Abstract
DNA methylation is known to regulate transcription and was recently found to be involved in exon recognition via cotranscriptional splicing. We recently observed that exon–intron architectures can be grouped into two classes: one with higher GC content in exons compared to the flanking introns, and the other with similar GC content in exons and introns. The first group has higher nucleosome occupancy on exons than introns, whereas the second group exhibits weak nucleosome marking of exons, suggesting another type of epigenetic marker distinguishes exons from introns when GC content is similar. We find different and specific patterns of DNA methylation in each of the GC architectures; yet in both groups, DNA methylation clearly marks the exons. Exons of the leveled GC architecture exhibit a significantly stronger DNA methylation signal in relation to their flanking introns compared to exons of the differential GC architecture. This is accentuated by a reduction of the DNA methylation level in the intronic sequences in proximity to the splice sites and shows that different epigenetic modifications mark the location of exons already at the DNA level. Also, lower levels of methylated CpGs on alternative exons can successfully distinguish alternative exons from constitutive ones. Three positions at the splice sites show high CpG abundance and accompany elevated nucleosome occupancy in a leveled GC architecture. Overall, these results suggest that DNA methylation affects exon recognition and is influenced by the GC architecture of the exon and flanking introns.
A mature mRNA is formed by the removal of introns from the mRNA precursor (pre-mRNA) and the ligation of exons through the process of splicing (Black 2003). Splicing occurs cotranscriptionally, which means that a large fraction of the introns are removed from the mRNA precursor while the transcript is still attached to the DNA by RNA polymerase II (Pol II) (Neugebauer 2002; Proudfoot et al. 2002; Luco et al. 2011). This fact sets the foundation for a cross-talk between the DNA and RNA levels, thereby providing new possibilities for splicing regulation by factors that are known to affect transcription. Pol II can affect splicing through kinetic coupling since the elongation rate of Pol II can control exon recognition by the splicing machinery (de la Mata et al. 2003; Schor et al. 2009; Ip et al. 2011).
One factor that is known to regulate transcription and could also regulate splicing is DNA methylation. DNA methylation is regarded as the “fifth base” and is defined as the addition of a methyl group to a cytosine base, predominantly when it is directly followed by guanine but can be found in other contexts as well (CG, CHG, CHH; where H = A, C, or T) (Bernstein et al. 2007). DNA methylation serves a role in many biological processes, including embryogenesis and development, genomic imprinting, and regulation of gene transcription (Li et al. 1992; Okano et al. 1999; Bestor 2000; Reik 2007). Due to dramatic advances in high-throughput DNA sequencing methods, several groups have recently mapped DNA methylation across the whole genome at single-base resolution (Meissner et al. 2008; Laurent et al. 2009; Lister et al. 2009; Gu et al. 2010; Li et al. 2010; Stadler et al. 2011; Smith et al. 2012; Ward et al. 2012; Xie et al. 2012). This new source of high-quality data represents a major step toward our understanding of the biological role of cytosine methylation and has already been used to establish a primary connection between DNA methylation and splicing. In their genome-wide methylation analysis, Lister et al. (2009) revealed that there is a higher level of CpG-methylation on exons than on flanking introns. However, this difference was dissolved when values were divided by cytosine composition since exons have a higher GC content, on average, than their flanking introns (Schwartz et al. 2009). A later genome-wide methylation analysis suggested that DNA methylation is a strong marker of exons and stronger yet for exon-boundaries, thus being a possible regulator of splicing (Laurent et al. 2009). In support of this suggestion, DNA methylation was found to be enriched in alternative splice sites and in splicing regulatory motifs (Anastasiadou et al. 2011), both of which are important regulatory regions for splicing. Furthermore, depletion of DNA methylation in Hox genes was found to remove Pol II stalling and facilitate transcriptional elongation and efficient splicing (Tao et al. 2010). Finally, mutually exclusive DNA methylation and transcriptional repressor CTCF protein binding were found to regulate exon inclusion by influencing Pol II elongation rate (Shukla et al. 2011).
Another epigenetic factor suspected to be involved in both transcription and splicing regulation is chromatin structure. The primary structure of chromatin involves DNA wrapped around nucleosomes. Approximately 147 base pairs of DNA are wrapped around a single nucleosome made up of an octamer of histone proteins and one linker histone (Kouzarides 2007). Positioning of nucleosomes along the genome is partially determined by the DNA sequence and GC content (Segal et al. 2006; Tillo and Hughes 2009; Tillo et al. 2009; Nikolaou et al. 2010) and is subject to modulation by chromatin remodelers (Vignali et al. 2000). DNA methylation, in itself, can act as a chromatin remodeler through two different pathways: either by inducing a rigid nucleosomal conformation that results in a more tightly wrapped nucleosome structure (Choy et al. 2010) or through the binding of methyl-binding proteins (MBPs) that silence transcription and modify surrounding chromatin (Sarraf and Stancheva 2004; Klose and Bird 2006). A reverse mechanism, in which nucleosome positioning or histone modifications can affect DNA methylation levels through the recruitment of DNA methyltransferase enzymes was also suggested (Robertson 2002). Several recent publications have shown a higher level of nucleosome occupancy on exons compared to the flanking introns, suggesting a link between nucleosome positioning and splicing regulation (Andersson et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009; Chen et al. 2010). The nucleosome is thought to act as a roadblock for Pol II elongation (Batsché et al. 2006; Hodges et al. 2009) as Pol II needs to pause and unwind the DNA double strand to release it from the nucleosome before continuing transcription elongation. This transcriptional pausing may allow cotranscriptional recognition of splicing signals in the pre-mRNA (de la Mata et al. 2003).
As nucleosome occupancy is strongly biased toward high GC content (Segal et al. 2006; Tillo and Hughes 2009; Tillo et al. 2009), which is in high abundance in exons (Schwartz et al. 2009). DNA methylation is also positively correlated with GC content (Bernardi et al. 1985; Jabbari et al. 1997; Varriale and Bernardi 2010). That being said, the level of methylated CpGs over the available CpGs decreases with increasing GC levels (Jabbari and Bernardi 1998; Oakes et al. 2007), thus painting a more complicated picture. If this is the case, could the roles of these epigenetic modifications in splicing be a side effect of the GC content of the exons? How could one isolate the measured levels of DNA methylation and nucleosome occupancy on exons and conclude a biological role unbiased by GC content?
To evaluate this, we use two populations of exons that are distinguished by their exon–intron GC-content pattern: differential GC, where the exon has higher GC content than do the flanking introns; and leveled GC, where there is no significant GC difference between the exon and introns. These groupings enabled us to examine DNA methylation on exon–intron architecture in an unbiased fashion and in a manner similar to our examination of nucleosome occupancy (Amit et al. 2012). We found DNA methylation to be a strong marker of exons regardless of GC content. Remarkably, nucleosomes are very selective in their GC-content requirements, marking exons with a differential GC content between exon and flanking introns but not exons with leveled GC content between exon and introns. In our analyses, we used mapped DNA-methylation data to calculate average DNA-methylation values: mCpG/CpG. Surprisingly, we found that when there is no GC differential between exon and introns, the mCpG/CpG level drops significantly in the intronic regions close to the splice sites. We then extended our analysis to look at the full human exome through “GC-content goggles” in the form of isochore maps (Costantini et al. 2006) and found the same patterns of epigenetic preferences to be a global genomic property. Moreover, we found that methylated CpGs in a leveled GC architecture accompanied higher inclusion of exons. Overall, our findings provide further support for the role of DNA methylation as a splicing regulator and reveal dynamic patterns of DNA methylation and chromatin organization upon exon–intron structure that are dependent upon the regional GC content and the GC differential between exon and introns.
