.2 (128–198) | 111 (9–213) | 117 (9–213) |
| PDGFRA Amplification | 0 | 4 | 4 |
| Institute of Cancer Research Cohort (28 patients) |
BRAFV600E | Wildtype BRAF | All |
|---|---|---|---|
| Diagnosis- AA | 0 | 6 | 6 |
| Diagnosis-GBM | 3 | 19 | 22 |
| Median Age at Diagnosis (months) |
146 (79–240) | 137 (5–272) | 138 (5–272) |
| PDGFRA Amplification | 0 | 3 | 3 |
AA = Anaplastic Astrocytoma; GBM = Glioblastoma. For individual patient data, refer to supplemental Tables 1 & 2.
To confirm sustainable tumor cell sources for BRAFV600E associated investigation, we examined 20 MA cell lines, 15 of which had been previously analyzed for BRAFV600E (22), for mutant sequence (supplemental Table 4s). This analysis confirmed V600E mutation in cell lines DBTRG-05MG, AM-38, NMC-G1, and KG-1-C. Three of 4 cell line mutations are heterozygous for BRAFV600E (supplemental Fig. 1s), with BRAFV600E homozygosity evident in AM-38 cells. Previously published data (22) combined with our results indicate a 10% incidence (4 of 41) of BRAFV600E in MA cell lines.
BRAF copy number and expression in MA cell lines and pediatric tumors
We examined the extent of variation in BRAF protein expression among MA cell lines, and in relation to BRAF expression in normal human astrocytes (NHA). Modest increases in BRAF protein were evident in 16 (80%) tumor cell lines relative to NHA (Figure 1). Only one cell line, 42MGBA, expressed appreciably less BRAF than NHA. Analysis of normalized BRAF expression values indicates that BRAFV600E status is not a significant determinant of tumor BRAF expression (supplemental Figure 2s, panel A), nor of BRAF copy number (supplemental Table 4s), for MA cell lines. Examination of BRAF mRNA expression in pediatric MAs (UCSF tumor series: see supplemental Table 3s) also indicates a lack of association between BRAFV600E status and BRAF expression (supplemental Fig. 2s, panel B), as well as between BRAFV600E and BRAF copy number (Table 3s).
We also assessed CRAF, p-ERK, and p-MEK levels in the MA cell lines, with results showing more variability in CRAF expression than was evident for BRAF (Fig. 1). Phosphorylation of ERK was readily detectable in all of the MA cell lines except 2 (8MGBA and 42MGBA: Fig. 1). Inspection of the phospho-ERK levels indicates no apparent association with either BRAF expression level or BRAFV600E status. In contrast, a clear association is indicated between BRAFV600E status and phospho MEK levels (Fig. 1).
Inhibition of ERK phosphorylation and in vitro growth of MA cell lines by BRAF shRNA
Lentivirus expressing BRAF shRNA were screened for ability to suppress BRAF protein expression. Construct 6289 was the most effective in this regard (Fig. 2A), and was used to develop isogenic derivatives from four BRAFV600E and four wild-type BRAF cell lines. Western blot results show that decreased BRAF protein is associated with suppression of ERK phosphorylation, as well as with inhibition of growth for all cell lines tested (Fig. 2A,B for representative results; additional results shown in supplemental Fig. 3s).
