Abstract
Granule neuron precursors (GNPs) are the most actively proliferating cells in the post-natal nervous system and mutations in pathways controlling their cell cycle can result in medulloblastoma. The transcription factor Atoh1 has been suspected to contribute to GNP proliferation, but its role in normal and neoplastic post-natal cerebellar development remains unexplored. We show that Atoh1 regulates the signal transduction pathway of Sonic Hedgehog, an extracellular factor that is essential for GNP proliferation, and we demonstrate that deletion of Atoh1 prevents cerebellar neoplasia in a mouse model of medulloblastoma. Our data shed light on the function of Atoh1 in post-natal cerebellar development and identify a new mechanism that can be targeted to regulate medulloblastoma formation.
Keywords: Atoh1, medulloblastoma, cerebellar granule neuron precursors, Sonic Hedgehog
Disruption of the delicate balance between proliferation and differentiation in cerebellar granule neuron precursors (GNPs) underlies medulloblastoma, the most common pediatric tumor of the nervous system (1, 2). Interestingly, a class of particularly aggressive medulloblastomas associated with very poor prognosis show high expression of Atoh1 (3), a transcription factor highly expressed in GNPs also known as Math1 (4), and recent in vitro studies proposed that Atoh1 might be involved in neoplastic proliferation (5, 6). Given the fact that deletion of Atoh1 in the mouse results in perinatal death (7), the function of this transcription factor in the developing post-natal cerebellum has remained opaque.
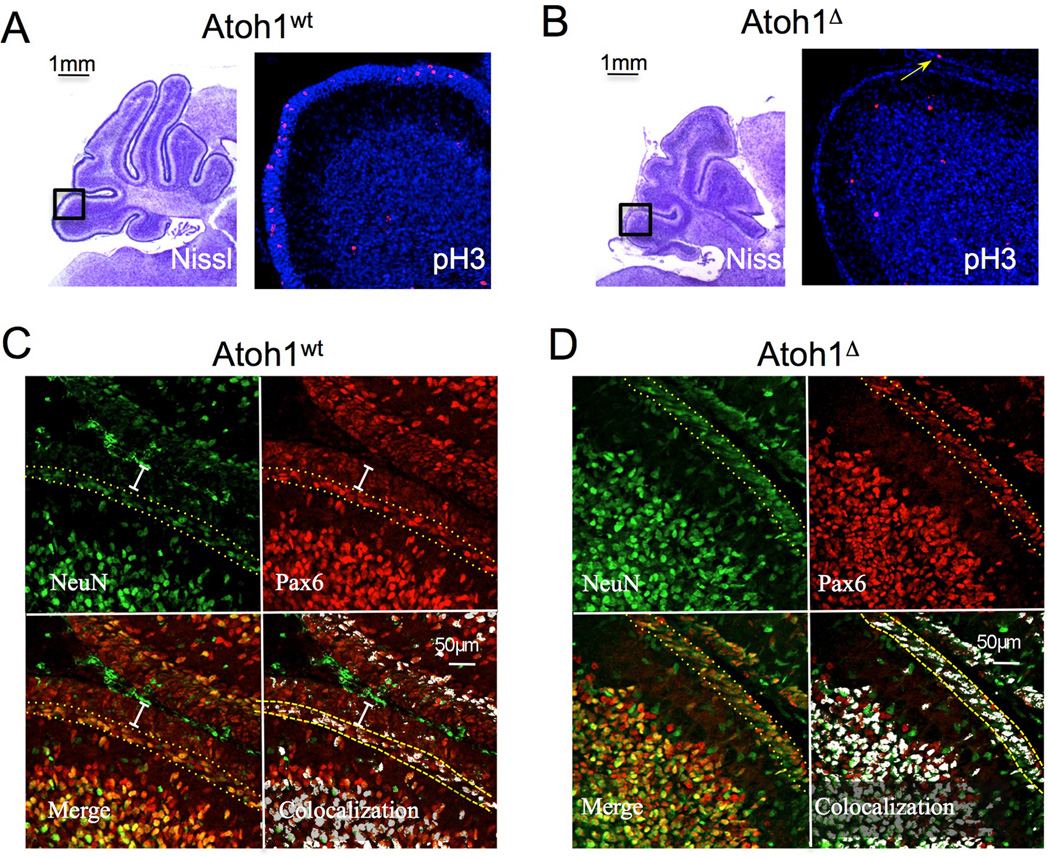
To delete Atoh1 in the post-natal developing cerebellum we crossed Atoh1flox/flox mice (8) with mice carrying the gene coding for a tamoxifen-inducible Cre recombinase in the Rosa locus (R26CreER) (9) and a null allele of Atoh1 (10). Following activation of Cre by tamoxifen, RosaCreER; Atoh1+/flox animals (designated herein as Atoh1wt) maintain one functional allele of Atoh1, whereas RosaCreER; Atoh1−/flox mice (designated herein as Atoh1▵) lose Atoh1 expression. We injected post-natal day 3 (P3) animals and analyzed their cerebella three days later. Nissl staining of matching sections of the external granule layer (EGL), the neuroepithelium formed by GNPs, revealed that Atoh1▵ animals had a much thinner EGL than their Atoh1wt littermates (Fig. 1S). Using phospo-histone H3 staining to visualize the M phase of the cell cycle and Tuj1 for neural differentiation, we found that the EGL of Atoh1▵ mice had been depleted of cycling immature precursors (Fig. 1A, B and 2S). Staining for active caspase 3 did not reveal any apoptosis in the EGL of Atoh1▵ mice (not shown). We thus investigated whether deletion of Atoh1 triggers the GNP differentiation to granule neurons or induces these cells to trans-differentiate to other cell types. Figure 2S shows that cells still populating the surface of Atoh1▵ cerebellum that had deleted Atoh1 still expressed Zic1, a marker of differentiating post-mitotic EGL cells and mature granule neurons, making trans-differentiation unlikely. Proliferating GNPs express Pax6 at low levels, whereas differentiating post-mitotic precursors show high expression of Pax6 and turn on the neural differentiation marker NeuN. The cells residing on the surface of the cerebellum of Atoh1▵ expressed high levels of Pax6 and NeuN (Fig 1C and D), again suggesting that deletion of Atoh1 in the post-natal cerebellum activates the differentiation process of cerebellar granule neurons.
