Abstract
Adverse drug reactions (ADRs) are caused by interactions between drugs or their metabolites and specific proteins. Knowledge of these proteins is important for facilitating mechanistic research of ADRs and new drug discovery. Here, we identified 41 network modules from an ADR-protein network; analysed the function of each module; revealed the potential accompanying actions of the ADRs and the new ADR-related proteins (ADRPs) to a unique ADR and studied the characteristics of composition, subcellular location and tissue distribution of these ADRPs by comparing them with drug-related proteins (DRPs). The results indicated that ADRs are mainly caused by risk drug-related proteins (RDRPs) and that drug off-target effects are a secondary cause. Biological processes that enzymes involve are the main reason for the occurrence of ADRs. However, drug-related transporters have a higher risk of inducing ADRs than drug-related enzymes do, and ADRPs locating in the cell membrane tend to induce multiple ADRs.
Currently about 6700 drugs have been developed, including over 1400 FDA approved small molecule drugs and more than 5000 experimental drugs, additionally these drugs linked to about 4700 non-redundant proteins1. Among these drugs on the market, the mechanisms of many drugs in the clinic remain poorly characterised2. Thus, drugs even with known therapeutic targets often have unexpected and severe side effects, and some ADRs can't be explained by their interaction with their binding target. As a consequence, over the past ten years, more than 19 broadly used market drugs were withdrawn after exhibiting unexpected and severe side effects3. In order to make currently marketed drugs safer and more effective, understanding the mechanisms behind ADRs has become increasingly important. If we can predict ADRs in clinical settings, alternative medicines or combination therapies may be used to avoid ADRs. Furthermore, of the almost 5100 current experimental drugs, many could be prevented from entering the market if we could detect potential ADRs during the experimental phase.
In recent years, with the development of technologies for the study of drug action mechanisms, a large amount of biological data has been produced by experiments and computation, and some well-known drug databases have been established, such as DrugBank, the Human Protein Reference Database (HPRD), the Drug Adverse Reaction Target (DART) database, and the Comparative Toxicogenomics Database (CTD), etc. DrugBank (DrugBank1, www.drugbank.ca) is a comprehensive, open drug-related database that contains data regarding drug basic information, structural information, indications, etc. In addition, information provided concerning drug-related targets, carriers, enzymes, and transporters plays an important role in the mechanistic understanding of drug actions. The HPRD4 (www.hprd.org) is an authoritative database of human proteins that provides information on domains, motifs, gene ontology, pathways, and protein-protein interactions. The DART5 (http://bidd.nus.edu.sg/group/drt/dart.asp) database provides comprehensive information about adverse effects of drugs described in the literature, and also provides information regarding the related proteins of ADRs along with their functions, subcellular location, inducing adverse drug effects, ligands, and drugs. The CTD7 (http://ctd.mdibl.org/) includes curated data describing cross-species chemical-gene/protein interactions and chemical- and gene-disease associations to illuminate molecular mechanisms underlying variable susceptibility and environmentally influenced diseases. These data will provide insights into complex chemical-gene and -protein interaction networks and advance the understanding of the effects of environmental chemicals on human health. There are also other databases that contain information about ADRs, but these will not be discussed in this article. How to make better use of these data to study ADRs is our research focus.
Due to the complexity of ADRs occurrence, their pathogenesis is best studied at a systems level. Network analysis approaches in biology have proven useful for organising high-dimensional biological datasets and extracting meaningful information8. Network analysis is based mainly on the relationship between nodes, which include physical and/or chemical interactions, genetic regulatory interactions, gene co-expression, or some other shared property between nodes. Viewed from a global perspective, network analysis can help us to discover non-obvious, but intrinsically important nodes. To research the ADR occurrence mechanism, identifying drug off-target, studying the target related signal pathway or biological process have been a major strategy. Campillos et al. constructed a network of drugs by establishing connections between drugs with a degree of structural similarity and similar side effect profiles. By identifying pairs of drugs in this network with distinct targets, the authors were able to assign the targets of one drug to the drugs it was connected to and subsequently validated the binding of the drug to its predicted secondary target9. Although there have been some notable achievements, there is still much work on ADRs to be done, such as identifying ADR-ADR associations and new ADR-related proteins (ADRPs), analysing network topology properties, and identifying other differences between ADRPs and drug-related proteins.