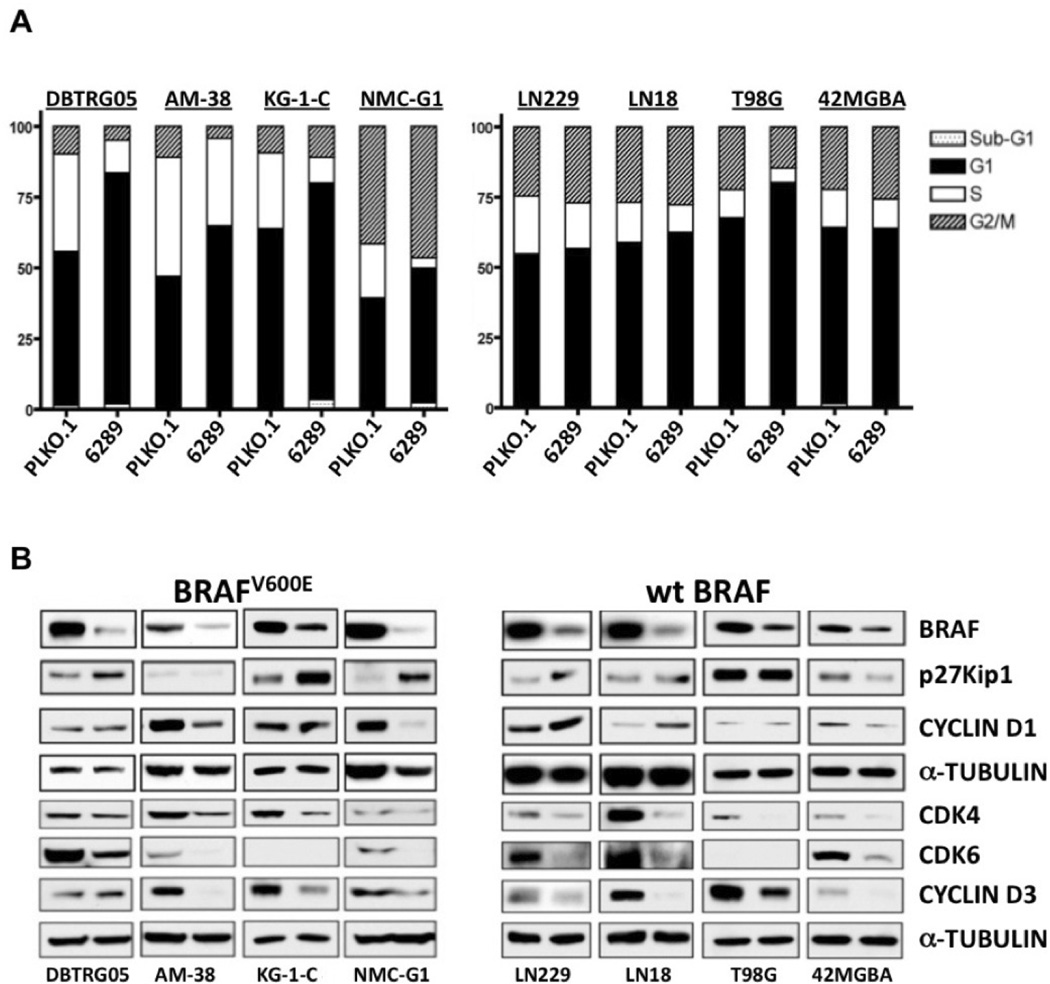
Figure 2.
shRNA suppression of BRAF expression inhibits ERK phosphorylation and in vitro growth of MA cell lines. A. DBTRG-05MG cells were infected with lentivirus expressing empty vector (PLKO.1), or with each of 4 BRAF shRNA lentivirus. Results show that lentivirus 6289 is the most effective at suppressing BRAF expression, as well as being the most effective in suppressing phosphorylation of ERK. B. In vitro growth analysis of modified DBTRG-05MG cells (left) shows decreased growth rate of cells transduced with 6289 lentivirus. Control and 6289 shRNA derivatives for wild-type BRAF cell line 42MGBA were similarly examined for effects of BRAF knockdown on p-ERK (A, right panel) and cell proliferation (B, right panel).
shRNA-mediated knockdown of BRAF results in G1 phase arrest and altered expression of cell cycle regulatory proteins
The anti-proliferative effects of BRAF suppression were further investigated by comparison of cell cycle distributions for BRAF shRNA and control shRNA cells. In each of the four BRAFV600E cell lines, BRAF knockdown increased G1 phase fractions, primarily at the expense of cells in S phase (Fig. 3A). A lesser extent of G1 increase from BRAF shRNA knockdown was evident in T98G cells, whereas shRNA knockdown resulted in little or no change in G1 fraction for the 3 additional wild-type BRAF cell lines. Although initial inspection of the cell cycle distributions could be interpreted as indicating that BRAF shRNA knockdown has little effect on the sub-G1 fraction of all MA cell lines, substantially higher sub-G1 increases were, in fact, evident in two of the four V600E cell lines (supplemental Figure 4s).
Fig 3.
