Abstract
Genome-wide chromatin annotations have permitted the mapping of putative regulatory elements across multiple human cell types. However, their experimental dissection by directed regulatory motif disruption has remained unfeasible at the genome scale. Here, we use a massively parallel reporter assay (MPRA) to measure the transcriptional levels induced by 145-bp DNA segments centered on evolutionarily conserved regulatory motif instances within enhancer chromatin states. We select five predicted activators (HNF1, HNF4, FOXA, GATA, NFE2L2) and two predicted repressors (GFI1, ZFP161) and measure reporter expression in erythroleukemia (K562) and liver carcinoma (HepG2) cell lines. We test 2104 wild-type sequences and 3314 engineered enhancer variants containing targeted motif disruptions, each using 10 barcode tags and two replicates. The resulting data strongly confirm the enhancer activity and cell-type specificity of enhancer chromatin states, the ability of 145-bp segments to recapitulate both, the necessary role of regulatory motifs in enhancer function, and the complementary roles of activator and repressor motifs. We find statistically robust evidence that (1) disrupting the predicted activator motifs abolishes enhancer function, while silent or motif-improving changes maintain enhancer activity; (2) evolutionary conservation, nucleosome exclusion, binding of other factors, and strength of the motif match are predictive of enhancer activity; (3) scrambling repressor motifs leads to aberrant reporter expression in cell lines where the enhancers are usually inactive. Our results suggest a general strategy for deciphering cis-regulatory elements by systematic large-scale manipulation and provide quantitative enhancer activity measurements across thousands of constructs that can be mined to develop predictive models of gene expression.
Genome-wide genetic association studies suggest that nearly 85% of disease-associated variants lie outside protein-coding regions (Hindorff et al. 2009), emphasizing the importance of a systematic understanding of regulatory elements in the human genome at the nucleotide level. In recent years, the prediction of human regulatory regions has benefited tremendously from advances in high-throughput experimental (Bernstein et al. 2010; Myers et al. 2011), computational (Berman et al. 2002; Sinha et al. 2008; Warner et al. 2008), and comparative (Bejerano et al. 2004; Moses et al. 2004; Xie et al. 2005; Kheradpour et al. 2007; Visel et al. 2008; Lindblad-Toh et al. 2011) methods, leading to a large number of putative regulatory elements (Pennacchio et al. 2006; Visel et al. 2009). The dissection of individual sequences and their evaluation in transient assays led to a greatly increased understanding of enhancer biology for human (Ney et al. 1990; Liu et al. 1992), fly (Zeng et al. 1994; Kapoun and Kaufman 1995), and worm (Jantsch-Plunger and Fire 1994). However, the dissection of regulatory motifs within enhancer elements has remained unfeasible at the genome scale (Baliga 2001; Patwardhan et al. 2009; Fakhouri et al. 2010). Moreover, the interplay of activators and repressors in establishing spatial domains of expression has been long studied, particularly in fly development (Stanojevic et al. 1991; Gompel et al. 2005).
In this work, we build on recent studies that have used genome-wide chromatin maps to predict thousands of candidate distal enhancer regions across multiple human cell types (Barski et al. 2007; Heintzman et al. 2009; Hesselberth et al. 2009; Ernst and Kellis 2010; Ernst et al. 2011), and we seek to characterize experimentally specific nucleotides within them that are important for their function. Regulatory element predictions typically span several hundred nucleotides, and their validation has also typically been at the level of regions spanning thousands of nucleotides (Pennacchio et al. 2006; Visel et al. 2009). Individual nucleotides were perturbed for only a handful of putative enhancers in a directed way (Ernst et al. 2011), limiting our understanding of the role of individual regulatory motifs and motif positions in establishing enhancer activity. This situation is remedied by recently developed massively parallel reporter assays (Melnikov et al. 2012; Patwardhan et al. 2012; Sharon et al. 2012; Arnold et al. 2013) that take advantage of large-scale sequencing to simultaneously measure the reporter activity of thousands of enhancer variants. However, these assays have only been used to dissect four human and one mouse enhancers, leaving open the question of what fraction of genome-wide regulatory predictions can be experimentally validated at the single-nucleotide level.
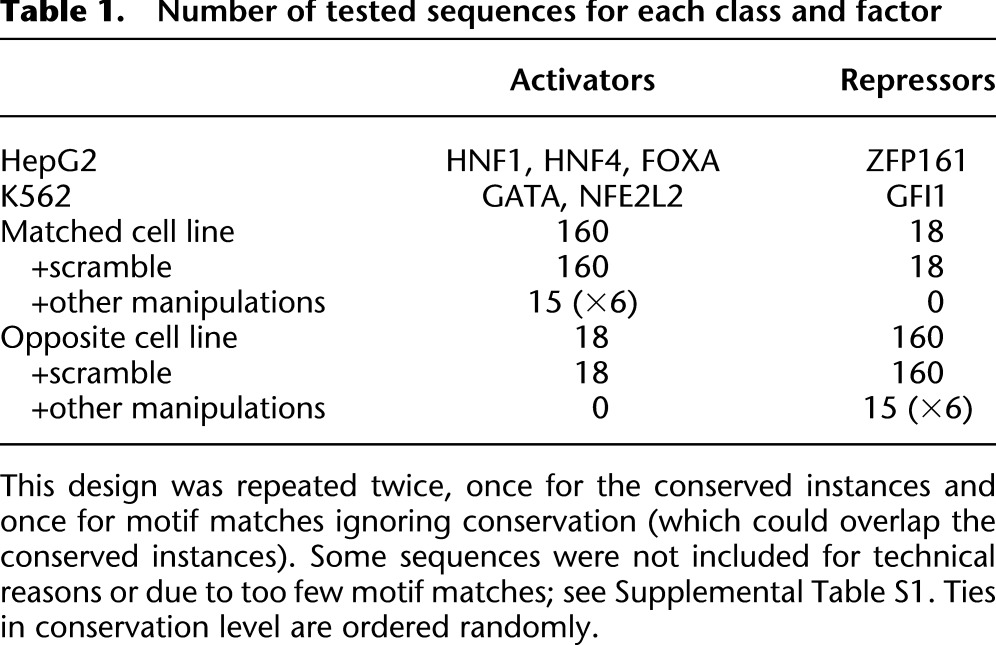
In order to match the genome-scale nature of regulatory predictions, we sought to experimentally test the role of regulatory motif predictions in 2104 candidate enhancers in two human cell lines (Ernst et al. 2011). We synthesized a library of enhancer constructs using microarray oligonucleotide synthesis, containing the wild-type enhancer sequences and specific variants (Table 1; Supplemental Table S1) that remove, disrupt, or improve the predicted causal regulatory motif instances for five predicted activators (HNF1, HNF4, FOXA, GATA, NFE2L2) and two predicted repressors (GFI1, ZFP161). For each variant, we tested 145 nucleotides of the enhancer element upstream of a SV40 promoter sequence and a luciferase ORF reporter coupled with a 10-nt unique tag. We transfected the resulting pool of plasmids into two human cell lines using 10 different tags for each construct, enabling us to measure the transcriptional levels induced by thousands of short DNA segments in vivo.
Table 1.
Number of tested sequences for each class and factor
Our study has several important implications. First, we demonstrate that short 145-bp enhancer segments can capture differences in reporter expression between erythroleukemia (K562) and liver carcinoma (HepG2) cell lines. Second, we report >21,672 distinct enhancer reporter assay measurements for thousands of distinct human enhancers, producing a resource in human cell lines nearly as big as the largest mouse enhancer resource (Visel et al. 2007). Third, while most previous approaches to systematic enhancer testing have been restricted to wild-type enhancers, we demonstrate the feasibility of directed mutations in thousands of distinct human enhancers. Lastly, our enhancer variants are engineered on the basis of predictive models of enhancer function, directly disrupting predicted activating and repressing regulatory motifs, and thus enabling the validation of a dramatically larger number of regulatory elements than what is permitted by exhaustive enumeration approaches. Our results lead to numerous new insights and systematic confirmations regarding gene regulation, including the central role of sequence specificity in enhancer activity, the role of repressor motifs in shaping enhancer tissue specificity, and a quantification of the relative role of context information in establishing wild-type enhancer activity.