Results
The level of methylated CpGs as a marker of exons
When attempting to understand the structure of DNA methylation around exons, one encounters a problem: GC content is usually higher in exons than in introns (Schwartz et al. 2009). This creates a difficulty when trying to assess relative values of DNA methylation in exons compared with flanking introns since DNA methylation requires CG dinucleotides, and those are more abundant when GC content is high. As a result, previous attempts that normalized DNA-methylation levels to cytosine abundance showed that the higher methylation marking of the exon dissolves and is similar to that in the flanking introns (Lister et al. 2009).
To assess whether higher DNA methylation in exons is due only to higher GC content in exons compared to the flanking sequences, we needed candidates that would enable us to simultaneously calculate DNA-methylation levels and control for GC content. We have recently identified two different approaches by which the splicing machinery is directed to exons and introns based on nucleotide composition of the genomic environment. In the first group, a differential GC content between exons and introns allows for better recognition of the exons by the splicing machinery. The second group is characterized by the same level of GC content in the exon and the flanking introns, and recognition by the splicing machinery is intron based (Amit et al. 2012). The latter group, with a leveled GC architecture, provided us with the candidates with GC content that enabled unbiased observation of DNA-methylation patterns.
The analyses of both GC groups enabled a deeper understanding as to their splicing regulation differences. Specifically, a total of 15,874 exons with higher GC in exons compared with flanking introns (differential GC group) and 16,269 exons with leveled GC between exon and flanking introns (leveled GC group) were retrieved based on the RefSeq tracks from the UCSC Genome Browser (http://genome.ucsc.edu/) as were published previously by Amit et al. (2012). We calculated per base DNA-methylation levels for each group based on genome-wide DNA-methylation data from human H1 embryonic stem (H1 ES) cells retrieved from Lister et al. (2009).
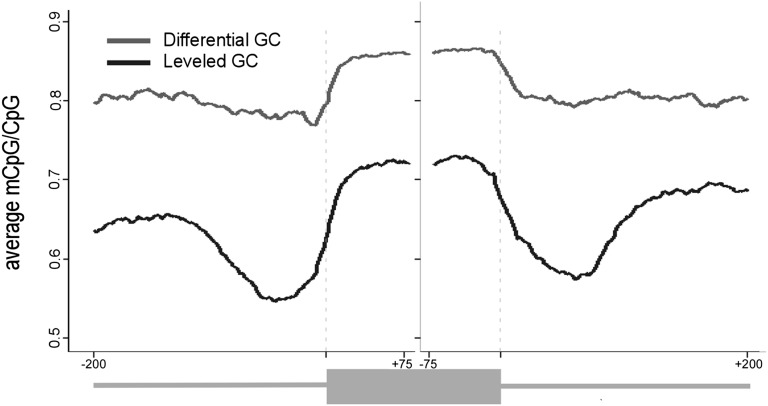
When controlling for DNA-methylation biases, the CpG abundance also needs to be taken into account since higher methylation levels can be a direct result of a higher abundance of the CG dinucleotide. For example, the sequence ATTGGGGCAC has 60% GC content but 0% CG dinucleotides, whereas the sequence ACGCCAATCG also has a GC content of 60% GC content but has 40% CG dinucleutides. To control for this potential bias, we calculated the average DNA-methylation level only at CpG sites (mCpG/CpG). This calculation is based on the raw DNA-methylation level, which is the methylation base calls at a reference coordinate divided by total base calls at that coordinate as reported by Lister et al. (2009). Next, we calculate the mCpG/CpG value as the sum of DNA-methylation level for each position relative to the 3′ or 5′ splice site of each exon divided by the number of exons with a CpG occurrence in that position (see Methods). This calculation was necessary to avoid the bias of higher CpG density on the exons as we show further on. Both groups of exon–intron structures show a significantly higher level of mCpG/CpG on the exons compared with the flanking introns (t-test, P-value < 2.2 × 10−16), and the overall level of methylated CpGs in the differential GC group is much higher than that of the leveled GC group (Fig. 1, P-value < 2.2 × 10−16). More specifically, in the group with differential GC content (Fig. 1, gray line), there is an increase from ∼80% methylated CpGs in the intronic sequences to ∼85% in the exon. Strikingly, in the leveled GC group (black line), there is a drop in CpG methylation in the intronic regions proximal to the exon (∼100 nt). If only the 100 intronic nucleotides flanking the exons are considered, there are ∼55% intronic methylated CpGs and >70% exonic methylated CpGs (>30% increase). This indicates that there is significantly more DNA methylation in exons than in introns, and this difference is most apparent when exons and introns with similar GC content are compared. It was previously reported that a lesser fraction of CpGs are methylated in regions of high CG content (such as that of the leveled GC group) than in lower GC regions (differential GC group) (Jabbari and Bernardi 1998; Oakes et al. 2007). Our results are in agreement with these findings.
Figure 1.
DNA-methylation levels in exons and flanking introns that differ by their GC content. Average of methylated CpGs along exon–intron structure with a differential GC content between the intron and the exon (gray) and along exon–intron structure in which the GC content is identical between the exon and the flanking introns (black). The average value was calculated per base for exons (75 nt from each splice site) and flanking intronic regions (200 nt). A running average of 20 was applied after omitting the following positions for having no CpG occurrences: 3′ss positions −4 to −1 and 5′ss positions +1 and +2.
In parallel with the analysis of mCpG/CpG, we also calculated the basic DNA methylation level (by base calls) and observed the expected stronger signal within exons relative to introns (Supplemental Fig. S1A). Another necessary analysis is that of the CpG abundance on the exon–intron structure since the human genome has a very low abundance of CG dinucleotides in general, but a much higher CG dinucleotides abundance in the coding sequence compared to intronic or intergenic regions (Karlin and Burge 1995; Gentles and Karlin 2001). In agreement with this, the results exhibit a strong signal of CpG abundance on the exons of both leveled and differential GC groups (Supplemental Fig. S1B). These results are a direct consequence of the CpG abundance bias in the coding sequence and prove the necessity in normalizing methylation levels to CpG abundance using the mCpG/CpG values.
We extended our analysis further to evaluate DNA-methylation levels in mouse ES cells and various human tissues based on data from Gu et al. (2010), who constructed genome-wide single-base DNA-methylation maps of the human and mouse samples using the reduced representation bisulfite sequencing (RRBS) protocol, which applies the DNA methylation-insensitive restriction enzyme MspI to map ∼5% of the CpG sites in the genome. We found that DNA-methylation levels in the different human and mouse tissues exhibit the same general pattern of higher exonic values in both GC groups (Supplemental Fig. S2). The noise level is much higher in these data, however, since the methylation data set is much smaller.