Effect of shRNA mediated BRAF suppression on MA cell cycle distributions and expression of cell cycle regulatory proteins. A. MA cell lines with (left) or without (right) BRAFV600E mutation were infected with lentivirus expressing empty control (PLKO.1) or BRAF shRNA (6289), then treated with puromycin to select infected cells. Pooled puromycin resistant cells were subjected to flow cytometry to determine cell cycle distributions. For all cell lines with BRAFV600E, suppression of BRAF expression resulted in increased proportion of G1 phase cells. For 3 of th 4 cell lines with wild-type BRAF, shRNA suppression had little effect on the G1 fraction of cells. B. Lysates from the same transduced cells as in “A” were subjected to Western Blot analysis to assess the effects of BRAF knockdown on the expression of proteins with known roles in regulating G1 phase cell cycle transit: CYCLIN D1 and CYCLIN D3, enzymatic subunits CDK4 and CDK6, and cyclin dependent kinase inhibitor p27Kip1. The left lane in each blot results from control shRNA and the right lane result is from use of the BRAF shRNA lentivirus. Comparison of results for any two cell lines shows that BRAF suppression effects are variable for the five G1 regulatory proteins examined, but are consistent in demonstrating that BRAF suppression results in decreased expression of one or more of the proteins that promote G1 transit, and/or increases the expression of the negative G1 transit regulator p27Kip1.
The RTK-RAS-RAF-MEK-ERK signaling pathway activates several key cell cycle regulatory proteins to promote G1 to S transition (10). In particular, ERK activation has been shown to induce cell proliferation by increasing CYCLIN D1 and decreasing p27KIP1 levels in various cell types (31–34). Western blot results for the 4 BRAFV600E cell lines showed that BRAF knockdown increased p27KIP1 or decreased CYCLIN D1, or both (Fig. 3B). For the MA cell lines with wild-type BRAF, shRNA suppression resulted in decreased CYCLIN D1 and increased p27KIP1 in 42MGBA and LN229, respectively. Investigation of additional cell cycle regulatory proteins revealed that BRAF knockdown significantly decreased CYCLIN D3 protein in all MA cell lines except DBTRG-05MG (Fig. 3B). Protein levels of CDK4 or CDK6, or both, were also reduced by BRAF shRNA in all of the cell lines.
PLX4720 suppresses MEK-ERK phosphorylation and cell proliferation in MA cells containing BRAFV600E mutation
As a complementary approach to investigating the suppression of BRAF expression in MA cells, we examined anti-proliferative effects of the small molecule inhibitor PLX4720, a tool compound that is closely related to the PLX4032 inhibitor currently being evaluated in clinical trials against melanoma harboring BRAFV600E (25). Dose-response curves for AM-38 and DBTRG05-MG (both BRAFV600E), and 42MGBA (wild-type BRAF) show that PLX4720 inhibits cell growth irrespective of cell line V600E status (Fig. 4A), but that the two lines with BRAFV600E are sensitive to much lower concentrations of the inhibitor: the EC50 for DBTRG-05MG and AM-38 is 1.75 and 6.19 µM respectively, whereas the EC50 of 42MGBA is 31.20 µM. Corresponding western blot results (Fig. 4B) show that PLX4720 inhibits phosphorylation of MEK and ERK in a dose dependent as well as a BRAFV600E dependent manner. In DBTRG-05MG and AM-38 cells, 2 µM PLX4720 decreased EGF-induced MEK and ERK phosphorylation, whereas 50 µM PLX4720 was needed to inhibit EGF induced MEK/ERK phosphorylation in 42MGBA cells. Importantly, the lowest dose of PLX4720 (2 µM) led to an activation of MEK/ERK signaling in 42MGBA, likely secondary to paradoxical activation of wild-type BRAF, as previously described (35–37).
Fig 4.
Effects of BRAF pharmacologic inhibition on cell proliferation, and on EGF stimulated signal transduction. A. DBTRG-05MG and AM-38, which harbor BRAFV600E, and 42MGBA, with wild-type BRAF, were incubated with increasing concentrations of PLX4720, with effects on growth inhibition indicated after 3 days treatment. EC50 values for DBTRG-05MG and AM-38 are 1.75 and 6.19 µM, respectively, whereas the EC50 for 42MGBA is much higher: 31.20 µM. B. Cultures of DBTRG-05MG, AM-38, and 42MGBA were incubated with serum free medium for 4 hours, then treated with the indicated concentrations of PLX4720 for 1 hour, followed by the addition of 10 ng/ml EGF. Ten min after the addition of EGF, cell lysates were harvested and analyzed by Western blot analysis using the indicated antibodies. Note that a negative effect on p-MEK and p-ERK is evident at 2 and 10 uM PLX4720 for BRAFV600E cells, but not for wild-type BRAF 42MGBA cells. C. GBM cells cultured in the presence of DMSO (0.1%), or 2 or 10 uM PLX4720, for 20 hours, and then harvested for cell cycle analysis with flow cytometry.