Results
Study design and enhancer selection
To multiplex enhancer validation assays, we leverage large-scale oligonucleotide array synthesis (LeProust et al. 2010) and high-throughput tag sequencing in a massively parallel reporter assay (Melnikov et al. 2012). Briefly, we constructed a pool of ∼54,000 distinct plasmids, each containing a candidate enhancer element upstream of a heterologous GC-rich promoter, and a reporter gene that contains a unique 10-bp tag (see Fig. 1C; Methods). We tested 145-bp elements, as the combined length of the tested enhancer, tag, and primer sequences is constrained to 200-bp oligonucleotides. We transfected the plasmid pool in vitro into human cell lines, isolated mRNAs transcribed from the plasmids, and then sequenced the PCR-amplified tags corresponding to each enhancer element. The resulting tag counts provided a reproducible digital gene expression-level readout of enhancer activity (Supplemental Fig. S1), enabling us to use this approach to test large numbers of candidate human enhancers. K562 cells are harder to transfect and consequently have a higher level of noise, leading to lower correlation values between replicates (r = 0.36 for K562 vs. 0.69 for HepG2).
Figure 1.
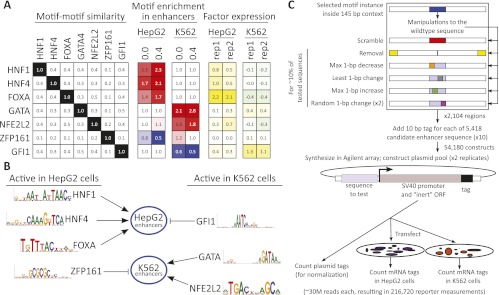
Selection of activator and repressor motifs. (A) Predicted activator and repressor motifs were chosen based on their lack of similarity to each other (left) (Supplemental Fig. S2); fold-enrichment for activators (red) and fold-depletion for repressors (blue) in the cell line of interest (middle); and microarray expression (Ernst et al. 2011) of the corresponding factor in the target cell line (log2, right). Black-white, red-blue, and green-yellow color gradients are used for emphasis, but all values are indicated. (B) Predicted activators and repressors for each cell type and corresponding motifs. HNF1, HNF4, and FOXA are predicted to act as activators of HepG2 enhancers in HepG2 cells. GATA and NFE2L2 are predicted to act as activators of K562 enhancers in K562 cells. GFI1 is predicted to act as a repressor of HepG2 enhancers in K562 cells, and ZFP161 is predicted to act as a repressor of K562 enhancers in HepG2 cells. Details on selection criteria and motif sources are available in Supplemental Figure S2. (C) For each of 2104 predicted enhancer regions, we designed between two and eight variants (colors as in Fig. 3A), each tested in two biological replicates in two cell lines, using 10 different tags per sequence. We also sequenced the plasmid library directly to provide tag counts used for normalization. A single Agilent array is thus used to obtain 54,180 reporter expression levels for 5418 enhancer variants.
We use this technology to validate predictive models of regulatory motif function within putative human enhancers. We focused on liver carcinoma (HepG2) and erythrocytic leukemia (K562) cell lines, for which rich experimental data sets are available due to their prioritized role in ENCODE (Myers et al. 2011). For both cell lines, we carried out genome-wide predictions of enhancer elements based on their chromatin states, defined by combinations of histone modifications (Ernst et al. 2011).
We then predicted relevant regulatory motifs for each cell line (Fig. 1A; Supplemental Fig. S2). Starting with a collection of 688 motifs (see Methods) we identified those that showed significant enrichment or depletion in cell line-specific enhancers for either HepG2 or K562 (Supplemental Fig. S2, middle). Notably, we found that when we only considered motif instances that were more highly conserved in 29 mammals (Lindblad-Toh et al. 2011), the enrichment or depletion levels tended to be more pronounced. Using motif–motif similarity as a guide and seeking motifs with higher levels of enrichment or depletion, we selected from this initial set of motifs a total of seven nonredundant motifs (Fig. 1A, left). When a motif was enriched in the enhancers for a cell line, we reasoned it may be involved in establishing enhancers and is likely an activator. We predicted three activators for HepG2 cells: HNF1, HNF4, and FOXA, all three known to regulate liver development (Courtois et al. 1987; Costa et al. 2003), and two for K562 cells: the hematopoiesis regulator family GATA (Weiss and Orkin 1995) and NFE2L2.
Conversely, we reasoned that motif depletion is a signature of a repressor because it suggests that motif absence is a condition for enhancer activity: GFI1 showed motif depletion in K562 enhancers and is indeed a known hematopoietic repressor (Hock and Orkin 2006); ZFP161, another known repressor (Sobek-Klocke et al. 1997; Orlov et al. 2007), showed motif depletion in HepG2 enhancers.
While the sharing of motifs across factors and post-translation modifications limit the interpretability of expression in this context, we found that for five of these seven motifs the corresponding factor had higher expression in the cell line, where motif enrichment or depletion was noted (Fig. 1A; Supplemental Fig. S2, right). The two exceptions are NFE2L2, which appears to be active in both cell lines, and ZFP161, which is the only factor we do not ultimately validate (see below).
Based on these regulatory predictions, we made specific hypotheses about the likely effect of individual motif disruptions for both activator and repressor motifs. For each regulator, we selected 178 enhancer regions centered on highly conserved motif occurrences in 29 mammals (Lindblad-Toh et al. 2011), and 178 enhancer regions centered on motif matches without regard to conservation (Table 1). In each case, 160 of the 178 were selected in enhancer chromatin states from the cell line with higher motif enrichment, and 18 were selected in enhancer states from the other cell line for control purposes. For each of 2104 wild-type enhancers, we tested one variant with a scrambled motif (Supplemental Fig. S3), and for a subset of 204 enhancers we also tested additional variants with diverse changes, including complete motif removal, single-nucleotide changes that maximally reduce, minimally change, or maximally increase the motif match score, and two random single-nucleotide changes. Except for the complete removal of the motif, which incorporates additional flanking genomic sequence to fill the 145 bp, none of the manipulations change the tested sequence outside the motif match. We tested a total of 5418 distinct sequences, which lacked systematic similarity to each other (see Methods), each using 10 different tags and two biological replicates in each cell type to provide a robust estimate of its activity, resulting in a total of 216,720 expression measurements (Supplemental Data S1).
Activator motifs
Our results support the role of activator motifs in enhancer function. For example, a HepG2-specific enhancer containing an HNF4 motif on chromosome 9 between ACTL7B and KLF4 (Fig. 2A,B) shows consistently high activity in HepG2 cells, as measured by all 20 tag replicates (Fig. 2C). The same region lies in a repressive chromatin state in K562 and, indeed, the reporter gene shows no expression when tested in K562 cells. The enhancer activity is abolished when the motif is scrambled, removed, or when highly informative motif positions 10 or 13 are mutated. The reporter expression remains consistently high in silent mutations that maintain or improve the position weight matrix (PWM) scores. These results were significant across 160 HNF4-containing enhancers in two cell lines (Fig. 3; Supplemental Fig. S4B), confirming that binding to the HNF4 motif as captured by the PWM score is required for enhancer activity specific to HepG2 cells.