In previous work (Amit et al. 2012), we found that when examining the differential GC group, there is a 50% increase in nucleosome occupancy on exons compared to flanking introns. However, there is a very small increase of the nucleosome occupancy signal (∼10% increase) on the exons belonging to the leveled GC group. Although the two GC-content groups differ in the occupancy of nucleosomes between exons and flanking introns, both have a strong methylation signal on the exons. Moreover, the group that is weakly marked by nucleosomes is more significantly marked by methylated CpGs than the group marked more strongly by nucleosomes. This suggests that DNA methylation in exons has biological relevance. The difference in epigenetic patterns may indicate differences in how the splicing machinery recognizes these two types of exons.
DNA methylation and exon recognition
To evaluate the role of DNA methylation in exon recognition, we examined whether the level of mCpG/CpG in alternatively spliced cassette exons differs from that of constitutively spliced exons. We performed an extensive analysis to identify alternative and constitutive exons using RNA-seq data obtained from Lister et al. (2009). The mRNA reads were extracted from the same cells used to obtain DNA methylation (H1 ES cells), allowing the integration of alternative splicing effects with DNA-methylation patterns. Next, we used the SpliceTrap tool (Wu et al. 2012) to quantify exon inclusion ratios based on the RNA-seq data (see Methods).
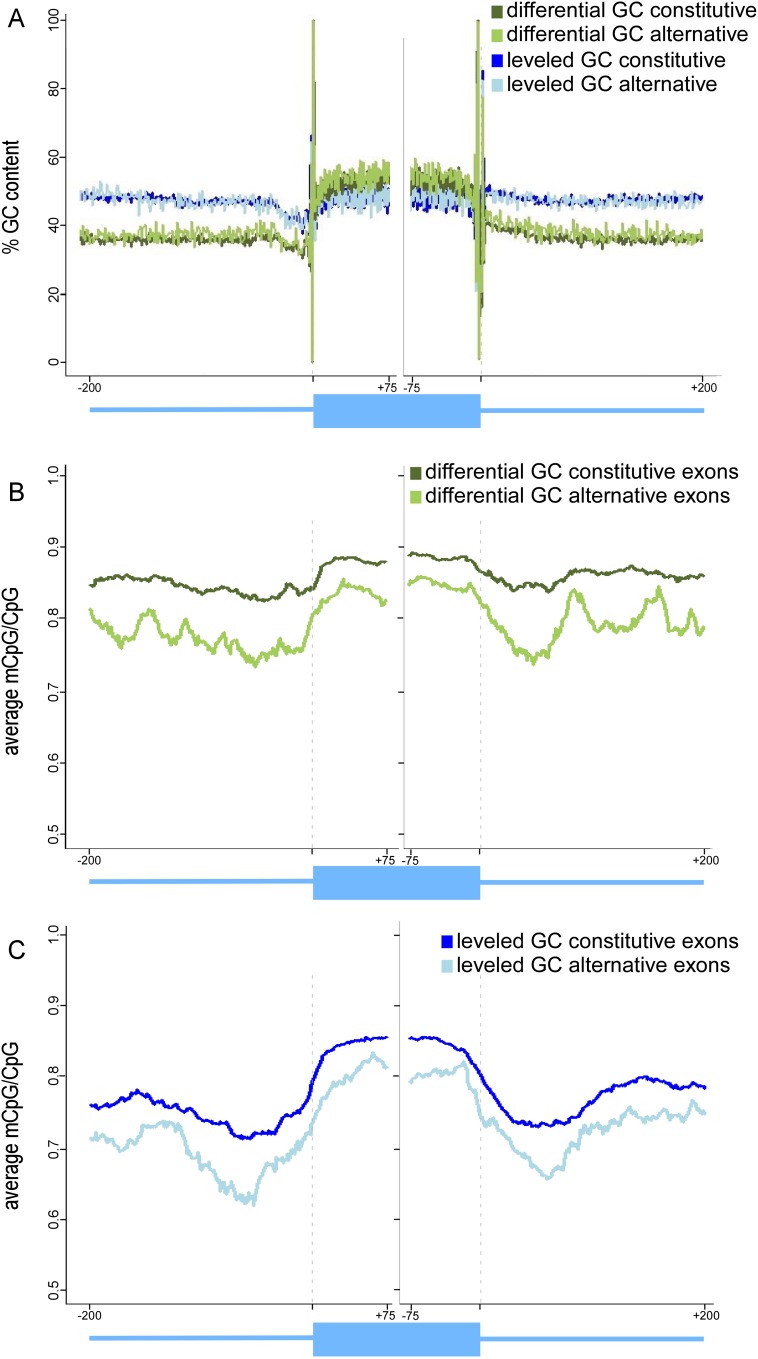
We identified 37,473 exons with canonical splice sites that we could statistically ensure their inclusion levels. We then divided these exons into two groups based on GC content: (1) leveled GC, which had no significant difference in GC content between exon and flanking introns; and (2) differential GC, in which exons are significantly higher in GC than flanking introns (P-value < 0.05). There were 7413 exons in the differential GC group (5734 constitutive exons and 1679 alternative exons) and 6037 exons in the leveled GC group (4936 constitutive exons and 1101 alternative exons). GC-content maps of both groups confirmed that the expected exon–intron GC structure was present in each group (Fig. 2A). The general pattern of mCpG/CpG signal around the exons remains the same for the differential GC constitutive exons and the leveled GC constitutive exons (multiple t-tests, P-values < 2.2 × 10−16) (Fig. 2B,C). Importantly, the levels of mCpG/CpG are significantly lower in alternative exons than in constitutive ones (multiple t-tests, P-value < 2.2 × 10−16) in both GC groups (Fig. 2B,C). These results further imply the conditions required for DNA methylation to play a part in the regulation of alternatively spliced exons.
Figure 2.
Average of methylated CpGs and GC content of constitutive and alternative exons. (A) Average of GC content percentage of differential GC content constitutive exons (dark green) and alternative exons (bright green) and of leveled GC content constitutive exons (dark blue) and alternative exons (bright blue). (B) Average of methylated CpGs of differential GC content constitutive exons (dark green) and alternative exons (bright green). (C) Average of methylated CpGs for leveled GC content constitutive exons (dark blue) and alternative exons (bright blue). The average value was calculated per base for exons (75 nt from each splice site) and flanking intronic regions (200 nt). A running average of 20 was applied for mCpG values after omitting the following positions for having no CpG occurrences: 3′ splice site positions −4 to −1 and 5′ splice site positions +1 and +2.
DNA-methylation levels by base calls, as well as CpG abundance were also calculated for these alternative and constitutive exons (Supplemental Fig. S3). Both present stronger signals on the exons than on the introns, yet the patterns are similar between alternative and constitutive exons of each group. These results emphasize, again, the need to observe the unbiased pattern of DNA methylation through the use of mCpG/CpG. When this value is used, differential DNA methylation still clearly defines the exons compared to their flanking intronic regions and also better marks constitutive exons compared to alternative ones.
Analysis of nucleosome occupancy was also done for these alternative and constitutive exons using the data used for the original GC-content groups (Amit et al. 2012). We found a strong signal of nucleosome occupancy for differential GC exons and a very weak signal for leveled GC exons (Supplemental Fig. S4).