Figure 1.
Atoh1 deletion disrupts GNP proliferation and induces differentiation. A, B) Phosphohistone H3 staining of cerebella of animals injected with tamoxifen. The arrow in B indicates a single cycling cell in the Atoh1▵ EGL. C and D) Pax6 and NeuN staining shows the immature GNPs (white bar in G) not expressing NeuN and the differentiating population (between the yellow dotted lined in G and H) co-expressing both markers. The co-localization pattern is shown on the bottom right. Scale bars are shown.
To investigate the molecular effects of Atoh1 deletion, we isolated GNPs from Atoh1flox/flox P5 animals and infected them with adenoviruses expressing either the GFP or the Cre recombinase gene. We cultured the transduced cells in the presence of Shh for three days (11), and evaluated their proliferative status by BrdU and phospo histone H3 staining. Atoh1 deletion led to a sharp decrease in cell proliferation (Fig. 3S A and B), suggesting that GNPs are unable to respond to Shh stimulation in the absence of Atoh1. We then purified total RNA from the transduced GNPs and performed quantitative RT-PCR analysis of the Shh target genes cyclin D1 and D2 (Ccnd1, Ccnd2), Mycn and Gli1 (Fig. 2A and data not shown). Surprisingly, all of these genes were down-regulated, as was Gli2, the main transcriptional effector of Shh signaling in GNPs (12, 13) (11, 14).
Figure 2.
Atoh1 controls GNP proliferation through regulation of Gli2 expression. A) QRT-PCR on RNA extracted from transduced GNPs cultured in presence of Shh for 3 days. The genes tested are shown below the bar graph. The amount of RNA for each gene was normalized over the GAPDH RNA level in each sample, and expressed as a percentage over the level of RNA of the GFP-transduced cells. B) Purified GNPs were infected either with a control retrovirus or a GLI2-expressing retrovirus and transduced with an Adenovirus expressing GFP or Cre. Cycling cells were labeled by BrdU and visualized by the red fluorescence signal. DAPI counterstaining (blue) reveals the nuclei. C) Quantification of the rescue effect by GLI2 on proliferation after deletion of Atoh1 in GNPs (BrdU positive cells/field). Student’s T-test results are shown. D) Luciferase assay showing that Atoh1 can activate transcription when it interacts with the Gli2 intronic region. Results are expressed as fold induction over the cells transfected with the empty expression vector. The reporter plasmids used in the experiments are shown below the graph. E) Chromatin-IP experiment indicating that Atoh1 is bound to the Gli2 genomic region in the postnatal cerebellum and that this sequence has the epigenetic hallmark of an active transcriptional enhancer (see text for details). Sample identities are shown above and the origin of the chromatin used in the reactions is indicated below the gel.