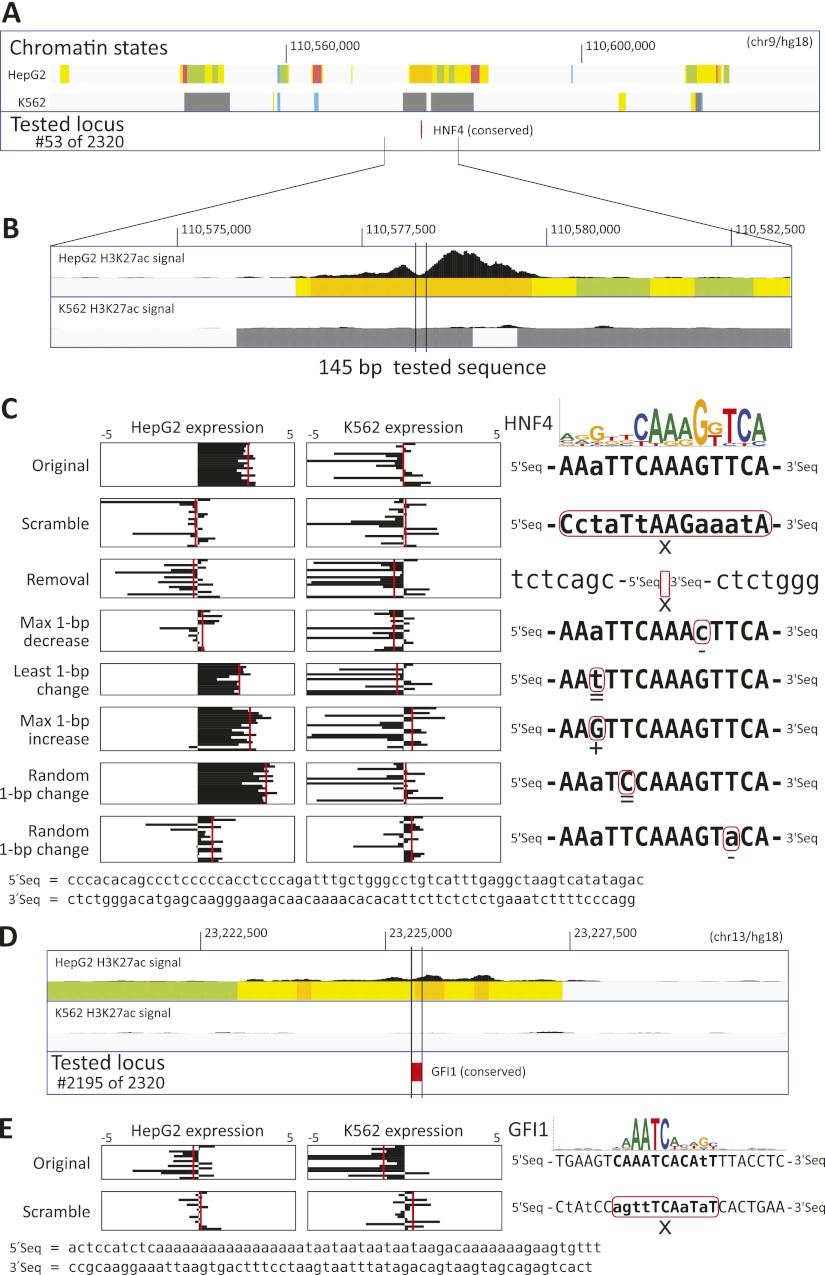
Figure 2.
Example activator and repressor motif manipulations (for all tested, see Supplemental Data S1). (A) HepG2 enhancer centered on a HNF4 motif (#53). Chromatin state tracks (Ernst et al. 2011) indicate promoters (red), poised promoters (purple), strong/weak enhancers (orange/yellow), insulators (blue), transcribed (green), repressed (gray), and low-signal/repetitive (light gray) regions. (B) The H3K27ac signal in HepG2 shows a dip on the HNF4 motif, consistent with nucleosome exclusion due to TF binding. (C) The original sequence shows expression (replicates in black, mean in red) in HepG2 but not K562, confirming the predicted cell-type specificity. Motif disruptions (scramble, removal, max 1-bp decrease, and the second random) eliminate HepG2 expression, while neutral and motif-improving changes do not, supporting the PWM model. The positions matching the motif consensus are indicated in uppercase. (D) HepG2 enhancer centered on a GFI1 instance (#2195), predicted to be repressed in K562 where GFI1 is active. (E) Expression for the original sequence in K562 is below baseline, confirming repression. Upon scrambling the motif, aberrant expression is seen in K562, where GFI1 is predicted to be a repressor, while no change is seen in HepG2.
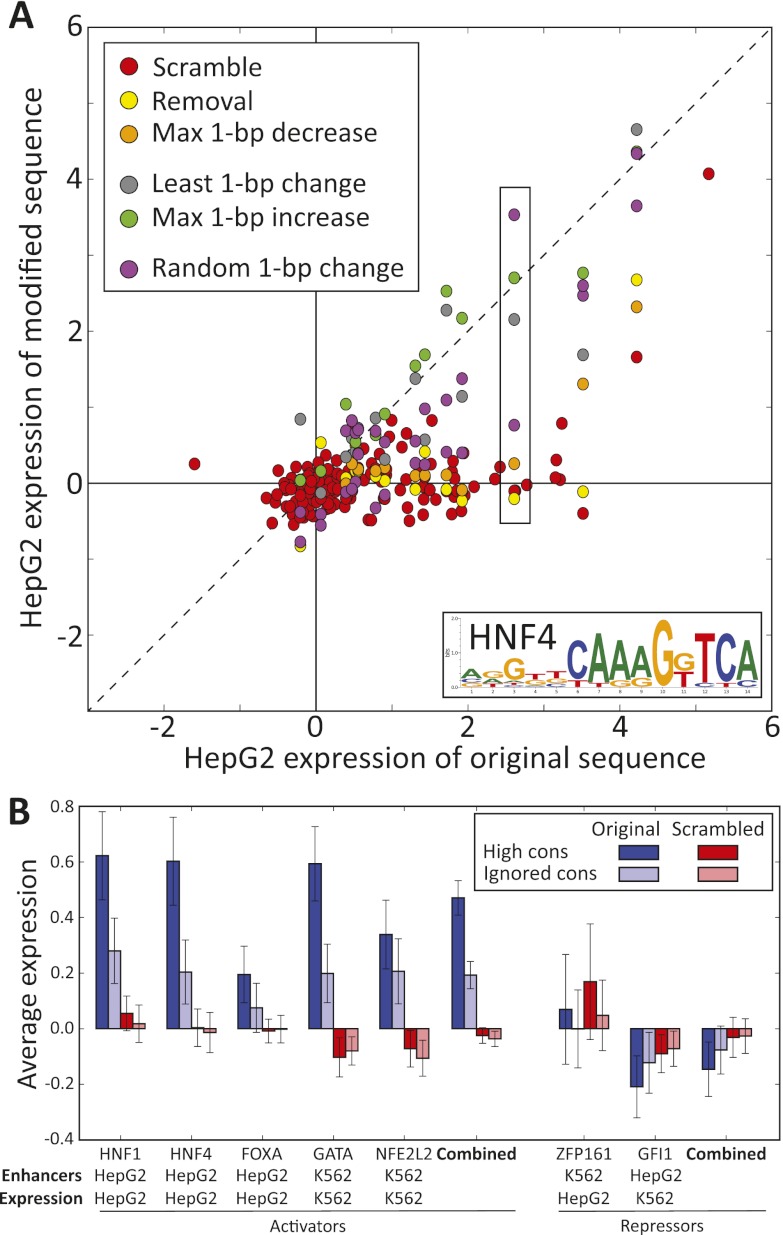
Figure 3.
Summary of motif manipulation results for all activators and repressors tested. (A) Average reporter gene expression for 160 predicted HepG2 enhancers centered on conserved HNF4 motifs for wild-type construct expression (x-axis) and modified construct expression (y-axis) for different modifications. A total of 160 constructs with scrambled motifs (red) consistently lie near the y-axis (no reporter expression), confirming the necessity of the conserved HNF4 motif. Five additional motif modifications were tested for the 15 most conserved HNF4 motifs. The preponderance of disruptive modifications (red, yellow, and orange points) showing decreased reporter expression (below the diagonal) demonstrate the dramatic reduction of enhancer activity for the most disruptive mutations, while the presence of neutral (gray) or motif-strengthening (green) modifications near and above the diagonal highlight the specificity of mutations to those that disrupt recognition of the motif. Box indicates example shown in Figure 2A–C. (B) Comparison of reporter expression for enhancers centered on five activators in the matched cell type and two repressors in the unmatched cell type. For the five predicted activators, wild-type reporter expression is higher for 160 enhancers centered on conserved motifs (dark blue) than for 160 enhancers centered on motifs ignoring conservation (light blue), and it is significantly reduced after motif scrambling (red, pink). For the two predicted repressors, motif scrambling results in increased reporter expression in the unmatched cell type (see model in Fig. 6). Error bars represent 95% confidence interval on the mean. Additional bar plots in Supplemental Figure S4. All statistics are shown in Supplemental Figure S2. All expression values in this figure are computed as described in the Methods.