Analysis of cell cycle distribution effects of 2 µM PLX4720 treatment revealed increased G1 fraction and reduced S phase component in BRAFV600E cells (Fig. 4C), with the opposite response observed in wild-type BRAF cells: i.e., decreased G1 and increased S phase cell components for 42MGBA. At 10 µM PLX4720 treatment, the G1 and S phase responses of 42MGBA were reversed relative to 2 µM responses, and despite indication of residual p-MEK/p-ERK (Fig. 4B). This reversal in effect is potentially associated with inhibitor off-target effects, as suggested by decreases in 42MGBA p-AKT at 10 and 50 µM PLX4720 treatments (Fig. 4B).
Because 42MGBA has reduced expression of BRAF compared to NHA one might predict these cells to be less representative of wild-type BRAF MA cell response to BRAF inhibition. To address whether the lack of effect of PLX4720 on wild-type BRAF glioma cells was unique to 42MGBA, a second wild-type BRAF cell line, U87, was also tested for PLX4720, with similar results obtained as for 42MGBA: i.e., low dose PLX4720 treatment results in paradoxical activation of p-MEK and p-ERK, with corresponding decreases and increases in U87 G1 and S phase components, respectively (supplemental Fig. 5s). Importantly, PLX4720 signaling pathway response for both wild-type BRAF (U87) and BRAFV600E (AM-38) cells were reproducible in serum-supplemented media (supplemental Fig. 6s).
Interestingly, 2 and 10 µM PLX4720 resulted in little change to the subG1 component of treated BRAFV600E cells (Fig. 4C). To address whether this lack of response was due to alternative signaling pathway protection from apoptosis, we examined the effect of concurrent BRAF + PI3K/mTOR inhibition. As shown in Figure 7s, combined PI3K/mTOR and BRAF blockade leads to a further increase in G1 arrest, but no significant increase in subG1 component. This result suggests that MA cells primarily depend on the PI3K/mTOR and BRAF pathways for proliferation.
To address whether observed cell line responses to RAF inhibition were specific to treatment with PLX4720, we additionally tested wild-type BRAF and BRAFV600E cells with sorafenib, a pan-RAF inhibitor. As shown in supplemental Figure 8s, whereas sorafenib induces a dose dependent decrease in p-MEK and p-ERK in both AM-38 and U87 cells, these signaling pathway changes occur without substantial change to the cell cycle distributions of either type of cell. Thus, the anti-proliferative effect of BRAF inhibition is specific to the use of the BRAFV600E selective-inhibitor PLX4720 against BRAFV600E cell lines.
PLX4720 represses growth of intracranial BRAFV600E MA xenografts
To investigate the effects of BRAF pharmacologic inhibition in vivo, we utilized an orthotopic xenograft model approach, in which tumor cells, modified with a luciferase reporter, are monitored for response to treatment, following intracranial injection in athymic mice. Mice with intracranial AM-38 showed reduced intracranial tumor growth from PLX4720 treatment, relative to corresponding vehicle-treated mice, resulting in significantly prolonged survival (Fig. 5A). The survival of mice with intracranial DBTRG-05MG was also significantly extended from treatment with PLX4720 (data not shown). AM-38 tumors in mouse brain, following one week of PLX4720 treatment, showed decreased Ki-67 staining, relative to tumor from a vehicle treated mouse (supplemental Fig. 9s panels A and B), corroborating the in vivo anti-proliferative treatment effect indicated by bioluminescence imaging. Western blot analysis of protein extract from AM-38 intracranial xenografts revealed that PLX4720 therapy decreases p-ERK (supplemental Fig. 9s, panel C), consistent with in vitro results (Fig. 4).
Figure 5.
Effect of PLX4720 on tumor growth and survival for mice with intracranial MA xenografts. A) Mice received intracranial injection of 300,000 luciferase modified BRAFV600E AM-38 cells, with intraperitoneal administration of vehicle (DMSO) or PLX4720 (20 mg/kg) daily for 2 weeks, beginning at day 7. Survival plot (left) shows that PLX4720 treatment significantly extends the survival of mice with intracranial BRAFV600E tumors, consistent with results from quantitative BLI which show significant anti-proliferative effect of PLX4720 at the first imaging time point subsequent to initiation of treatment (p = 0.018 for control vs. PLX4720 BLI values: middle panel). Bioluminescence images (right) of intracranial tumor in control and PLX4720 treatment group mice show lesser AM-38 bioluminescence signal in the mouse administered PLX4720 following the 5th day of PLX4720 treatment (mice shown had median treatment group luminescence values at time of initiation of therapy). B) In contrast, no PLX4720 survival advantage nor anti-proliferative effect is evident for mice with wild-type BRAF intracranial xenografts (U87).