CpG abundance and chromatin organization
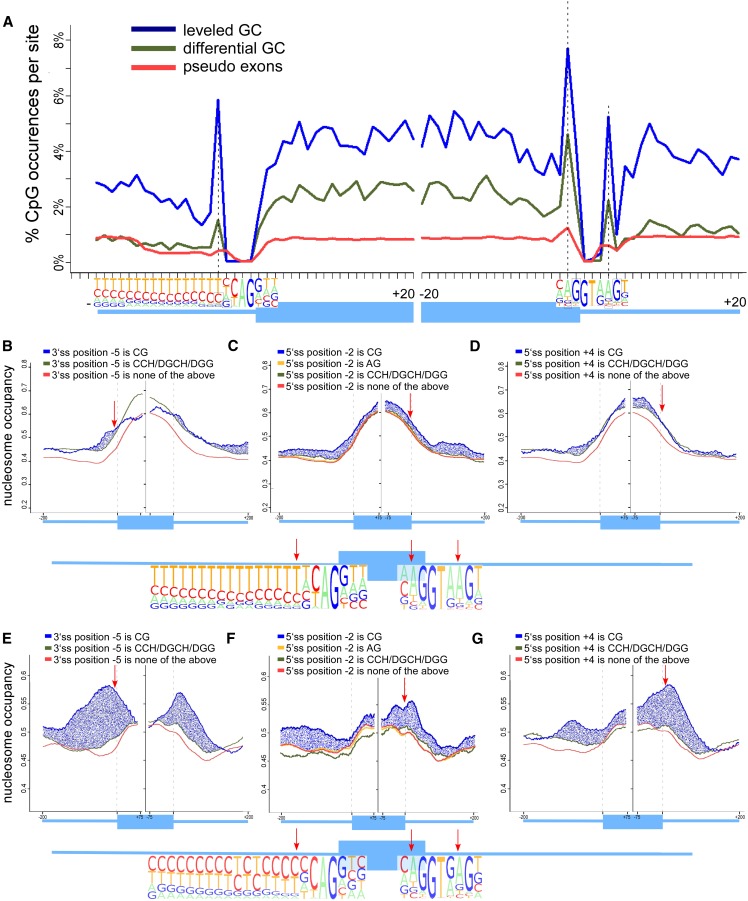
When examining methylation levels in regions immediately adjacent to exons, we observed extremely high methylation values near both the 3′ and the 5′ splice sites. This may be due to the presence of the CG dinucleotide as part of the consensus splice signal. To determine the relevance of this methylation, we first focused on deciphering the specific patterns of CpG abundance at positions −20 to +20 nt relative to each splice site. As a control, we used a data set of approximately 130,000 pseudo exons constructed by Ke et al. (2011). The GC content of these pseudo exons is not biased to any GC differential between exon and introns (Supplemental Fig. S5). In Figure 3A, the CpG abundance at the splice sites without application of a running average is shown. First, as previously observed, we found that CpG abundance is higher in the exons than in flanking introns in both GC groups but not in the group of pseudo exons. Second, we identified three positions with a significantly higher CpG percentage than the rest of the evaluated positions. All three are part of the splice site signal: position −5 of the 3′ splice site, position +4 of the 5′ splice site, and position −2 of the 5′ splice site. When followed by G, the cytosines in all three positions are methylated in at least 70% of the cases. These same positions show very minor increase (if at all) in the group of pseudo exons. Interestingly, a CG dinucleotide is not part of the consensus splice site signal in these positions. However, the nucleotide adjacent to two of the positions (5′ splice site −2 and +4) in the consensus sequence is a G, which partially accounts for the high CpG percentage in those two positions. These peaks in CpG abundance that are mostly methylated demonstrate very high methylation levels within both splice site regions and might point to a regulatory role for those positions when methylated. The fact that these peaks are diminished in the group of pseudo exons further implies on a possible role for these positions in the splicing process.
Figure 3.
CpG-abundance peaks at splice sites and their effect on nucleosome occupancy. (A) Percentage of methylated CpGs around the 3′ and 5′ splice sites of the differential GC exon–intron group (green), leveled GC content exon–intron group (blue), and pseudo exons (red). The percentage was calculated per base for exons (20 nt from each splice site) and flanking intronic regions (20 nt), and the number of exons with a CpG at each position was divided by total exons. The structure of the exon–intron junctions are shown in the bottom with pictogram depictions of the splice sites based on Gelfman et al. (2012). Specific positions with high levels of DNA methylation are marked in black boxes and dashed lines. (B–D) Average per base nucleosome occupancy levels for differential GC exons. Nucleosome occupancy levels are presented for three positions within the splice sites: (B) position −5 of the 3′ splice site; (C) position −2 of the 5′ splice site; and (D) position +4 of the 5′ splice site. Nucleosome occupancy levels are given based on dinucleotide composition: (1) CG dinucleotides (blue); (2) CCH/DGCH/DGG (green); and (3) any other composition (red). Position −2 of the 5′ splice site is compared to the AG dinucleotide composition, which represents the consensus dinucleotide at this position. Structure of the differential GC exon–intron junctions are shown in the bottom of these panels with pictogram depictions of the splice sites. (E–G) Average per base nucleosome occupancy levels for leveled GC exons. Nucleosome occupancy levels are presented for three positions within the splice sites: (E) position −5 of the 3′ splice site; (F) position −2 of the 5′ splice site; and (G) position +4 of the 5′ splice site. Structure of the leveled GC exon–intron junctions are shown in the bottom of these panels with pictogram depictions of the splice sites.
We next constructed several analyses to examine the role of these peak methylated positions as possible chromatin remodelers. It has been shown that certain DNA sequences with high affinity binding to the histone can direct nucleosome positioning (Lowary and Widom 1998), and that this in turn acts as a barrier to transcription (Bondarenko et al. 2006; Bintu et al. 2012). Furthermore, DNA methylation may have an intrinsic effect on nucleosome positioning on the DNA (Chodavarapu et al. 2010; Cedar and Bergman 2012). Thus, a change in nucleosome occupancy that accompanies DNA methylation might have an indirect influence on Pol II elongation rate and cotranscriptional splicing. We are not aware of any method that enables a quantification of the effect of a single base change in CpG-methylation upon chromatin organization. Thus, we decided to perform an analysis that was independent of the previous methylation analyses that relied on data from H1 ES cells and examined nucleosome occupancy as a result of CpG dinucleotide compositions at several positions relative to the splice sites using the data compiled by Schones et al. (2008). In H1 ES cells, a mean of 82.3% of the CpG sites are methylated (Lister et al. 2009), and 70%–88% of the CpG dinucleotides in both GC groups are methylated (see Fig. 1); therefore, we assumed that a CpG dinucleotide could act as a representative of a methylated position. Our next step was to divide the exons into subgroups based on dinucleotide composition at the three methylation peak positions: (1) CG dinucleotide at the peak position (CG composition subgroup); (2) all other dinucleotide with C and G that is not CG (GC composition subgroup); (3) all other dinucleotide compositions (i.e., AA, AT, AC, not-C composition subgroup); and (4) AG dinucleotide (AG subgroup). The GC composition subgroup was used as a control for the CG composition since the GC content is identical but CpG-methylation is not possible. For the analysis of the −2 position of the 5′ splice site, we used the AG subgroup, which represents the consensus dinucleotide at position −2 of the 5′ss (this subgroup is contained in the larger not-C subgroup). We constructed nucleosome occupancy data sets for the four exon subgroups based on their composition at the three peak positions for both leveled and differential GC exons.