The motif scrambling analysis strongly confirmed the central role of all predicted causal motifs for all five activators for establishing enhancer activity in their respective cell line (Fig. 3B). Reporter expression was consistently reduced to background levels when the predicted activator motifs were scrambled. HNF1, HNF4, GATA, and NFE2L2 were individually significant, both for conserved motifs (each Wilcoxon P-value PW < 10−10) and for motifs ignoring conservation (each PW < 10−3). Summed across all five activators, the results were striking for both conserved (combined PW = 2.9 × 10−54) and nonconserved motifs (combined PW = 5.1 × 10−17).
Each additional modification was consistent with the predicted affinity of each TF motif (Supplemental Figs. S4A, S5A). Similarly, we found significant reduction when the motif was removed (combined PW = 1.5 × 10−4) and when the single most informative base was mutated (PW = 1.7 × 10−6). Moreover, single-nucleotide modifications that increase the motif match score resulted in a significant increase in expression (PW = 5.6 × 10−3). Neutral changes that do not affect the motif-binding affinity showed no significant change in expression from the wild-type enhancer (PW = 0.08) but were significantly more expressed than the scramble (PW = 3.4 × 10−7). Lastly, for random manipulations, we confirmed that changes in expression correlated with the change in motif match score (permutation PP = 2.8 × 10−3 for wild-type expression score > 0.5; Supplemental Fig. S6). The strong agreement with the PWM-predicted changes is consistent with the accuracy of the PWM models (Benos et al. 2002) and suggests that reporter activity is correlated with binding affinity when all else is maintained unchanged.
We estimated the proportion of enhancers that are functional in the matched cell line using two complementary approaches. First, we compared the fraction of sequences whose reporter expression decreased upon motif scrambling to what we would expect if no sequences were functional. We found that 71% of the 799 sequences we tested with conserved activator motifs had a reduction in reporter expression upon motif scrambling (Supplemental Fig. S7). We expect the fraction of functional enhancers that depend on their motif instances, f, to satisfy the equation f + (1 − f)/2 = 71%, because conservatively all of the functional instances and half of the nonfunctional instances should reduce in expression upon motif scrambling. Solving this equation gives us an estimate of f = 42% of sequences with conserved activator motifs being functional. Conversely, only 61% of sequences where motif instances were chosen ignoring conservation had reduced expression upon motif scrambling, leading to an estimate of f = 23%. These estimates are conservative, however, because they expect that scrambling a functional motif always leads to a detectably lower level of expression, never producing a better binding site (e.g., for another factor) by chance.
In our second approach, we computed an expression P-value (one-tailed Mann-Whitney) for each tested sequence by comparing its replicate values to those of all scrambled sequences, which we took as a baseline (Supplemental Tables S2, S3). At a P-value threshold of 0.05, 41% of the 793 sequences tested with conserved activator motifs had significant expression in the matched cell line, compared with only 9% of the same sequences with scrambled motifs. For sequences selected ignoring motif conservation, 25% were significant compared with 8% of the scrambled counterparts. Moreover, the fraction of sequences that are detected for each manipulation generally agrees with the expected effect of the manipulation (Supplemental Table S2). This second approach has the additional advantage that it can pinpoint which of the tested sequences is functional.
Both of these estimates likely underestimate the true number of functional enhancers, because some enhancers may require additional context not captured in the 145 bp we tested, and because some enhancers may be incompatible with the SV40 promoter.
Enhancer context
We also used our experimental results to gain insights into the sequence determinants of wild-type enhancer activity, which continues to be an unsolved challenge in genomics (King et al. 2005; Su et al. 2010). For example, the exact same NFE2L2 motif match sequence associated with different enhancer context information led to dramatically different wild-type expression levels (Supplemental Fig. S8), emphasizing the importance of the ∼135-nt sequence context. We sought features that distinguished the most versus least expressed 25% tested sequences (described here), and also the sequences showing the greatest reduction versus the least reduction upon motif scrambling (Supplemental Fig. S10).
When restricting our analysis only to those sequences that were chosen without respect to motif conservation in order to avoid confounding issues, we found several properties that distinguish the most expressed from least expressed enhancers (Fig. 4). Evidence of nucleosome exclusion based on dips in the H3K27 acetylation signal (He et al. 2010; Ernst et al. 2011) and DNase I hypersensitivity (Song et al. 2011) were seen coincident with the highly expressed sequences (Mann-Whitney PU = 6 × 10−12 and PU = 2 × 10−9, respectively). A stronger PWM score was also predictive of more highly expressed sequences (PU = 5 × 10−3). Moreover, a greater number of matching motifs with additional TFs were found in the enhancer context (3.7 vs. 2.8 factors on average, PU = 2 × 10−4), but very few of the tested sequences had additional occurrences of the tested motif (average number of instances: nine vs. four per hundred for the top vs. bottom 25%; PU = 0.34).
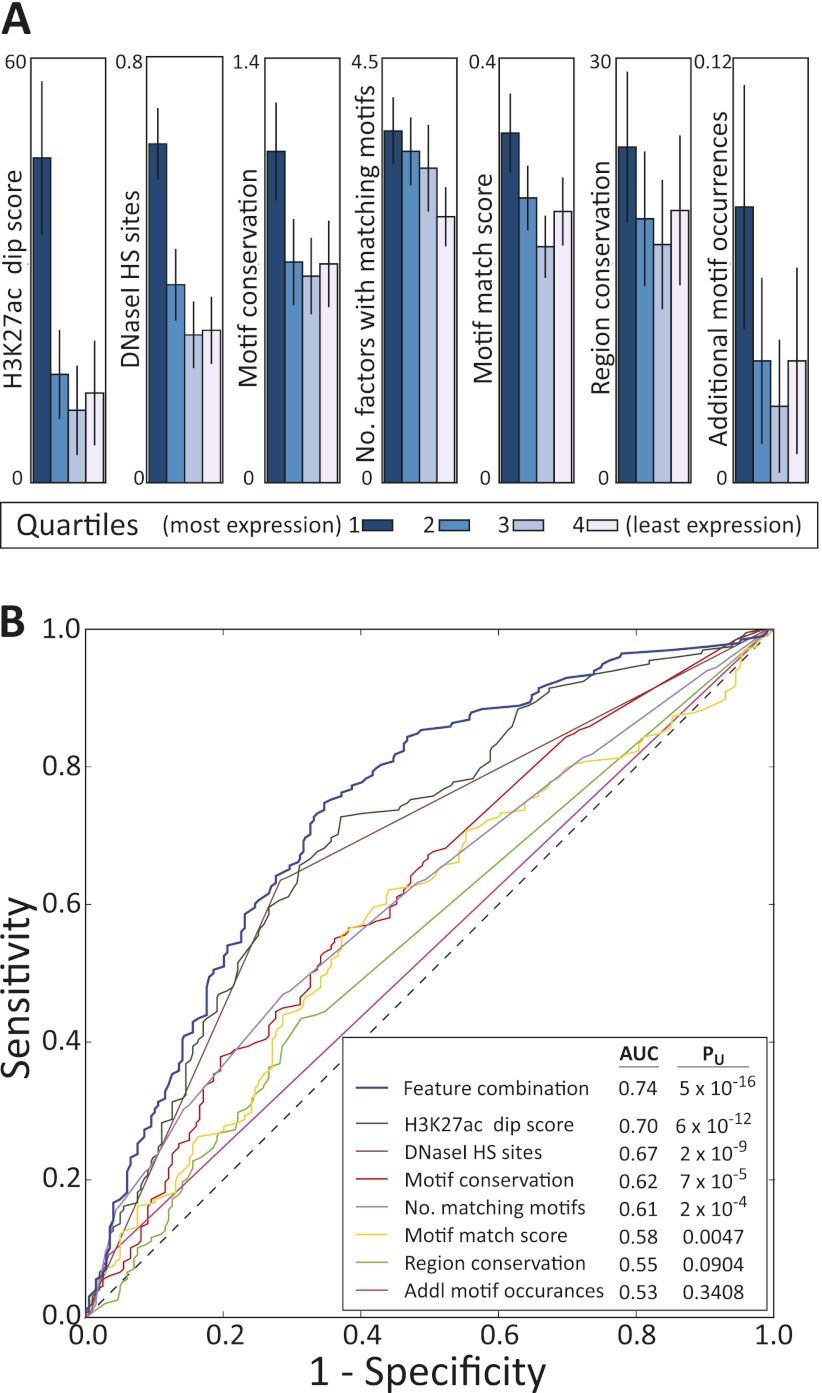
Figure 4.