To determine whether PLX4720 shows activity against wild-type BRAF xenografts, we established intracranial tumors with U87 cells that had been previously modified with luciferase lentivirus and used for investigation of response to cdk4/6 inhibition (29). PLX4720-treated mice bearing wild-type BRAF U87 xenografts showed no delayed tumor growth or survival advantage, consistent with in vitro results of U87 which indicated low sensitivity to PLX4720 (U87 EC50 of 48.8 µM, data not shown). In fact, results from bioluminescence monitoring indicated increased mean U87 xenograft growth rate, as well as reduced survival from PLX4720 treatment (Fig. 5B). Results from Ki-67 staining of PLX4720-treated U87 tumors showed consistency with bioluminescence imaging results: Ki-67 positive cells were more abundant in PLX4720 treated tumor (supplemental Fig. 9s, panels D and E).
Discussion
Patients with malignant astrocytomas have a poor prognosis. Despite aggressive therapy, relapse rates are high, with median survival being 14 months in adults (1), and the majority of pediatric MA patients succumbing to cancer within 3 years of diagnosis (2). There is a relative dearth of information regarding recurrent molecular characteristics in pediatric MAs that can be used for therapeutic hypothesis testing. PDGFRA amplification, though noted in a fraction of these tumors (13, 14, 38), has yet to be successfully exploited for improved MA patient outcomes from the use of PDGF receptor inhibitors (39). This is also the case for EGFR amplification in adult MA’s, for which the use of EGF receptor small molecule inhibitors has yet to confer a clear survival benefit (40, 41). The lack of success in targeting EGF and PDGF receptors in treating brain tumor patients is perhaps a result of receptor tyrosine kinase functional redundancy that compensates for the blockade of just one type of RTK (42). In principle, pathway blockade downstream of RTKs may be more successful, since numerous RTKs converge on a few key signaling nodes in promoting cell proliferation. In this study, we found that BRAF may represent such a point of signal convergence, since the growth of all tested MA cell lines was inhibited by suppression of BRAF expression (Fig. 2).
Although gain of function mutation of KRAS, NRAS, and BRAF are among the most frequently occurring oncogene alterations in cancer, such mutations are uncommon in adult MAs (17, 18). In spite of the low mutation incidence, RTK-RAS-RAF-MEK-ERK signaling is upregulated in most of these tumors, and high ERK activation correlates with poor prognosis for MA patients (43). In this report, we confirm the finding of BRAF activating mutations (BRAFV600E) in a subset of pediatric MAs. BRAFV600E was identified in 6 of 58 (10%) pediatric MAs, from two distinct series of tumors, as well as in 4 MA cell lines. In vitro, BRAFV600E mutation was associated with elevated p-MEK levels. Because BRAF is located on chromosome 7 (q35), and gain of chromosome 7 is frequently detected in MAs, BRAF copy number is commonly increased in these tumors (44). We also found copy number gains of BRAF in most MA cell lines and primary tumors, although no focal or high-level BRAF amplifications were evident (Tables 3s, 4s). In addition to BRAF copy number increases and mutation, we found elevated expression of BRAF protein, relative to normal human astrocytes, in a majority of MA cell lines (Fig. 1).
Our results show that inhibition of BRAF expression by shRNA knockdown leads to decreased p-ERK levels as well as reduced proliferation of MA cell lines, independent of BRAFV600E status (Fig. 2; supplemental Figure 3s). However, inspection of flow cytometry results from BRAF shRNA modified cells (Fig. 3A), for cell cycle effects of BRAF shRNA, reveals a more rapid anti-proliferative effect of BRAF knockdown on BRAFV600E cells, suggesting increased cellular proliferation dependence on, and addiction to BRAF signaling for BRAFV600E cells. This dependence is further indicated by results from BRAF pharmacologic inhibition with PLX4720, which show specificity of anti-proliferative effect for tumor cell lines and xenografts with V600E (Figs. 4 and 5). PLX4720 BRAF blockade resulted in MAPK pathway inhibition, decreased growth rate, and increased G1 arrest in cell lines with BRAFV600E, whereas BRAF wild-type cells, when treated with low doses of PLX4720, show increased MAPK activation and a decrease in G1 phase cells. Observation of a stimulatory effect of BRAF inhibition on MAPK activation in cells with wild-type BRAF has been reported by others (35–37). It is important to note that BRAFV600E inhibition did not promote substantial apoptotic response of BRAF-mutant cells, and that combined BRAF + PI3K/mTOR blockade led only to further G1 arrest, without increased subG1 component (Fig. 7s). These data suggest that MA cells primarily depend on the PI3K/mTOR and BRAF pathways for proliferation, and not so much for survival.