There was a substantial increase in nucleosome occupancy when any of the three peak positions is a CG in the group of leveled GC exons (Fig. 3E–G). More specifically, we observe significantly higher levels of occupancy in the vicinity of all three positions when the composition is CG compared with any other composition examined (multiple t-tests, P-value < 1.76 × 10−13). This effect is diminished in the group of differential GC exons (Fig. 3B–D), where we observe a depleted signal on the exons when position −5 of the 3′ splice site is a CG (Fig. 3B, blue line) rather than GC composition (P-value < 0.006) (Fig. 3B, green line) and a small increase in signal for positions −2 and +4 of the 5′ splice site (P-value < 7.8 × 10−7 and P-value < 0.043, respectively) (Fig. 3C–D). We extended this analysis to several other positions along the exon and flanking introns. In general, we observe a trend of increased nucleosome occupancy when a CG is present for the group of leveled GC exons, but the effect is smaller than at the peak positions (Supplemental Fig. S6). This effect was not observed in differential GC exons (Supplemental Fig. S7); in these exons, nucleosome occupancy is not significantly affected by dinucleotide composition near the splice site. We also validated that a CG at the three peak positions does not have a fundamental effect on the overall GC content of the subgroup. The GC content analysis of exons with the four dinucleotide compositions exhibit very minor changes, if at all, between the different composition subgroups (Supplemental Fig. S8) that cannot explain the strong nucleosome occupancy signal of the CG composition subgroup that is observed in Figure 3.
Two observations are apparent from these analyses: First, there is a significant change in the occupancy of nucleosomes in leveled GC exons when CG dinucleotides are present in the region of the splice sites; this difference is significant relative to any other composition at positions −5 of the 3′ splice site and positions +4 and −2 of the 5′ splice site and relative to the AG consensus sequence composition at position −2 of the 5′ss. Since most CG dinucleotides at these positions are methylated, we assume that this may help to direct nucleosome binding to exons that can, in turn, be mediated by methyl-binding proteins. Second, these results again highlight differences between the leveled and differential GC groups with regards to the effect of DNA methylation. In the differential GC group, there is little if any effect of CG dinucleotides near the exon–intron junctions. In contrast, in the leveled GC group the effect is a strong nucleosomal signal. This difference suggests that the mechanisms of exon recognition may differ depending on the GC-content environment of a particular exon, the whole gene, or the genomic location itself, as we show further on.
Epigenetic patterns on exons based on genomic location
The major differences in epigenetic patterns between the leveled and differential GC groups led us to consider that perhaps genomic location impacted epigenetic pattern since some chromosomes are known to contain higher GC content than others (Costantini et al. 2006). Thus, if the groups belong in different chromosomes, it could explain the difference in basic GC-content architecture. It is also possible that the genomic location results in a particular epigenetic pattern and is another factor in exon recognition.
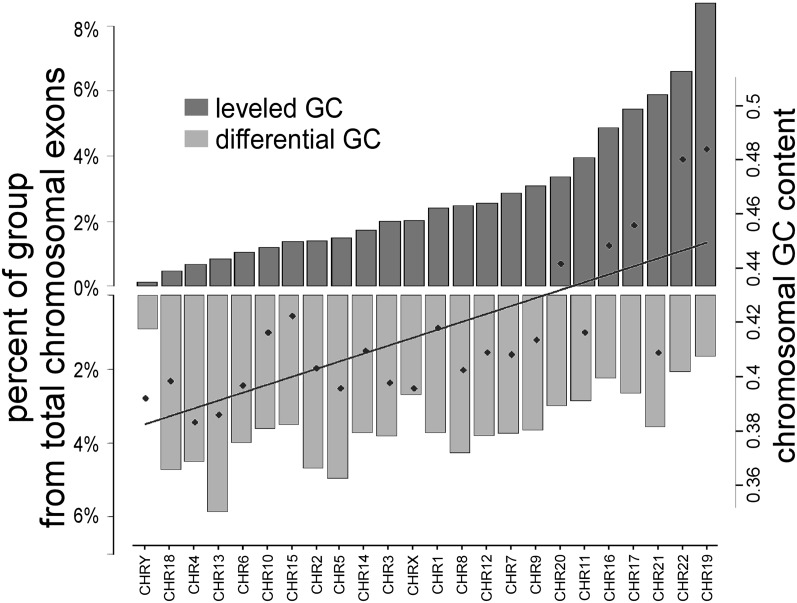
Analysis of the distribution of each GC group (leveled or differential GC) along the chromosomes shows that the groups are not equally distributed along the genome. We observed that the exons from the differential GC group (Fig. 4, light gray) are more abundant in lower GC-content chromosomes, whereas the leveled GC exons (dark gray) are more abundant in high GC-content chromosomes. This being the case, the patterns of epigenetic modifications that are observed along the exon–intron structure (Figs. 1, 2) might be a general property that is dependent on regional GC content. To evaluate this, we used human isochore maps that were constructed by Costantini et al. (2006). These maps divide the entire human genome into regions no smaller than 300K bases that are largely homogeneous in GC content. Costantini et al. (2006) divided those regions into two gene spaces that they called “core” isochores, where GC content is >46%, and “desert” isochores, where GC content is <46%. We divided all available internal exons into core and desert groups and obtained 53,102 core exons and 85,979 desert exons.
Figure 4.
Distribution of differential and leveled GC exons by chromosomal location. Genomic distribution by chromosome of differential GC (light gray) and leveled GC (dark gray) exons. The left y-axis represents the percentage of each group in each chromosome. The right y-axis represents the general GC content of the chromosomes.
As expected based on our previous results, DNA methylation is a strong marker of an exon when these ∼140,000 human exons are considered (Supplemental Fig. S9A). The methylated CpGs were constructed for alternative and constitutive exons that are expressed sequence tag (EST)-based since the RNA-seq data provided reliable inclusion level for ∼37,000 exons, which was insufficient for this analysis. We observe a general marking of the exons by a higher level of methylated CpGs (Supplemental Fig. S9B) that also exhibits a slightly larger mean increase in core exons (+24%) than in desert exons (+19%). Also, the level of methylated CpGs drops in alternative exons and their flanking introns in both desert and core exons relative to constitutively spliced exons as was observed using RNA-seq-based groupings. The differences in patterns and levels of methylated CpGs when exons are grouped using EST-based data rather than RNA-seq data are a consequence of the more heterogeneous GC differential of the isochore exons as they are grouped based on general GC content and not based on exon–intron GC differential.
Analysis of nucleosome occupancy revealed that nucleosomes are more often present on exons rather than introns when regional GC content is low (Supplemental Fig. S9C, green lines). Thus, when examining sequences that reside in low GC regions, one can expect a strong occupancy of nucleosomes on the exons. However, when GC content is regionally high, nucleosomes are spread more evenly along introns and exons (Supplemental Fig. S9C, blue lines).