Importance of sequence context for enhancer function. (A) Association of top scoring enhancers with: the average H3K27ac signal value in the matched cell type 200 bp away, minus the value centered on the motif (in 25-bp windows); overlap with DNase I annotations in the matched data (Song et al. 2011); the raw motif conservation score (Kheradpour et al. 2007; Lindblad-Toh et al. 2011); the number of factors with matching motifs in regions outside of the motif match in the tested sequence; the strength of the motif match; the number of bases indicated as conserved by SiPhy-ω 12-mers (Garber et al. 2009); and the number of matches to the tested motif within the tested sequence. (B) Predictive power for recognizing enhancers that are likely to show high wild-type reporter expression based on each of these individual features and a combination of features using logistic regression (Hall et al. 2009).
Evolutionary conservation of the motif and region tested was also predictive of reporter activity, consistent with evidence of functionality. The tested motif had a higher conservation level (Kheradpour et al. 2007; Lindblad-Toh et al. 2011) for enhancers with higher reporter activity (PU = 7 × 10−5). However, overall conservation of the entire sequence (Lindblad-Toh et al. 2011) did not provide significant discriminative power (Fig. 4). This is likely indicative of our strategy for selecting candidate enhancers based on chromatin state and regulatory motif conservation, which leads to a very narrow region of high conservation (Supplemental Fig. S11), in contrast to previous strategies that initially focused on high regional conservation (Pennacchio et al. 2006; Visel et al. 2008). Interestingly, amongst candidates with conserved sequence motifs, the highest reporter expression was associated with lower neighboring sequence constraint (64.7 conserved bases for the top 25% vs. 75.3 for the bottom 25%, PU = 2 × 10−5; Supplemental Fig. S12). This suggests that the specificity of sequence conservation to the motif is informative of likely enhancer function, perhaps because high overall conservation is due to reasons independent of the motif occurrence.
Overall, none of these seven tested features explains a large portion of the variance in the expression values (e.g., R2 = 9.1 to 16.4% for H3K27ac dip across the five activators), indicating that reporter gene expression levels strongly depend on additional features that remain to be characterized. Because the wild-type sequences have very similar sequence biases as their motif scrambled counterparts, we reason that experimental biases play a relatively small role in explaining differential expression. A logistic regression combination of these features led to a modest increase in performance compared with the best individual feature, suggesting that no one feature completely captures the likelihood of activity (Supplemental Fig. S9).
Repressor motifs
We next turned to the two predicted repressors, GFI1 and ZFP161, whose motifs were depleted in K562 and HepG2 enhancers, respectively (Fig. 1A), suggesting that they act as repressors in the corresponding cell type. We designed experiments that test enhancer repression in a cell line where the enhancer is not usually active (Supplemental Fig. S9), reasoning that mutating repressor motifs would lead to aberrant expression by abolishing repression.
Indeed, we found that HepG2 enhancers containing conserved GFI1 motif instances showed a significant increase in K562 reporter expression after scrambling of the GFI1-predicted repressor motif (PW = 3.7 × 10−2) (Fig. 2D,E), supporting our model that GFI1 acts as a repressor of HepG2-specific enhancers in K562 cells (Fig. 3B). Also as predicted, we found no change in enhancer activity when HepG2 enhancers with scrambled GFI1 motifs were tested in HepG2 cells (PW = 0.58), as the GFI1 repressor was only predicted to act in K562 cells (Fig. 2). Repressor activity was not validated for ZFP161, possibly because it was erroneously identified as a HepG2 repressor, or because we tested an insufficient number of functional sites to produce statistical significance. Alternatively, additional signals may maintain HepG2 repression even without repression by ZFP161.
Lastly, we confirmed that manipulation of activator motifs only led to expression changes in the matched cell lines where the corresponding activator protein was expressed. This was true for four of the five activators (Supplemental Fig. S4B), with the notable exception of NFE2L2, suggesting that it is also active in HepG2, which has indeed been previously reported (Gong and Cederbaum 2006). This suggests that the techniques used here may be useful more broadly for identifying factors active in a cell line, although the corresponding motifs would have to be known.
Luciferase validation with longer constructs and diverse promoters
In order to study the extent to which our results were affected by the promoter used in the assay and the 145-bp length of the tested sequences, we used a lower throughput experimental approach to validate the motif disruptions on 10 loci. We selected sequences with conserved motifs at a range of massively parallel reporter assay (MPRA) expression values, and generated constructs with the wild-type and motif scrambled sequences tested in MPRA, and an additional 177 bp upstream and 178 bp downstream (total of 500 bp). We measured the enhancer activity of these sequences with a luciferase assay using both the original SV40 promoter used with MPRA and also a minimal TATA promoter (see Methods).
We found a strong correlation between high-throughput and low-throughput assays and across promoter types (Fig. 5). Using the original SV40 promoter, the change in expression observed after motif scrambling for MPRA and luciferase showed r = 0.84 correlation (permutation PP = 8 × 10−6), confirming that the measured effects are robust to the length of the construct (145 vs. 500 bp) and the reporter technology (MPRA vs. luciferase). We also found a strong correlation between the TATA promoter and the SV40 promoter in the luciferase assays (r = 0.85, PP = 2 × 10−6), confirming that the choice of promoter region did not profoundly affect our results.
Figure 5.
Robustness to tested sequence length and promoter type. (A) Comparison of MPRA (blue) vs. luciferase reporter assays (green/red) using 500-bp sequences instead of 145-bp and alternate promoters. For each of the 10 candidate enhancers, we list the predicted regulator, the enhancer ID (Supplemental Data S1), and the cell type in which the element was tested (matched cell type for predicted activators, unmatched for predicted repressors). Each bar indicates the expression of the original sequence and the effect of motif scrambling (direction of the arrow). MPRA experiments used 145-bp sequences centered on the motifs and a strong SV40 promoter (blue), and luciferase experiments used 500-bp sequences centered on the motifs with either a strong SV40 promoter (green) or TATA promoter (red). Data is normalized by subtracting from each expression value the mean for scrambles in that cell line across these 10 sequences. Asterisks indicate significance values using a t-test on the individual replicate values for the sequences (*) P < 0.05, (**) P < 0.01, (***) P < 0.001; see Methods (Mann-Whitney P-values are available in Supplemental Table S5). (B) Results for each of the sequences tested in A for the reverse cell type where the factor was not predicted to be active. A significant and large change was seen for NFE2L2 (#66), consistent with MPRA results. In addition, we observe significant, albeit smaller, luciferase changes for HNF1 (#129), ZFP161 (#1476), HNF1 (#1929), and GFI1 (#2302). Luciferase SV40 values for HNF1 (#1929) in K562 are absent due to a sample tracking error (see Supplemental Table S5).
Specifically for activator disruptions tested in matched cell types, we found that each sequence that showed a significant drop in reporter expression upon motif scrambling with MPRA also showed a significant expression drop with luciferase reporters with both the SV40 and TATA promoters (all t-test PT < 0.05) (Fig. 5A). Similarly, for the predicted repressor GFI1, we found a significant increase in luciferase expression upon motif scrambling with the SV40 promoter (PT = 1.1 × 10−3) (Fig. 5A), confirming our MPRA results. The increase in expression was more modest with the TATA promoter, consistent with our interpretation of relative reporter expression increase being due to the higher basal expression of the SV40 promoter. In the unmatched cell type, the significant changes we observed generally had a modest effect for both MPRA and luciferase assays (Fig. 5B), supporting the predicted cell-type specificities. The only exception was for NFE2L2, which showed a large and significant reduction in MPRA and luciferase reporter expression upon motif scrambling in both cell types, consistent with an activating role for NFE2L2 in both cell types, as discussed above (Fig. 5B; Supplemental Fig. S4B).