In this study, based on the data of ADRs induced by approved drugs and their related proteins, we constructed an ADR-protein network by incorporating relationships between ADRs and their related proteins (ADRPs), and the directed interactions among ADRPs from human PPIs. Furthermore, we identified ADR-ADR associations, predicted new ADRPs, analysed the characteristics of the topology of ADRPs in the human protein-protein interaction (PPI) network, and studied the composition, subcellular localisation, and tissue distribution of ADRPs. Increased knowledge of these proteins is important for research into ADR mechanisms, rational drug design, and safety evaluation.
Results
ADR-protein network
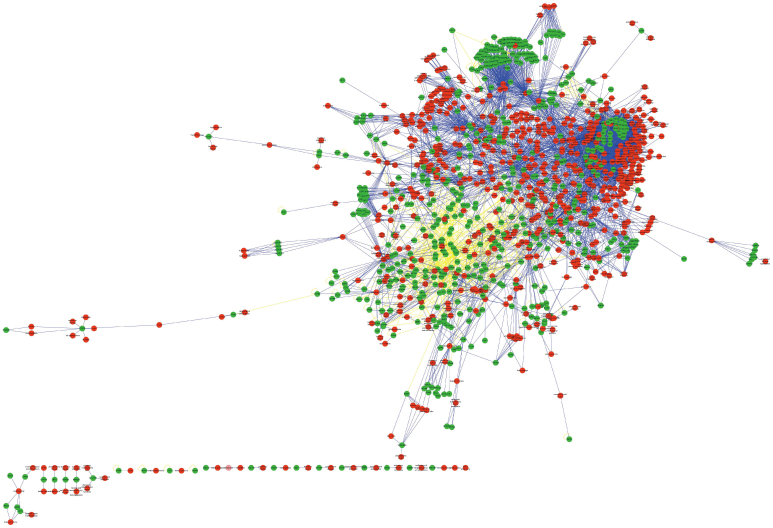
If most ADRs specifically targeted a single protein, then the ADR-protein network would consist of isolated nodes with few or no edges between them. In fact, the ADR-protein network displays many connections among different ADRs. The ADR-protein network consists of 1,169 nodes and 9,613 edges, with a highly connected network component comprising 1,116 nodes. Among these nodes, there were 622 ADRs and 547 ADRPs. Among the edges, there were 8,923 ADR-ADRP interactions and 690 ADRP-ADRP interactions. Figure 1 (Supplementary Fig. 1) shows a global view of the ADR-protein network with the following colour-coded nodes and edges: ADR (red), ADRP (green), ADR-ADRP connections (blue), and ADRP-ADRP connections (light yellow). Supplementary Figure 2 shows the degree distribution of all the nodes in the ADR-protein network, with the degree ranging from 1 to 110; most nodes display few connections, and a few nodes are highly connected. In addition, the distribution follows a power-law distribution, which was fitted by applying the Fit Power Law of Network Analysis, where (D, k) = a*k−γ, γ = 1.413, correlation = 0.942, and R-squared = 0.909. Supplementary Table 1 shows the differences among the metrics described in the Methods section between the ADR-protein network and the random network. This result indicates that the ADR-protein network revealed associations between ADRs and their related proteins that were not seen in the random network. Now we are taking cluster coefficient as the example. It was found in this study that the average cluster coefficient of the ADR-protein network was larger than that of the random network. Clustering coefficient, as the proportion of the observed connections between the neighbours against the maximum number of possible connections6, assesses the subnetwork modularity. So higher cluster coefficient suggests that the ADR-protein network consists of close-connected modules.
Figure 1. A global view of the ADR-protein network.
The network contains ADRs (red), ADRPs (green), ADR-ADRP connections (blue), and ADRP-ADRP connections (light yellow). A higher-quality image file of Fig. 1 is supplied in Supplementary Fig. 1.