To determine whether Gli2 down-regulation was responsible for the failure to proliferate in the absence of Atoh1, we infected GNPs with a retroviral vector expressing Gli2 before deleting Atoh1. Cells infected with the Gli2-expressing retrovirus were still able to proliferate in response to Shh (Fig. 2B and C), showing that down-regulation of Gli2 following Atoh1 deletion plays a major role in GNP withdrawal from the cell cycle. Since Gli2 transcription does not require Shh (15), we reasoned that down-regulation of its mRNA is unlikely to be a secondary effect of the blockade of the Shh signal and that Gli2 might be a direct transcriptional target of Atoh1—which would explain how deletion of Atoh1 inhibits Shh signaling.
Atoh1 activates its target genes through binding to regions enriched in specific sites called E-boxes (16). Computational scanning identified a DNA sequence highly enriched in E-boxes (eight E-boxes) in the second intron of Gli2 (Fig. 4S A). We designed an oligonucleotide spanning two E-boxes in the cluster that are conserved in mouse and human and used it as a probe for electrophoretic mobility shift assay (EMSA) experiments (See Figure 4S A, boxed sequence). After incubating the probe with nuclear extracts of Neuro2a cells transfected with an Atoh1-Flag expression plasmid, we observed the appearance of a specific complex which was supershifted by an anti-Flag antibody (Fig. 4S B, lane 2, arrow and 3, arrowhead) and competed by the wild type (wt) unlabelled oligonucleotide but not by an oligonucleotide bearing a point mutation in each of the two E-boxes (lanes 4 and 5). Next, we cloned the entire sequence spanning the E-box cluster in front of a minimal promoter driving the expression of the luciferase gene. Co-transfection of Neuro2a cells with the Gli2 reporter construct and a vector expressing Atoh1 resulted in strong luciferase expression (Figure 2D), showing that Atoh1 acts as a positive regulator through binding to this sequence. The control reporter vector containing only the minimal promoter had no effect.
To verify our findings in vivo, we took advantage of a knock-in mouse model in which the endogenous Atoh1 protein is tagged with three flag peptides on its C-terminus (Fig. 5S). Atoh1flag/flag P5 cerebella were collected and subjected to Chromatin Immuno-Precipitation (ChIP). As a negative control, we took cortex (which does not express Atoh1) from the same animals. As shown in figure 2E, we obtained a strong amplified band in the cerebellar sample but virtually no signal in the negative control, implying that Atoh1 binds to this DNA region during cerebellum development. To confirm that this sequence is a transcriptional enhancer, we carried out additional chromatin IP experiments using an antibody recognizing the monomethylated form of histone H3 on lysine 4 (me1-H3), a well-defined marker of enhancers (17). An antibody recognizing the trimethylated form of histone H3 on lysine 4 (me3-H3), a marker of promoter sequences (17), served as a negative control. As expected, we recovered the DNA sequence corresponding to the Gli2 intergenic region only when we used anti me1-H3 antibody, confirming that the region bound by Atoh1 in the Gli2 gene is an active transcriptional enhancer. Finally, using an unbiased approach (ChIP followed by deep sequencing) we identified the sites bound by Atoh1 in vivo in GNPs. Analysis of the results confirmed that Atoh1 binds to the intergenic Gli2 E-box cluster in the developing cerebellum (T.J.K. and H.Y.Z., manuscript in preparation). We next evaluated whether the two genes are coexpressed in the neural progenitors that give rise to GNPs. Starting at E14.5 we observed strong expression of Gli2 and Atoh1 in the cells that are populating the surface of the developing cerebellum (Fig 6S A). To visualize the Atoh1 protein, we used a mouse model expressing an Atoh1-GFP fusion protein from the endogenous Atoh1 locus (18). At E18.5, a stage of cerebellar development that immediately precedes the post-natal expansion of the GNP population in the EGL, the cells expressing Atoh1 in the EGL also expressed Gli2. This is consistent with the possibility that Gli2 is a transcriptional target of Atoh1 (Fig 6S B).