Lastly, we also tested two predicted enhancer elements whose expression change in the matched cell type was not found to be significant using MPRA (HNF4 #344 and HNF1 #1929), and in both cases, we found that the 500-bp constructs tested with the lower-throughput luciferase assays resulted in a significant reduction in expression in at least one of the two promoters. We conclude that in some cases, MPRA may have failed to validate enhancer activity due to promoter incompatibility, insufficient flanking sequence, or lack of power, suggesting that our estimates of the fraction of functional sequences may be conservative.
Discussion
We performed a systematic, regulatory motif-driven assay of the activity of more than 2000 cell type-specific enhancers, a number comparable to the largest collection of enhancers experimentally tested in vivo in all mammalian systems (Visel et al. 2007), and constitutes, to our knowledge, the first resource of hundreds of experimentally validated enhancer manipulations in human cells (Supplemental Table S3; Supplemental Data S1). We strongly confirmed the enhancer activity and cell-type specificity of enhancer chromatin states across thousands of loci, the ability of 145-bp segments to recapitulate activity and cell-type specificity in two human cell lines, the necessary role of regulatory motifs in enhancer function, and the complementary roles of activator and repressor motifs (Fig. 6).
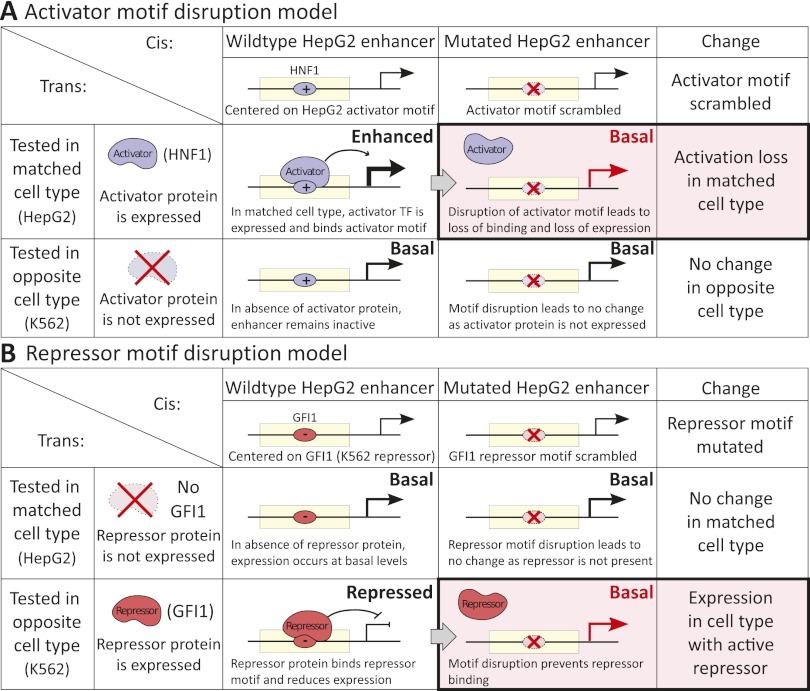
Figure 6.
Enhancer activator and repressor models. In our model of enhancer activity, the cell-type specificity of enhancers is maintained by the combined action of activators (such as HNF1 and HNF4 for HepG2 enhancers) that are expressed and bind in the matched cell type (HepG2), and the action of repressors (such as GFI1) that are expressed and bind in the unmatched cell type (K562). (A) Predicted enhancer activators are expressed in the cell type of enhancer activity, and their motifs are enriched within active enhancers. Disruption of the predicted activator motif leads to reduced reporter expression as the activator no longer binds its target motifs. (B) Predicted enhancer repressors are expressed in the other cell type and serve to reduce expression of the reporter gene, by preventing activator binding in the enhancer region or neighboring promoter. Disruption of the repressor motifs shows an effect only in the unmatched cell type, where binding of the repressors is disrupted, thus leading to derepression.
Our regulatory model made specific predictions regarding activator and repressor function, and cell-type specificity. We found these predictions largely confirmed, consistent with results for individual enhancers on a much smaller scale. We find statistically robust evidence that scrambling, removing, or disrupting the predicted activator motifs reduces enhancer function to baseline, while silent or motif-strengthening changes maintain or increase enhancer activity. Together, these provide strong, systematic evidence for the position weight matrix model of binding and the rule-based function of enhancers. In contrast to recent reports of many “ultraconserved” enhancers that apparently tolerate no mutations, our results are consistent with a modular and motif-centric definition of enhancer elements.
Conversely, we find that for one of the two tested repressors (GFI1) scrambling motifs leads to aberrant reporter expression in the cell line, where the enhancers are usually not active. We did not observe a significant change in expression for ZFP161, the other repressor we tested. This may have been because the motif may have improperly been identified as a repressor, an insufficient number of enhancers were tested, or that the action of additional regulators is necessary to activate enhancers from K562 in HepG2. The positive result with GFI1 highlights the importance of repressor motifs in confining the activity of enhancer elements. Moreover, we confirm that enhancer context plays a large role in determining enhancer activity, possibly due to synergistic or antagonistic effects between multiple regulators.
The elements we tested were capable of driving enhancer activity, despite including only 145 bp of ∼900 bp on average for chromatin-based enhancer predictions (Ernst et al. 2011). Moreover, we found that nucleosome exclusion signals at the endogenous enhancer location were the features most predictive of wild-type enhancer activity, even though the elements were tested outside their endogenous chromatin context. Together, these properties suggest that DNA sequence features contained within the tested elements are partly responsible for establishing the endogenous chromatin state of nucleosome depletion, either through nucleosome positioning motifs (Segal et al. 2006) that may have a role in our constructs if they are chromatinized, or by recruitment of sequence-specific regulators that also alter the nucleosome landscape at the endogenous locations. However, additional experiments will be required to determine the relative contribution of chromatin vs. primary sequence information, and to elucidate the sequence elements responsible for establishing the regulatory potential of endogenous enhancers. While we focused here on distal enhancers by selecting putative sequences at least 2 kb from any annotated TSS, we tested all sequences proximally to a common SV40 promoter region, and additional studies will be necessary to evaluate the ability of these sequences to activate transcription from varying distances including downstream from the TSS and with promoters other than SV40. Lastly, it is possible that the 10-bp-long tags used as barcodes at the 3′ of the mRNA sequences may have a small effect on the expression levels of the reporter genes, but we expect this effect to be mitigated by the use of 10 randomly chosen distinct tags for each tested sequence.
The methodology presented here provides an effective means for large-scale enhancer validation with diverse applications. In this study we focused on directed experimental manipulations of a large number of enhancers, and large numbers of disruptions for individual cis-regulatory motifs. However, the current methodology is also well-suited to exhaustive manipulation of small numbers of elements, the systematic testing of pairs or sets of elements, and even de novo enhancer design. The ability to test larger sequences, to ensure genome integration, and to maintain the original genomic context, will likely further expand the range of possible applications of the technology. Moreover, while the ∼5000 enhancers that we can test per experiment is still smaller than the ∼35,000 predicted enhancers for each cell type (Ernst et al. 2011), future experimental advances could permit an exhaustive testing of enhancer elements. Overall, we expect the wealth of quantitative enhancer activity measurements provided here, across thousands of wild-type and engineered constructs, and future applications of this technology to have a great impact in generating and testing predictive models of gene expression in the human genome.
Methods
Selection of enhancer regions
We define cell-type specific enhancers as the union of states 4 and 5 (“strong enhancers”) from our ENCODE study (Ernst et al. 2011) excluding regions within 2 kb of a TSS using GENCODE v2b (Harrow et al. 2006). A total of 688 motifs were collected from several databases (Matys et al. 2003; Sandelin et al. 2004; Badis et al. 2009), matched to the genome at a P-value stringency of 4−8 (the frequency at which a fully specified 8-mer matches a uniformly random genome), and evaluated for conservation using 29 mammals (Lindblad-Toh et al. 2011), as previously described (Kheradpour et al. 2007). We do not include motif instances in coding exons, 3′UTRs, or repeats. Specific motifs and the number of matches in each cell line are chosen as described in the Results section under experimental design. Each unique 145-mer sequence was tested only once (e.g., if an instance ignoring conservation is also selected as a conserved one or if a random mutation matches a 1-bp disruption).