Supplementary Fig. 3 shows the degree distribution of all ADRPs, where the degree describes the number of ADRs associated with a protein. Supplementary Fig. 4 shows the degree distribution of all ADRs, where the degree of an ADR describes the number of its connected proteins. Supplementary Figs. 3 and 4 show that most ADRs and their related proteins have a low degree of connectedness, while a few are highly connected, and follow a power-law distribution. The average degrees of ADRs and ADRPs were 7.43 and 8.48, respectively, while the median degrees were 3 and 4, respectively.
The information above suggested that ADRs and ADRPs presented complex regulatory relationships, reflecting one-to-one, one-to-many and many-to-many relations.
For one-to-one, that is to say, one ADRP induces a single ADR, or an ADR is mediated by one ADRP. Although there were only 13 pairs such relations in the ADR-protein network (Supplementary Table 2), clearly understanding this kind of relations can not only control the ADR occurrence by regulating the abnormal function of this single protein but also make these ADRPs as biomarkers for new drugs safety evaluation during new drugs early development stages.
We found that 396 ADRs (63.7% of all ADRs) linked at least two ADRPs, and 113 (18.2%) more than 10 ADRPs (Supplementary Table 3), which suggested that most ADRs were mediated by more than 2 ADRPs, indicating the complicated occurring mechanisms of these ADRs, especially those with higher degree . If any ADRP was affected by drugs, a corresponding ADR symptom occurred, increasing the frequency of these ADRs in clinical drugs practice. Likewise, regulating any abnormal protein associated with corresponding ADR into normal state may not completely reverse the ADR. Therefore, comprehensively exposition of the pathological process of these ADRs occurrence will help to prevent and treat these ADRs.
The degree distribution of all ADRPs showed that 500 ADRPs (91.4% of all ADRPs) were associated with at least 2 ADRs and 140 (25.6%) more than 10 ADRs (Supplementary Table 4), which reflected that these ADRs with higher degree tended to mediate a variety of adverse reactions. Due to sharing the same proteins, these ADRs occurred accompanying with each other. The preventive and therapeutic methods of these ADRs are the same and similar. It is, thus, very necessary to find out these ADRs that are likely to occur simultaneously.
Network modules
We identified 52 modules from the ADR-protein network by applying the Cytoscape plugin MINE. After excluding modules that only contained ADRPs or ADRs, 41 modules remained with sizes ranging from 3 (module 37–41) to 34 (module 9) (Supplementary Fig. 5). To study the mechanisms of ADR occurrence, we detected associations among different modules using gene ontology (GO) annotation10 based on biological processes, cellular components, and molecular functions (Supplementary Table 3). Using BinGO (p < 0.05), there were 39 modules enriched for GO biological processes, with 15 modules highly related to 4 GO terms: GO 23033 (signalling pathway), GO 23046 (signalling process), GO 23060 (signal transmission), and GO 42221 (response to chemical stimulus). Furthermore, 26 modules were enriched for GO cellular components, with the highest enrichment seen for GO 5887 (integral to plasma membrane) and GO 31226 (intrinsic to plasma membrane). Moreover, all the modules were related to GO molecular functions, and 14 modules had the highest enrichment for GO 4871 (signal transducer activity) and GO 60089 (molecular transducer activity). These highly enriched GO terms indicate that ADRPs are mainly involved in the structure and function of the cell membrane.