To extend in vitro observations to the in vivo setting, we generated intracranial xenografts using MA cell lines with and without BRAFV600E. Our results (Fig. 5) demonstrate that BRAF inhibition by PLX4720 significantly decreases tumor growth rate (as indicated by quantitative BLI), with corresponding increase in survival for mice carrying BRAF-mutant xenografts, and to an extent showing reasonable consistency with length of treatment. Experiments are ongoing to determine maximal extent of PLX4720 survival benefit, given sustained daily treatment, and using multiple MA models. Molecular analysis of PLX4720 treated BRAFV600E tumors shows decreased p-ERK in vivo (supplemental Fig. 9C), as well as decreased Ki-67 staining (supplemental Fig. 9A, 9B). These results are consistent with the decreased tumor cell proliferation indicated by BLI monitoring. In contrast, U87 xenografts, harboring wild-type BRAF, did not respond to PLX4720, and, in fact, showed a trend towards increased tumor growth rate and decreased survival (Fig. 5B, supplemental Fig. 9D, 9E).
BRAFV600E is now a validated therapeutic target in metastatic melanoma, with high response rates reported using both the Plexxikon BRAFV600E specific inhibitor PLX4032 (25) and the Glaxo Smith Kline inhibitor GSK2118436 (45). Phase III studies suggest a survival advantage for patients treated with PLX4032, when compared to standard chemotherapy (46). Taken together with the results reported here, there is a highly supportive rationale for using BRAF specific pharmacological inhibition in treating MAs with BRAFV600E.
Supplementary Material
Statement of Translational Relevance.
Malignant astrocytoma (MA) is the most common histopathologic subclassification for primary central nervous system cancer, and the outcome for patients with MA is dismal. In the current study we demonstrate a 10% incidence of BRAFV600E in pediatric MAs, and demonstrate that a BRAF small molecule inhibitor has substantial activity against intracranial xenografts established from BRAFV600E MA cells, while being ineffective against intracranial xenografts established from wild-type BRAF MA cells. This finding suggests that patients with BRAFV600E MA can be effectively treated with BRAF-specific small molecule inhibitors. Because there is a clinically approved analogue for the BRAF inhibitor we have used in our studies, we anticipate these results to have immediate impact with regard to stimulating clinical trial evaluation of BRAF small molecule inhibitors for treating patients with BRAFV600E MA.
Acknowledgements
We thank David Ellison (St Jude) and Sabine Mueller (UCSF) for help in compiling clinical data, and Brian West and Gideon Bollag at Plexxikon Inc. for providing drug and technical assistance. This study was supported by grants from the Pediatric Brain Tumor Foundation (DAR, WAW, CDJ), McDonnell Foundation (CDJ, DAR), NCI grants 5P50CA097257-10 (WAW, CDJ) and CA096832 (SJB), the Rally Foundation for Childhood Cancer Research (TN), the Campini Foundation (TN), NINDS grant K08NS065268 (TN), and the NIHR Biomedical Research Centre (CJ).