Selection of motifs and factors
We ensured that all seven motifs show no sequence similarity to each other (Fig. 1A), but as we manipulate cis-acting regulatory motifs, not trans-acting TFs, we did not seek to distinguish the specific family member recognizing each motif in a given condition, and referred to the motif by the TF family name.
Wild-type sequence diversity
We produced alignments of every pair of the tested 145-bp wild-type sequences using MUSCLE v3.8 (Edgar 2004) with default parameters on both relative strands. We found that 116 of the 2104 tested sequences had >70% sequence identity with another tested wild-type sequence. For comparison, 2104 randomly selected 145-bp sequences were taken from the chromatin-based HepG2/K562 enhancers and a similar number (130) had >70% sequence identity. We conclude that the selection procedure does not significantly enrich for putative enhancer sequences that are highly similar.
Generation of motif manipulations
The various motif manipulations were performed based on the position weight matrix (PWM) for each motif (Supplemental Data S2). Each match for a given motif was scrambled using the same permutation (Supplemental Fig. S3). This permutation was determined by creating 100 random scrambles and choosing the one with the lowest correlation (Pietrokovski 1996) to the original motif. Other manipulations involved choosing the single base-pair change that reduces, improves, or makes the smallest change to the PWM match score where the specific change depends on both the motif and the specific sequence that it matches. Two random manipulations were performed by choosing two positions (without replacement) and changing them to one of the other three bases regardless of the effect it has on the PWM match score. The complete removal of the motif is the only modification that changed the tested sequence outside the position of the motif (additional nucleotides from the flanking genomic sequence were added to the borders to fill 145 bp).
Oligonucleotide library design and synthesis
Oligonucleotide libraries were designed to contain, in order, the universal primer site ACTGGCCGCTTCACTG, the variable 145-bp test sequence, KpnI/XbaI restriction sites (GGTACCTCTAGA), a variable 10-bp tag sequence, and the universal primer site AGATCGGAAGAGCGTCG (Melnikov et al. 2012). Each sequence was tested with 10 unique tags in order to reduce variance due to stochastic rates of amplification of specific plasmids. If a putative enhancer or any of its manipulations contained the recognition sequence for any restriction enzyme (GGTACC, TCTAGA, or GGCCNNNNNGGCC), then that putative enhancer was excluded and an additional one was chosen. The resulting 54,000-plex 200-mer oligonucleotide libraries were synthesized by Agilent, Inc.
MPRA plasmid construction
Full-length oligonucleotides were isolated using 10% TBE-Urea polyacrylamide gel (Invitrogen) and then amplified by 20–26 cycles of emulsion PCR as described by Schutze et al. (2011) using Herculase II Fusion DNA Polymerase (Agilent) and primers GCTAAGGGCCTAACTGGCCGCTT-CACTG and GTTTAAGGCCTCCGTGGCCGACGCTCTTCCGATCT containing SfiI sites. Purified PCR products were then digested with SfiI (NEB) and directionally cloned into the SfiI-digested MPRA vector pGL4.10M (Melnikov et al. 2012) using One Shot TOP10 Electrocomp E. coli cells (Invitrogen). To preserve library complexity, the efficiency of transformation was maintained at >3 × 108cfu/μg. The isolated plasmid pool was digested with KpnI/XbaI to cut between the tested sequence and tag, ligated with a synthetic KpnI–XbaI fragment containing the SV40 early enhancer/promoter (derived from pGL4.73, Promega) and the luc2 luciferase ORF (derived from pGL4.10, Promega) (Ernst et al. 2011) and then transformed into E. coli as described above. Finally, to remove the vector background, the resultant plasmid pool was digested with KpnI, size selected on a 1% agarose gel, self-ligated and retransformed into E. coli.
Cell culture and transfection
HepG2 cells (ATCC HB-8065) were maintained in Eagle's Minimum Essential Medium supplemented with 10% fetal bovine serum (FBS) penicillin (50 units/mL) and streptomycin (50 μg/mL). For HepG2 transfections, 5 × 106 cells were plated in 15-cm plates. Transfections were performed 24 h after plating using Fugene HD (Promega) according to the manufacturer's instructions. In each transfection we used 15 μg of DNA and a Fugene:DNA ratio of 7:2. K562 cells (ATCC CCL-243) were cultured in RPMI-1640 supplemented with 10% FBS and 1% GIBCO Antibiotic-Antimycotic (Invitrogen). For K562 transfections, 20 μg of DNA was introduced into 4 × 106 cells using a Nucleofector II device with Nuclefector Kit V and program T-016; 24 h post-trasfection/nucleofection, cells were lysed in RLT buffer (Qiagen) and frozen at −80°C. Total RNA was isolated from cell lysates using RNeasy kit (Qiagen). We chose the transfection method for each cell line that maximized efficiency while minimizing cell death.
Tag-seq
mRNA was extracted from 100 μg of total RNA using MicroPoly(A)Purist kits (Ambion) and treated with DNase I using the Turbo DNA-free kit (Ambion). First-strand cDNA was synthesized from 400 to 700 ng of mRNA using the High Capacity RNA-to-cDNA kit (Applied Biosystems). Tag-seq sequencing libraries were generated directly from 10% of a cDNA reaction or 50 ng of plasmid DNA by 26 cycle PCR using Pfu Ultra II HS DNA polymerase 2X master mix (Agilent) and primers AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCG-ATCT and CAAGCAGAAGACGGCATACGAGAT-LIB-GTGACTGGAGTTCAGACG-TGTGCTCTTCCGATCTCGAGGTGCCTAAAGG (where -LIB- is a library-specific 8-nt index sequence). The resultant PCR products were size-selected using 2% agarose E-Gel EX (Invitrogen). The libraries were sequenced in indexed pools of eight or individually using 36-nt single-end reads on an Illumina HiSeq 2000 instrument.
Data processing and normalization
To infer the tag copy numbers in each Tag-seq library, all sequence reads were examined, regardless of their quality scores. If the first 10 nucleotides of a read perfectly matched one of the 54,000 designed tags, and the remaining nucleotides matched the expected upstream MPRA construct sequence, this was counted as one occurrence of that tag. All reads that did not meet this criterion were discarded. This procedure was repeated separately for the plasmid, HepG2 mRNA, and K562 mRNA pools. The plasmid and mRNA counts for each tag was normalized by the total number of counts from the respective source, and a ratio of the mRNA to plasmid counts was then generated for each tag. A single value was produced for each tested sequence by taking the mean over the tags/replicates, excluding any that had fewer than 40 plasmid reads. The log2 of this value divided by the median was used throughout (this normalization is monotonic and consequently does not affect the P-values for the statistical tests used). Because only a small portion of our tested sequences corresponded to what we later determined to be a functional wild-type enhancer or a nondisruptive mutation, we estimate the 0 baseline level to be approximately the background level of expression for our promoter. Consistent with this, the 2098 sequences with scrambled motifs (and thus no expected expression) have a mean normalized expression of −0.0054 for HepG2 cells and −0.06 for K562 cells. Five probes had 0 RNA counts and their log2 values were replaced by −7 (the smallest non-zero mean had a log2 of −6.82).
Low-throughput luciferase validation
To validate the MPRA findings, we synthesized 10 pairs of 500-nt gBlocks (IDT) that each contained a wild-type or scrambled motif, the corresponding genomic flanking sequences, the constant 5′ end TCGCTAGCCTCGAGG, and the constant 3′ end ATATCAAGATCTGGC. Each gBlock was directly cloned into PCR linearized vectors pGL4[SV40-luc2] (Ernst et al. 2011) and pGL4.23 (Promega), and the resulting reporter constructs were verified by Sanger sequencing. Transfections into HepG2 and K562 cells were performed as for MPRA (see above) with four replicates per sequence pair. Luciferase activities were measured 24 h post-transfection using the Dual-Glo Luciferase Assay (Promega) and an EnVision 2103 Multilabel Plate Reader (PerkinElmer). We report expression values for each sequence as the log2 ratio of the signals from the gBlock plasmid over a control plasmid.