If Atoh1 plays a central role in the regulation of Gli2, its expression might be required for the genesis of medulloblastomas induced by constitutive activation of the Shh pathway. To test this hypothesis, we used a mouse model bearing an inducible allele coding for a mutant form of the Smoothened protein, an activator of the Shh pathway, fused with YFP (RosaSmoM2). The expression of the mutant Smoothened allele can be activated by Cre recombinase, resulting in the formation of medulloblastomas in the developing cerebellum (19). We generated RosaSmoM2/CreER; Atoh1+/flox animals (designated SmoAtoh1wt) and RosaSmoM2/CreER; Atoh1lacZ/flox mice (designated SmoAtoh1▵). Cre-mediated recombination activates the Shh pathway in all of the animals by inducing expression of the SmoM2 gene, but SmoAtoh1wt mice still express Atoh1 from the functional allele, whereas SmoAtoh1▵ lack Atoh1 expression. After injecting P4 animals, we harvested the brains ten days later. Non-injected animals served as controls.
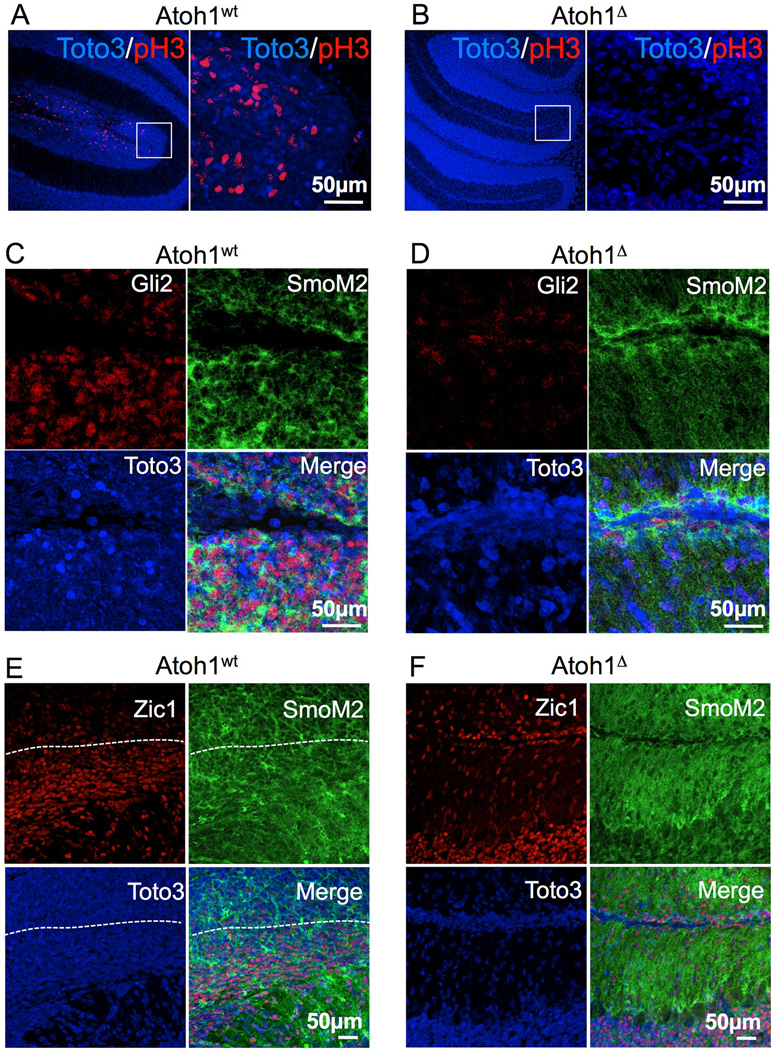
In normal P14 animals the vast majority of GNPs have already stopped cycling and the EGL is reduced to a few rows of cells (Fig. 3A), whereas SmoAtoh1wt mice (15/15 animals) showed clearly hypertrophic EGLs (compare Fig3A and 3B). All these animals developed pre-neoplastic lesions in the external layer of the cerebellum (Fig. 3B, middle and right panels), where the EGL lost the typical layered organization (20, 21). Strikingly, none of the SmoAtoh1▵ animals showed any evidence of over-proliferation in the EGL (0/12 animals and had atrophic cerebella (Fig 3C), strongly suggesting that Atoh1 deletion blocks the mitogenic activity of Smoothened in GNPs. Moreover, the EGL of SmoAtoh1▵ animals was much thinner than that of control littermates (compare Figure 3A and C, right panels, arrows). We investigated the proliferative status of the GNPs of SmoAtoh1wt and SmoAtoh1▵ animals by phospho-histone H3 staining. The ectopic cell masses in SmoAtoh1wt mice showed numerous mitotic cells (Figure 4A), whereas the EGL of SmoAtoh1▵ animals was negative for the presence of cycling cells (Fig. 4B). Staining for caspase 3 revealed no sign of apoptosis in SmoAtoh1▵ (not shown). As expected, the outgrowths in SmoAtoh1wt mice were positive for the YFP staining, an indication of SmoM2 expression, and showed high levels of Gli2 (Fig. 7S , arrow and 4C). We also observed strong staining with the YFP antibody in SmoAtoh1▵ animals (Fig. 7S B), but Gli2 levels were drastically reduced (Fig. 4D), confirming that Atoh1 expression is required for Gli2 expression in the EGL even in the presence of constitutively active Shh