Footnotes
We declare no conflict of interest (or relationship that would be suspected of constituting conflicts) at the time of submission.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Cohen KJ, Heideman RL, Zhou T, Holmes EJ, Lavey RS, Bouffet E, Pollack IF. Temozolomide in the treatment of children with newly diagnosed diffuse intrinsic pontine gliomas: a report from the Children's Oncology Group. Neuro Oncol. 2011;13:410–416. doi: 10.1093/neuonc/noq205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollack IF, Hamilton RL, James CD, Finkelstein SD, Burnham J, Yates AJ, et al. Rarity of PTEN deletions and EGFR amplification in malignant gliomas of childhood: Results from the children's cancer group 945 cohort. J Neurosurg. 2006;105:418–424. doi: 10.3171/ped.2006.105.5.418. [DOI] [PubMed] [Google Scholar]
- 6.Ganigi PM, Santosh V, Anandh B, Chandramouli BA, Sastry Kolluri VR. Expression of p53, EGFR, pRb and bcl-2 proteins in pediatric glioblastoma multiforme: A study of 54 patients. Pediatr Neurosurg. 2005;41:292–299. doi: 10.1159/000088731. [DOI] [PubMed] [Google Scholar]
- 7.Pollack IF, Finkelstein SD, Burnham J, Hamilton RL, Yates AJ, Holmes EJ, et al. Age and TP53 mutation frequency in childhood malignant gliomas: Results in a multi-institutional cohort. Cancer Res. 2005;61:7404–7407. [PubMed] [Google Scholar]
- 8.Pollack IF, Finkelstein SD, Woods J, Burnham J, Holmes EJ, Hamilton RL, et al. Expression of p53 and prognosis in children with malignant gliomas. N Engl J Med. 2003;346:420–427. doi: 10.1056/NEJMoa012224. [DOI] [PubMed] [Google Scholar]
- 9.Sure U, Ruedi D, Tachibana O, Yonekawa Y, Ohgaki H, Kleihues P, et al. Determination of p53 mutations, EGFR overexpression, and loss of p16 expression in pediatric glioblastomas. J Neuropathol Exp Neurol. 1997;56:782–789. [PubMed] [Google Scholar]
- 10.Sebolt-Leopold JS, Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat Rev Cancer. 2004;4:937–947. doi: 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]
- 11.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Gines C, Gil-Benso R, Benito R, Mata M, Pereda J, Sastre J, Roldan P, et al. The activation of ERK1/2 MAP kinases in glioblastoma pathobiology and its relationship with EGFR amplification. Neuropathol. 2008;28:507–515. doi: 10.1111/j.1440-1789.2008.00911.x. [DOI] [PubMed] [Google Scholar]
- 13.Schiffman JD, Hodgson JG, VandenBerg SR, Flaherty P, Polley MY, Yu M, et al. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70:512–519. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paugh BS, Qu C, Jones C, Liu Z, Adamowicz-Brice M, Zhang J, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bax DA, Mackay A, Little SE, Carvalho D, Viana-Pereira M, Tamber N, et al. A distinct spectrum of copy number aberrations in pediatric high-grade gliomas. Clin Cancer Res. 2010;16:3368–3377. doi: 10.1158/1078-0432.CCR-10-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGillicuddy LT, Fromm JA, Hollstein PE, Kubek S, Beroukhim R, De Raedt T, et al. Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis. Cancer Cell. 2009;16:44–54. doi: 10.1016/j.ccr.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knobbe CB, Reifenberger J, Reifenberger G. Mutation analysis of the ras pathway genes NRAS, HRAS, KRAS and BRAF in glioblastomas. Acta Neuropathol. 2004;108:467–470. doi: 10.1007/s00401-004-0929-9. [DOI] [PubMed] [Google Scholar]
- 18.Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121:397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 19.Morice C, Nothias F, Konig S, Vernier P, Baccarini M, Vincent JD, et al. RAF-1 and BRAF proteins have similar regional distributions but differential subcellular localization in adult rat brain. Eur J Neurosci. 1999;11:1995–2006. doi: 10.1046/j.1460-9568.1999.00609.x. [DOI] [PubMed] [Google Scholar]
- 20.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 21.Marais R, Light Y, Paterson HF, Mason CS, Marshall CJ. Differential regulation of raf-1, A-raf, and BRAF by oncogenic ras and tyrosine kinases. J Biol Chem. 1997;272:4378–4383. doi: 10.1074/jbc.272.7.4378. [DOI] [PubMed] [Google Scholar]
- 22.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 23.Tsavachidou D, Coleman ML, Athanasiadis G, Li S, Licht JD, Olson MF, et al. SPRY2 is an inhibitor of the ras/extracellular signal-regulated kinase pathway in melanocytes and melanoma cells with wild-type BRAF but not with the V599E mutant. Cancer Res. 2004;64:5556–5559. doi: 10.1158/0008-5472.CAN-04-1669. [DOI] [PubMed] [Google Scholar]