In these results, we observe two genome-scale exon populations that vary based on their GC content. We find them to differ in their epigenetic patterns upon exon–intron structure, and this difference could mean a different mechanism of splicing regulation that is based on genomic location.
Discussion
In this study, we analyzed DNA methylation at single-base resolution and evaluated nucleosome occupancy patterns along exon–intron structures. To eliminate biases generated by differential GC content between exon and introns, we analyzed two groups of exons with and without a GC-content differential between exon and flanking introns. This enabled us to examine the pattern of DNA methylation across the exon–intron structure, taking into account, for the first time, GC-content differential. We previously showed that the GC differential between exon and intron allows better recognition of exons by the splicing machinery; yet it was unclear how exons are recognized in the leveled GC architecture, with similar GC content in exon and introns. The use of these groups for analysis of the effect of DNA methylation on splicing has two advantages: (1) it makes a control possible for a GC content bias between exon and introns (that is evident when analyzing DNA methylation); and (2) it reveals epigenetic differences between the groups that might explain differences in exon recognition mechanisms.
Division of exons into groups characterized by GC differentials between exons and flanking introns provides a tool to analyze epigenetic modifications that are generally influenced by GC content (Bernardi et al. 1985; Jabbari et al. 1997; Segal et al. 2006; Tillo and Hughes 2009; Varriale and Bernardi 2010). However, CpG density in itself could determine DNA-methylation levels (Choi et al. 2009). In order to view DNA methylation while taking into account CpG content, we compared the number of methylated CpGs to total CpGs, thus creating the mCpG/CpG ratio. We previously showed that DNA methylation levels can help predict alternative cassette exons (Gelfman et al. 2012); a closer inspection of our current results reveals that it is not the absolute methylation value that distinguishes constitutive exons from alternative ones but the differential in the ratio of mCpG/CpG between exon and introns, which is also dependent in the general GC environment. We find a significantly higher level of methylated CpGs in exons compared with introns, regardless of GC-content differential. However, differential mCpG/CpG is much higher (∼50%) in the leveled GC exon–intron architecture, where DNA methylation drops significantly in the intronic regions proximal to the exon. Remarkably, in this group there is no well-defined nucleosome marking the location of the exon relative to the flanking introns; nucleosomes mark exons significantly only when there is a substantial difference in GC content between exon and introns (Amit et al. 2012). Also, we previously showed that the exons exhibiting differential GC content and higher nucleosome occupancy compared to flanking introns are better recognized by the splicing machinery. Thus, CpG methylation can presumably be a part of the code that allows the splicing machinery to locate exons that have no GC differential. A possible explanation may be that the drop in DNA-methylation levels in proximal regions to the exon allows binding of certain proteins that can affect exon inclusion, such as is the case with transcriptional repressor CTCF protein (Shukla et al. 2011). On the other hand, by dividing exons based on RNA-seq data from H1 ES cells into alternative and constitutive, we identified a strong decrease in methylated CpGs in alternative exons as well as their flanking sequences compared with constitutive exons. This suggests that lower levels of DNA methylation of the whole intron–exon–intron strip are associated with suboptimal recognition of alternatively spliced exons.
The effect on splicing of a strong DNA-methylation signal in a leveled GC architecture may be indirectly through Pol II stalling. Since DNA methylation is a chromatin remodeler (Sarraf and Stancheva 2004; Klose and Bird 2006; Choy et al. 2010) and nucleosome occupancy causes Pol II stalling (Batsché et al. 2006; Hodges et al. 2009), these two mechanisms could be interlaced. Thus, it may be that peak CpG abundance at the splice sites might influence nucleosomal positioning. Through the use of the dinucleotide composition analyses, we found that in a leveled GC architecture nucleosome occupancy is strongly affected by the presence of CG dinucleotides. We consider the differences between the two GC groups as an indicator of different epigenetic interplays around exons in different GC environments.
It was previously shown that DNA sequence with high histone-binding affinity can direct nucleosome positioning and create a barrier to Pol II (Lowary and Widom 1998; Bondarenko et al. 2006). A recent paper by Bintu et al. (2012) suggested a mechanism by which there is an increase in pause density of Pol II in the central region of the nucleosomes (H3/H4 tetramer center) that is lessened in the entry and exit regions (first and last H2A/H2B dimmers, respectively). Taken together with the observed effect of CG composition on nucleosome occupancy, we propose that perhaps these findings point to a similar structure upon the exons that is governed by CpG methylation, where the peaks in CpG methylation at the 5′ and 3′ splice sites act as the central area binding for the nucleosomal barrier, whereas the drop in methylated CpGs in intronic flanking regions of leveled GC exons point to the entry/exit regions of the nucleosome. However, further investigation is required to better understand and tackle this hypothesis.
In conclusion, our findings provide an extensive, genome-wide overview of DNA methylation and nucleosome occupancy relative to exon–intron structures. The analyses here support a role for DNA methylation in splicing and in chromatin remodeling. Our data suggest that the regional location of exons within the genome impacts their GC environment and that this, in turn, has a significant effect on how the splicesomal machinery recognizes the exons. This should be taken into account when examining the effects of other epigenetic markers, such as histone modifications, upon exon–intron structures.
Methods
Construction of EST-based exon and intron data sets
Data sets of human exons for the differential GC group and the leveled GC group were retrieved based on the RefSeq tracks from the UCSC Genome Browser (http://genome.ucsc.edu/) as described previously by Amit et al. (2012). The differential GC group contained 15,874 exons and flanking introns, and the leveled GC group contained 16,269 exons and flanking introns. GC-content-based exonic groups for the mouse genome were also retrieved as described by Amit et al. (2012). The mouse differential GC group included 15,758 exons and flanking introns, and the mouse leveled GC group contained 16,583 exons and flanking introns. All exons examined are internal exons that are flanked on both sides by introns.
Data sets of human exons for the “desert” and “core” isochore groups were retrieved based on the RefSeq tracks from the UCSC Genome Browser (http://genome.ucsc.edu/). All internal exons of the human exome were divided into two groups based on the GC content of the isochore. Isochore maps were retrieved from Costantini et al. (2006). Exons that reside in regions of >46% GC content are considered core exons, and exons that reside in lower GC regions are considered desert exons. Overall, 85,979 exons and flanking introns construct the “desert” isochore group, of which 9649 are alternative exons; and 53,102 exons and flanking introns construct the “core” isochore group, of which 6443 are alternative exons.
Construction of RNA-seq-based leveled and differential GC exonic groups
For construction of constitutive and alternative exon data sets, we used RNA-seq raw reads that were obtained from Lister et al.( 2009). The mRNA-seq reads were aligned to the human genome (hg18) using the Bowtie alignment tool (Langmead et al. 2009). Next, we applied the SpliceTrap software (Wu et al. 2012) to quantify exon inclusion ratios from the mapped RNA-seq reads. SpliceTrap is a statistical tool built to quantify exon inclusion ratios from RNA-seq data. SpliceTrap quantifies for every exon the extent to which it is included, skipped, or subjected to size variations due to alternative 3′/5′ splice sites or Intron Retention. We identified 37,473 exons with canonical splice sites for which inclusion levels could be measured reliably with a minimum number of three reads per junction for each exon isoform. Next, we divided the identified exons into leveled and differential GC exons. For the differential GC group, we searched for exons with significantly higher GC content (t-test, P-value < 0.05) than their flanking introns, and for the leveled GC group we searched for exons with no significant differences in GC content between exons and flanking introns. We identified a total of 7413 exons in the differential GC group (5734 constitutive exons and 1679 alternative exons) and 6037 exons in the leveled GC group (4936 constitutive exons and 1101 alternative exons).