signaling. Zic1 staining showed that the cells that underwent recombination in SmoAtoh1▵ activated the granule neuron differentiation program, despite the constitutive activation of the Shh pathway (Fig. 4F). Once again, these data confirmed that Gli2 expression in GNPs requires Atoh1 and that its absence abolishes the response to Shh. Moreover, the fact that deletion of Atoh1, a gene expressed only in GNPs, is able prevent medulloblastoma formation strongly suggests that these cells are the cell-of-origin of tumors caused by constitutive activation of Shh signaling in the developing cerebellum. We provide genetic and molecular evidence that Atoh1 regulates proliferation of GNPs in the post-natal cerebellum by controlling the activity of the Shh pathway expression through the direct transcriptional regulation of Gli2. Consistent with this role, Atoh1 is also required for the formation of medulloblastomas induced by constitutive activation of the Shh pathway. A recent report suggests that Atoh1 acts as a tumor suppressor in colorectal cancer and Merkel cell carcinomas by inducing cell differentiation and apoptosis through expression of tyrosine kinase receptor type I (Ntrk1), which is opposite the function we describe in this paper (22). We propose that Atoh1 might activate the transcription of both Gli2 and Ntrk1. The activation of Gli2 makes the cells competent to transduce the mitogenic signal of Shh, whereas expression of Ntrk1 allows the cells to respond to differentiative or apoptotic stimuli. If the cells are exposed to Shh, as in the case of GNPs in the EGL, the outcome of Atoh1 expression is proliferation. On the other hand, if the cells are exposed to a differentiative signal, such as factors that activate tyrosine kinase receptors in the gut, Atoh1 expression allows the cells to respond by exiting the cell cycle and differentiating.
Figure 3.
Atoh1 deletion prevents tumor formation in a mouse model of Shh-induced medulloblastoma. Pictures of A) P14 cerebella of a control animal that did not receive tamoxifen, and thus doesn't express the activated form of Smoothened; of B) an Atoh1wt and of C) an Atoh1▵ animal injected with tamoxifen to activate the expression of the constitutively active Smoothened (see text). The treatment is shown on the side of each image series. The cerebellum is outlined with a dotted line in the left panels; the arrowheads point out two neoplastic outgrowths. A sagittal section around the midline is shown in the middle panels; neurons and neuronal precursors are visualized by Nissl staining. The boxed regions are shown at high magnification in the right panels; arrows indicate the EGL.
Figure 4.