- 24.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic BRAF kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torchia EC, Boyd K, Rehg JE, Qu C, Baker SJ. EWS/FLI-1 induces rapid onset of myeloid/erythroid leukemia in mice. Mol Cell Biol. 2007;27:7918–7934. doi: 10.1128/MCB.00099-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim JS, Lee C, Bonifant CL, Ressom H, Waldman T. Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol Cell Biol. 2007;27:662–677. doi: 10.1128/MCB.00537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 29.Michaud K, Solomon DA, Oermann E, Kim JS, Zhong WZ, Prados MD, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70:3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hashizume R, Gupta N, Berger MS, Banerjee A, Prados MD, Ayers-Ringler J, et al. Morphologic and molecular characterization of ATRT xenografts adapted for orthotopic therapeutic testing. Neuro Oncol. 2010;12:366–376. doi: 10.1093/neuonc/nop033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhatt KV, Hu R, Spofford LS, Aplin AE. Mutant BRAF signaling and cyclin D1 regulate Cks1/S-phase kinase-associated protein 2-mediated degradation of p27Kip1 in human melanoma cells. Oncogene. 2007;26:1056–1066. doi: 10.1038/sj.onc.1209861. [DOI] [PubMed] [Google Scholar]
- 32.Schiappacassi M, Lovat F, Canzonieri V, Belletti B, Berton S, Di Stefano D, et al. p27Kip1 expression inhibits glioblastoma growth, invasion, and tumor-induced neoangiogenesis. Mol Cancer Ther. 2008;7:1164–1175. doi: 10.1158/1535-7163.MCT-07-2154. [DOI] [PubMed] [Google Scholar]
- 33.Lavoie JN, L'Allemain G, Brunet A, Müller R, Pouysségur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–20616. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 34.Gysin S, Lee SH, Dean NM, McMahon M. Pharmacologic inhibition of RAF-->MEK-->ERK signaling elicits pancreatic cancer cell cycle arrest through induced expression of p27Kip1. Cancer Res. 2005;65:4870–4880. doi: 10.1158/0008-5472.CAN-04-2848. [DOI] [PubMed] [Google Scholar]
- 35.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 37.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zarghooni M, Bartels U, Lee E, Buczkowicz P, Morrison A, Huang A, et al. Whole-genome profiling of pediatric diffuse intrinsic pontine gliomas highlights platelet-derived growth factor receptor alpha and poly (ADP-ribose) polymerase as potential therapeutic targets. J Clin Oncol. 2010;28:1337–1344. doi: 10.1200/JCO.2009.25.5463. [DOI] [PubMed] [Google Scholar]
- 39.Pollack IF, Jakacki RI, Blaney SM, Hancock ML, Kieran MW, Phillips P, et al. Phase I trial of imatinib in children with newly diagnosed brainstem and recurrent malignant gliomas: A pediatric brain tumor consortium report. Neuro Oncol. 2007;9:145–160. doi: 10.1215/15228517-2006-031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prados MD, Chang SM, Butowski N, DeBoer R, Parvataneni R, Carliner H, et al. Phase II study of erlotinib plus temozolomide during and after radiation therapy in patients with newly diagnosed glioblastoma multiforme or gliosarcoma. J Clin Oncol. 2009;27:579–584. doi: 10.1200/JCO.2008.18.9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown PD, Krishnan S, Sarkaria JN, Wu W, Jaeckle KA, Uhm JH, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North central cancer treatment group study N0177. J Clin Oncol. 2008;26:5603–5609. doi: 10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–290. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 43.Mawrin C, Diete S, Treuheit T, Kropf S, Vorwerk CK, Boltze C, et al. Prognostic relevance of MAPK expression in glioblastoma multiforme. Int J Oncol. 2003;23:641–648. [PubMed] [Google Scholar]
- 44.Jeuken J, van den Broecke C, Gijsen S, Boots-Sprenger S, Wesseling P. RAS/RAF pathway activation in gliomas: The result of copy number gains rather than activating mutations. Acta Neuropathol. 2007;114:121–133. doi: 10.1007/s00401-007-0239-0. [DOI] [PubMed] [Google Scholar]
- 45.Long GV, Kefford RF, Carr PJA, et al. Phase 1/2 Study Of Gsk2118436, A Selective Inhibitor Of V600 Mutant (Mut) Braf Kinase: Evidence Of Activity In Melanoma Brain Metastases (Mets). European Society of Medical Oncolgy Congress 2010. Late Breaking Abstract. Ann Oncol. 2010;21(suppl 8) NP Abstract LBA27. [Google Scholar]
- 46.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O'Day SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J, Nelson B, Hou J, Lee RJ, Flaherty KT, McArthur GA BRIM-3 Study Group. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.