Construction of basic DNA methylation maps by base calls
Genome-wide single-base resolution data on DNA methylation was obtained from Lister et al. (2009), who conducted MethylC-seq analyses to map genome-wide DNA methylation for two human cell lines, H1 human embryonic stem cells and IMR90 fetal lung fibroblasts. In this method, they used bisulfite-conversion to convert cytosine to uracil (which is later transformed by PCR amplification to thymidine), while methylated cytosines are not affected. Next, high-throughput sequencing was processed using the Illumina analysis pipeline. The Bowtie alignment tool (Langmead et al. 2009) was used to align the results to the human reference genome (hg18). The base calls per reference position on each strand were used to identify methylated cytosines at 1% FDR.
To map the methylated cytosines, we cast single-base resolution values upon sets of genomic intervals (in this case, exons and introns), resulting in three matrices for each set of exons: upstream intron, exon, and downstream intron. Next, we construct two databases for the two splice sites. One database we centered on the 3′ slice site (intron positions are below zero and exon positions are above zero). The other database was centered on the 5′ splice site. All sequences were aligned to the plus strand; thus, methylation values of an exon that is coded on the minus strand are reversed. Once all exons and introns were positioned relative to the splice site, we calculated per base statistical measures of methylation values for all intervals in a genomic set. The exon–intron strip was constructed from 75-nt exonic nucleotides and 200 intronic nucleotides. Exons shorter than 75 nt received a null value for the remaining positions, and the same is done for introns shorter than 200 nt. For exons that are shorter than 150 nt, there is a duplication of the data at 5′ and 3′ splice sites. For example, for an exon with the length of 100 nt, positions 1–75 will be used for display at the 3′ end of the exon, and positions 25–100 will be used for display at the 5′ end of the exon. We used this method in order to be faithful to the true values with regard to each splice site and not to the middle of the exon. All scripts were written using the perl script language and the R statistical computing program (Team 2006), the latter was also used for both statistical analysis and graphical display.
DNA-methylation maps for the different human tissues and mouse ES cells were constructed using data retrieved from Gu et al. (2010), who performed genome-wide single-base DNA-methylation mapping of clinical samples using the RRBS protocol. The tissues examined were primary colon tumor, normal colon, blood cells from a healthy individual, and blood cells from an individual with colon cancer.
Calculation of the mCpG/CpG score
Calculation of the value of mCpG/CpG was done to take into account the original base calls of DNA methylation measured by Lister et al. (2009) and the number of CpGs in a certain position relevant to the exon–intron junction of all measured exons. This calculation is based on the base call methylation data sets of exons and introns that are positioned relative to the splice sites. For each nucleotide relative to the splice site, we extracted only the exons/introns that have CG dinucleotide in that position. Next, we calculated the average methylation level only in those cases. For example, in a data set of 20 exons, 10 exons in the differential GC group have a CpG at position −1 of the 5′ splice site. If methylation values are 0.8*4, 0.7*3, and 0.9*3, the mCpG/CpG value will be (0.8*4 + 0.7*3 + 0.9*3)/10 = 0.8. The same calculation in the leveled GC group will include more exons with methylation values (15/20) and more CpG positions (15). This method takes into account cases in which a certain exonic position is a CpG but the methylation base calls are zero (numerator can be zero). However, the method does not take into account cases in which the denominator is zero (there is no CpG at examined position of an examined exon).
CpG abundance around splice sites
The CpG abundance is the percentage of exons that have a CpG at an examined position. For example, in a data set of 100 exons, if two exons have a CpG at position +1 of the 3′ splice site and three exons have a CpG at position +2 of the 3′ splice site, the CpG abundances will be 0.02 and 0.03, respectively. CpG abundance was also calculated on a data set of 134,935 pseudo exons that was obtained from Ke et al. (2011). The pseudo exons were defined as intronic sequences having lengths between 50 and 250 nt and a splice site score of above 75 for 3′ splice site and above 78 for 5′ splice site, using the position specific scoring matrix (PSSM) method of Shapiro and Senapathy (1987). The consensus values are in the range of 0 to 100; the median value for a “real” exon 3′ splice site is 80, and that for a “real” exon 5′ splice site is 82. Pseudo exons that contain interspersed repeats were removed using RepeatMasker.
Construction of nucleosome occupancy maps for exons
We obtained nucleosome-occupancy levels using nucleosome score profiles generated by Schones et al. (2008). The maps were constructed using Illumina high-throughput sequencing of DNA fragments attached to nucleosomes in activated T cells, following micrococcal nuclease (MNase) digestion (Schones et al. 2008). In order to map nucleosome positioning upon the exon–intron genomic regions, we cast single-base values upon genomic intervals as was done for DNA-methylation data. Next, we applied a running average of 20 nt on the vector of average values for the exon–intron strip.
Nucleosome occupancy maps based on positional nucleotide composition were constructed using the same methods. We constructed nucleosome occupancy maps for exons based on their dinucleotide composition in 20 different positions along the exon–intron sequence. For the 3′ splice site, we constructed nucleosome occupancy data sets at positions −150, −20, −10, −6, −5, +1, +2, +3, +10, and +20 relative to the exon–intron junction. For the 5′ splice site, we constructed nucleosome occupancy data sets at positions −20, −10, −5, −3, −2, +4, +5, +10, +20, and +150 relative to the exon–intron junction. For each position, we constructed sub-data sets based on the following dinucleotide composition: (1) CG, all exons with a C at the position examined and G at the next position; (2) other C and G combinations: CC, GG, and GC; exons where the dinucleotide was either preceded by a CG dinucleotide (as in “CGC” or “CGG,” i.e., DGC\G) or followed by a CG dinucleotide (as in “CCG” or “GCG,” i.e., C\GCH) were excluded to prevent a case in which a methylated CpG in the vicinity to the position at hand will affect the result; (3) all other possible dinucleotide compositions at the requested and consecutive positions (AA, AT, AG, AC, etc.); and (4) AG, all exons with a C at the position examined and G at the next position; this subset of group 3 was used only for evaluation of position −2 of the 5′ splice site since it represents the consensus sequence at that position. We then constructed average nucleosome occupancy maps for each subgroup of exons separately. Since there is no necessity for inclusion level liability in this analysis yet there is a strong statistical need for large subgroups, we analyzed the larger exonic data sets that are based on the work by Amit et al. (2012) and not the smaller groups that originate from RNA-seq experiments.