Disruption of Atoh1 blocks Shh-induced proliferation and Gli2 expression in a mouse model of medulloblastoma. A and B) The M phase of the cell cycle was visualized by phosphohistone H3 staining in the cerebellum of Atoh1wt A) or Atoh1▵ (B) animals injected with tamoxifen. The red signal shows actively cycling cells, whereas the blue signal reveals cell nuclei. The boxed portion of the left panels is magnified at the right. The scale and genotype are shown. C) Zic1 staining of a cerebellar sample from an Atoh1wt animal and D) from an Atoh1▵ mouse. The cells above the white line in E are the Zic1-negative undifferentiated cells of the neoplastic tissue.
The central role of Atoh1 in the regulation of Gli2 has important implications for the modulation of normal and neoplastic proliferation in the developing cerebellum. The possibility to exploit this developmental mechanism to control Shh-induced proliferation makes Atoh1 a possible target for therapeutic strategies to inhibit medulloblastomas.
Supplementary Material
Acknowledgments
We thank Hsiao-Tuan Chao, Xiuyun Liu and Richard Atkinson for experimental support and Vicky Brandt for helping in editing the manuscript. Thanks to Hugo Bellen, Gianpietro Dotti, Meenakshi B. Bhattacharjee, Elena Battaglioli, Jeff Neul, and members of the Zoghbi lab for critical reading of the manuscript. The confocal microscopy was supported by the IDDRC at Baylor College of Medicine (5 P30 HD024064). H.Y.Z. is an investigator and T.J.K. is a postdoctoral research associate with the Howard Hughes Medical Institute.
Footnotes
The transcription factor Atoh1 regulates sonic hedgehog signaling in the developing cerebellum and is required for medulloblastoma formation.
References
- 1.Polkinghorn WR, Tarbell NJ. Nat Clin Pract Oncol. 2007 May;4:295. doi: 10.1038/ncponc0794. [DOI] [PubMed] [Google Scholar]
- 2.Gilbertson RJ, Ellison DW. Annu Rev Pathol. 2008;3:341. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- 3.Salsano E, Pollo B, Eoli M, Giordana MT, Finocchiaro G. Neurosci Lett. 2004 Nov 11;370:180. doi: 10.1016/j.neulet.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 4.Bertrand N, Castro DS, Guillemot F. Nat Rev Neurosci. 2002 Jul;3:517. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- 5.Briggs KJ, et al. Genes Dev. 2008 Mar 15;22:770. doi: 10.1101/gad.1640908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF. Genes Dev. 2008 Mar 15;22:722. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Arie N, et al. Nature. 1997 Nov 13;390:169. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 8.Shroyer NF, Wallis D, Venken KJ, Bellen HJ, Zoghbi HY. Genes Dev. 2005 Oct 15;19:2412. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badea TC, Wang Y, Nathans J. J Neurosci. 2003 Mar 15;23:2314. doi: 10.1523/JNEUROSCI.23-06-02314.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Arie N, et al. Development. 2000 Mar;127:1039. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- 11.Wechsler-Reya RJ, Scott MP. Neuron. 1999 Jan;22:103. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 12.Blaess S, Corrales JD, Joyner AL. Development. 2006 Mar 29; doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- 13.Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL. Development. 2004 Nov;131:5581. doi: 10.1242/dev.01438. [DOI] [PubMed] [Google Scholar]
- 14.Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP. Dev Biol. 2004 Jun 15;270:393. doi: 10.1016/j.ydbio.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Development. 1999 Sep;126:3915. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 16.Krizhanovsky V, Soreq L, Kliminski V, Ben-Arie N. J Mol Neurosci. 2006;28:211. doi: 10.1385/JMN:28:2:211. [DOI] [PubMed] [Google Scholar]
- 17.Robertson AG, et al. Genome Res. 2008 Dec;18:1906. doi: 10.1101/gr.078519.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rose Matthew F., Ren Jun, Ahmad Kaashif, Chao Hsiao-Tuan, Klisch Tiemo J., Flora Adriano, Greer John J., Zoghbi Huda Y. Neuron. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuller U, et al. Cancer Cell. 2008 Aug 12;14:123. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliver TG, et al. Development. 2005 May;132:2425. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- 21.Kessler JD, et al. Genes Dev. 2009 Jan 15;23:157. doi: 10.1101/gad.1759909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bossuyt W, et al. PLoS Biol. 2009 Feb 24;7:e39. doi: 10.1371/journal.pbio.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.