Statistical analysis
The paired Wilcoxon signed-rank test is used for comparing different versions of the same set of sequences (e.g., original to scramble). The unpaired Mann-Whitney U-test is used to compare two different sets of sequences (e.g., conserved versus ignoring conservation). Combined P-values are calculated, when indicated, by taking the expression values across multiple factors and using them together for the corresponding statistical test by treating them as one list of values. Where replicates for two sequences are directly compared, we use the individual log replicate values with the unpaired, unequal variance Student's t-test (Mann-Whitney P-values are also included in Supplemental Table S5). Correlations are computed using Pearson's r, and corresponding permutation P-values are computed as the percentile of the absolute correlation amongst 10 million absolute correlations between the vectors randomly shuffled. P-values are computed in a two-tailed manner, unless otherwise specified. Additional P-values including for individual factors can be found in Supplemental Figure S5 and Supplemental Tables S3–S5.
Data access
Data sets are available at the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) (accession number GSE33367) and in the Supplemental Material.
Acknowledgments
We thank E.M. LeProust and S. Chen of Agilent, Inc. for oligonucleotide library synthesis and the staff of the Broad Institute Genome Sequence Platform for assistance with data generation. This work was supported by NIH grants HG004037 to M.K., including supplemental funds HG004037-S1 for the experimental work, and by the Broad Institute and Harvard Stem Cell Institute to T.S.M.
Author contributions: P.K. and M.K. designed the sequences and analyzed the data with J.E. and T.S.M. providing substantial input. A.M., P.R., X.Z., and T.S.M. performed the molecular biology experiments. L.W. and J.A. performed the cell culture experiments. M.K. oversaw the computational aspects of this work and T.S.M. oversaw the experimental aspects. P.K. and M.K. wrote the paper with substantial input from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.144899.112.
Freely available online through the Genome Research Open Access option.
References
- Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339: 1074–1077 [DOI] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. 2009. Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga NS 2001. Promoter analysis by saturation mutagenesis. Biol Proced Online 3: 64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D 2004. Ultraconserved elements in the human genome. Science 304: 1321–1325 [DOI] [PubMed] [Google Scholar]
- Benos PV, Bulyk ML, Stormo GD 2002. Additivity in protein-DNA interactions: How good an approximation is it? Nucleic Acids Res 30: 4442–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci 99: 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Holterman AX, Wang X 2003. Transcription factors in liver development, differentiation, and regeneration. Hepatology 38: 1331–1347 [DOI] [PubMed] [Google Scholar]
- Courtois G, Morgan JG, Campbell LA, Fourel G, Crabtree GR 1987. Interaction of a liver-specific nuclear factor with the fibrinogen and α1-antitrypsin promoters. Science 238: 688–692 [DOI] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri WD, Ay A, Sayal R, Dresch J, Dayringer E, Arnosti DN 2010. Deciphering a transcriptional regulatory code: Modeling short-range repression in the Drosophila embryo. Mol Syst Biol 6: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X 2009. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25: i54–i62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433: 481–487 [DOI] [PubMed] [Google Scholar]
- Gong P, Cederbaum AI 2006. Transcription factor Nrf2 protects HepG2 cells against CYP2E1 plus arachidonic acid-dependent toxicity. J Biol Chem 281: 14573–14579 [DOI] [PubMed] [Google Scholar]
- Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH 2009. The WEKA data mining software: An update. SIGKDD Explor 11: 10–18 [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D et al. 2006. GENCODE: Producing a reference annotation for ENCODE. Genome Biol (Suppl 1) 7: S4.1–S4.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. 2010. Nucleosome dynamics define transcriptional enhancers. Nat Genet 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. 2009. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci 106: 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Orkin SH 2006. Zinc-finger transcription factor Gfi-1: Versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr Opin Hematol 13: 1–6 [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Fire A 1994. Combinatorial structure of a body muscle-specific transcriptional enhancer in Caenorhabditis elegans. J Biol Chem 269: 27021–27028 [PubMed] [Google Scholar]
- Kapoun AM, Kaufman TC 1995. A functional analysis of 5′, intronic and promoter regions of the homeotic gene proboscipedia in Drosophila melanogaster. Development 121: 2127–2141 [DOI] [PubMed] [Google Scholar]
- Kheradpour P, Stark A, Roy S, Kellis M 2007. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res 17: 1919–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC 2005. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res 15: 1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeProust EM, Peck BJ, Spirin K, McCuen HB, Moore B, Namsaraev E, Caruthers MH 2010. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res 38: 2522–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478: 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Chang JC, Moi P, Liu W, Kan YW, Curtin PT 1992. Dissection of the enhancer activity of β-globin 5′ DNase I-hypersensitive site 2 in transgenic mice. Proc Natl Acad Sci 89: 3899–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. 2003. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31: 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG Jr, Kinney JB, et al. 2012. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol 30: 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses AM, Chiang DY, Pollard DA, Iyer VN, Eisen MB 2004. MONKEY: Identifying conserved transcription-factor binding sites in multiple alignments using a binding site-specific evolutionary model. Genome Biol 5: R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ney PA, Sorrentino BP, McDonagh KT, Nienhuis AW 1990. Tandem AP-1-binding sites within the human β-globin dominant control region function as an inducible enhancer in erythroid cells. Genes Dev 4: 993–1006 [DOI] [PubMed] [Google Scholar]
- Orlov SV, Kuteykin-Teplyakov KB, Ignatovich IA, Dizhe EB, Mirgorodskaya OA, Grishin AV, Guzhova OB, Prokhortchouk EB, Guliy PV, Perevozchikov AP 2007. Novel repressor of the human FMR1 gene—identification of p56 human (GCC)n-binding protein as a Krüppel-like transcription factor ZF5. FEBS J 274: 4848–4862 [DOI] [PubMed] [Google Scholar]
- Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J 2009. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol 27: 1173–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee SI, Cooper GM, et al. 2012. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol 30: 265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444: 499–502 [DOI] [PubMed] [Google Scholar]
- Pietrokovski S 1996. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res 24: 3836–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B 2004. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 32: D91–D94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze T, Rubelt F, Repkow J, Greiner N, Erdmann VA, Lehrach H, Konthur Z, Glokler J 2011. A streamlined protocol for emulsion polymerase chain reaction and subsequent purification. Anal Biochem 410: 155–157 [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J 2006. A genomic code for nucleosome positioning. Nature 442: 772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E, Kalma Y, Sharp A, Raveh-Sadka T, Levo M, Zeevi D, Keren L, Yakhini Z, Weinberger A, Segal E 2012. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol 30: 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S, Adler AS, Field Y, Chang HY, Segal E 2008. Systematic functional characterization of cis-regulatory motifs in human core promoters. Genome Res 18: 477–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobek-Klocke I, Disque-Kochem C, Ronsiek M, Klocke R, Jockusch H, Breuning A, Ponstingl H, Rojas K, Overhauser J, Eichenlaub-Ritter U 1997. The human gene ZFP161 on 18p11.21-pter encodes a putative c-myc repressor and is homologous to murine Zfp161 (Chr 17) and Zfp161-rs1 (X Chr). Genomics 43: 156–164 [DOI] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. 2011. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res 21: 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanojevic D, Small S, Levine M 1991. Regulation of a segmentation stripe by overlapping activators and repressors in the Drosophila embryo. Science 254: 1385–1387 [DOI] [PubMed] [Google Scholar]
- Su J, Teichmann SA, Down TA 2010. Assessing computational methods of cis-regulatory module prediction. PLoS Comput Biol 6: e1001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA 2007. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA 2008. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet 40: 158–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JB, Philippakis AA, Jaeger SA, He FS, Lin J, Bulyk ML 2008. Systematic identification of mammalian regulatory motifs' target genes and functions. Nat Methods 5: 347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MJ, Orkin SH 1995. GATA transcription factors: Key regulators of hematopoiesis. Exp Hematol 23: 99–107 [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Pinsonneault J, Gellon G, McGinnis N, McGinnis W 1994. Deformed protein binding sites and cofactor binding sites are required for the function of a small segment-specific regulatory element in Drosophila embryos. EMBO J 13: 2362–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]