Pictograms
Graphical representations of PSSMs were composed using a BioPerl (Stajich et al. 2002) module for generating Scalable Vector Graphics (SVG) output of Pictogram display for consensus motifs, as was described by Burge et al. (1999). The height of each letter is proportional to the frequency of the corresponding base at the given position, and bases are listed in descending order of frequency from top to bottom. Pictograms were constructed for the 5′ splice site of differential and leveled GC exons and also for a subgroup of exons with a CG dinucleotide composition at position −2 of the 5′ splice site.
Acknowledgments
This work was supported by grants from the Israel Science Foundation (ISF 61/09, ISF Bikura 838/10, and ISF Morasha 64/12) and by the Deutsche-Israel Project (DIP 6403). This study was supported in part by a fellowship from the Edmond J. Safra Center for Bioinformatics at Tel Aviv University.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.143503.112.
References
- Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S, Lev-Maor G, Burstein D, Schwartz S, Postolsky B 2012. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep 1: 543–556 [DOI] [PubMed] [Google Scholar]
- Anastasiadou C, Malousi A, Maglaveras N, Kouidou S 2011. Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers. DNA Cell Biol 30: 267–275 [DOI] [PubMed] [Google Scholar]
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J 2009. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res 19: 1732–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsché E, Yaniv M, Muchardt C 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol 13: 22–29 [DOI] [PubMed] [Google Scholar]
- Bernardi G, Olofsson B, Filipski J, Zerial M, Salinas J, Cuny G, Meunier-Rotival M, Rodier F 1985. The mosaic genome of warm-blooded vertebrates. Science 228: 953–958 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES 2007. The mammalian epigenome. Cell 128: 669–681 [DOI] [PubMed] [Google Scholar]
- Bestor TH 2000. The DNA methyltransferases of mammals. Hum Mol Genet 9: 2395–2402 [DOI] [PubMed] [Google Scholar]
- Bintu L, Ishibashi T, Dangkulwanich M, Wu YY, Lubkowska L, Kashlev M, Bustamante C 2012. Nucleosomal elements that control the topography of the barrier to transcription. Cell 151: 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM 2006. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell 24: 469–479 [DOI] [PubMed] [Google Scholar]
- Burge CB, Tuschl T, Sharp PA 1999. Splicing of precursors to mRNAs by the spliceosomes. In The RNA World (Cold Spring Harbor Monograph Series 37), pp. 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Cedar H, Bergman Y 2012. Programming of DNA methylation patterns. Annu Rev Biochem 81: 97–117 [DOI] [PubMed] [Google Scholar]
- Chen W, Luo L, Zhang L 2010. The organization of nucleosomes around splice sites. Nucleic Acids Res 38: 2788–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. 2010. Relationship between nucleosome positioning and DNA methylation. Nature 466: 388–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Bae JB, Lyu J, Kim TY, Kim YJ 2009. Nucleosome deposition and DNA methylation at coding region boundaries. Genome Biol 10: R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Wei S, Lee JY, Tan S, Chu S, Lee TH 2010. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc 132: 1782–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini M, Clay O, Auletta F, Bernardi G 2006. An isochore map of human chromosomes. Genome Res 16: 536–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- Gelfman S, Burstein D, Penn O, Savchenko A, Amit M, Schwartz S, Pupko T, Ast G 2012. Changes in exon–intron structure during vertebrate evolution affect the splicing pattern of exons. Genome Res 22: 35–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Karlin S 2001. Genome-scale compositional comparisons in eukaryotes. Genome Res 11: 540–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Bock C, Mikkelsen TS, Jager N, Smith ZD, Tomazou E, Gnirke A, Lander ES, Meissner A 2010. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods 7: 133–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C 2009. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325: 626–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe B 2011. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res 21: 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari K, Bernardi G 1998. CpG doublets, CpG islands and Alu repeats in long human DNA sequences from different isochore families. Gene 224: 123–127 [DOI] [PubMed] [Google Scholar]
- Jabbari K, Caccio S, Pais de Barros JP, Desgres J, Bernardi G 1997. Evolutionary changes in CpG and methylation levels in the genome of vertebrates. Gene 205: 109–118 [DOI] [PubMed] [Google Scholar]
- Karlin S, Burge C 1995. Dinucleotide relative abundance extremes: A genomic signature. Trends Genet 11: 283–290 [DOI] [PubMed] [Google Scholar]
- Ke S, Shang S, Kalachikov SM, Morozova I, Yu L, Russo JJ, Ju J, Chasin LA 2011. Quantitative evaluation of all hexamers as exonic splicing elements. Genome Res 21: 1360–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP 2006. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. 2009. Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu J, Tian G, Li N, Li Q, Ye M, Zheng H, Yu J, Wu H, Sun J 2010. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol 8: 669–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J 1998. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276: 19–42 [DOI] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T 2011. Epigenetics in alternative pre-mRNA splicing. Cell 144: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer KM 2002. On the importance of being co-transcriptional. J Cell Sci 115: 3865–3871 [DOI] [PubMed] [Google Scholar]
- Nikolaou C, Althammer S, Beato M, Guigó R 2010. Structural constraints revealed in consistent nucleosome positions in the genome of S. cerevisiae. Epigenetics Chromatin 3: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes CC, La Salle S, Smiraglia DJ, Robaire B, Trasler JM 2007. A unique configuration of genome-wide DNA methylation patterns in the testis. Proc Natl Acad Sci 104: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ 2002. Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2006. R: A language and environment for statistical computing (R Foundation for Statistical Computing, Vienna, Austria). http://www.R-project.org/
- Reik W 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447: 425–432 [DOI] [PubMed] [Google Scholar]
- Robertson KD 2002. DNA methylation and chromatin—unraveling the tangled web. Oncogene 21: 5361–5379 [DOI] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I 2004. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell 15: 595–605 [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR 2009. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci 106: 4325–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G 2009. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol 16: 990–995 [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P 1987. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res 15: 7155–7174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S 2011. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479: 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484: 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell 36: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495 [DOI] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JGR, Korf I, Lapp H 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12: 1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xi S, Briones V, Muegge K 2010. Lsh mediated RNA polymerase II stalling at HoxC6 and HoxC8 involves DNA methylation. PLoS ONE 5: e9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R 2009. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol 16: 996–1001 [DOI] [PubMed] [Google Scholar]
- Tillo D, Hughes TR 2009. G + C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Field Y, Lieb JD, Widom J, Segal E, Hughes TR 2009. High nucleosome occupancy is encoded at human regulatory sequences. PLoS ONE 5: e9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varriale A, Bernardi G 2010. Distribution of DNA methylation, CpGs, and CpG islands in human isochores. Genomics 95: 25–28 [DOI] [PubMed] [Google Scholar]
- Vignali M, Hassan AH, Neely KE, Workman JL 2000. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol 20: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MC, Wilson MD, Barbosa-Morais NL, Schmidt D, Stark R, Pan Q, Schwalie PC, Menon S, Lukk M, Watt S, et al. 2012. Latent regulatory potential of human-specific repetitive elements. Mol Cell 49: 262–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Akerman M, Sun S, McCombie WR, Krainer AR, Zhang MQ 2012. SpliceTrap: A method to quantify alternative splicing under single cellular conditions. Bioinformatics 27: 3010–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B 2012. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148: 816–831 [DOI] [PMC free article] [PubMed] [Google Scholar]