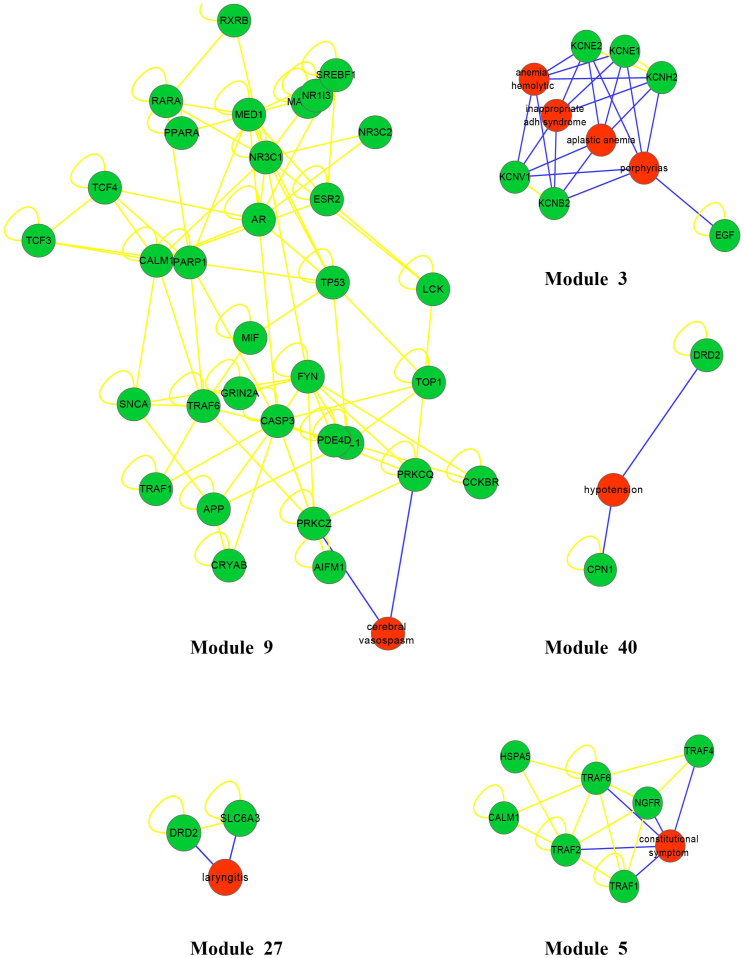
ADRs connected with the same proteins generally accompany one another when a given drug affects these proteins. These associations exist not only within one module but also among different modules. For example, in module 3 (Fig. 2), there are 4 ADRs, including haemolytic anaemia, porphyrias, inappropriate ADH syndrome, and aplastic anaemia. According to the disease categories in the International Classification of Diseases (ICD-10), haemolytic anaemia and aplastic anaemia are blood diseases, inappropriate ADH syndrome is an endocrine disease, and porphyrias are metabolic diseases. Although these ADRs belong to different disease categories, all of them show relationships with the same potassium channel proteins. Such phenomena also arise across modules; for instance, DRD2 appears in module 27 (Fig. 2) related to laryngitis, and in module 40 (Fig. 2) connected to hypotension, and symptoms of laryngitis and hypotension may appear simultaneously when drugs affect DRD2. Thus, based on the ADR-protein network, it is possible to identify ADRs that are likely to occur together.
Figure 2. The example of modules 3, 5, 9, 27, and 40 in the ADR-protein network.
The higher-quality image file of all modules is supplied in Supplementary Fig. 5.
In some modules, we were able to uncover known ADR-ADRP associations, but we also found unconfirmed ADR-protein associations in the ADR-protein network, such as in modules 5 and 9, etc. In these modules, there are two types of proteins: ADRPs that are directly connected to a unique ADR and ADRIPs that are indirectly connected to ADRs via other ADRPs. The ADRIPs were not confirmed to relate with a unique ADR but rather to proteins within a module that typically share the same biological functions. Our research therefore shows that ADRIPs are tightly connected with unique ADRs in terms of function, although this must be confirmed by additional experiments.
Characteristics of ADRPs
Researching the characteristics of ADRPs and the differences between ADRPs and DRPs will not only promote research of the mechanisms underlying ADRs but will also promote rational drug design and safety evaluation. We therefore analysed the composition, subcellular location, tissue distribution, and network properties in PPIs of ADRPs.
Characteristics of composition
Of 547 ADRPs and 1,445 approved drug-related proteins (DRPs), 54% of all ADRPs were risk drug-related proteins (RDRPs), of which 91% of RDRPs were drug targets. In additional, 91% of all RDRPs are drug targets as well as drug off-targets. Therefore, ADRs directly induced by risk drug-related proteins account for the majority of all ADR occurrences, whereas drug off-target effects represent the secondary reason.
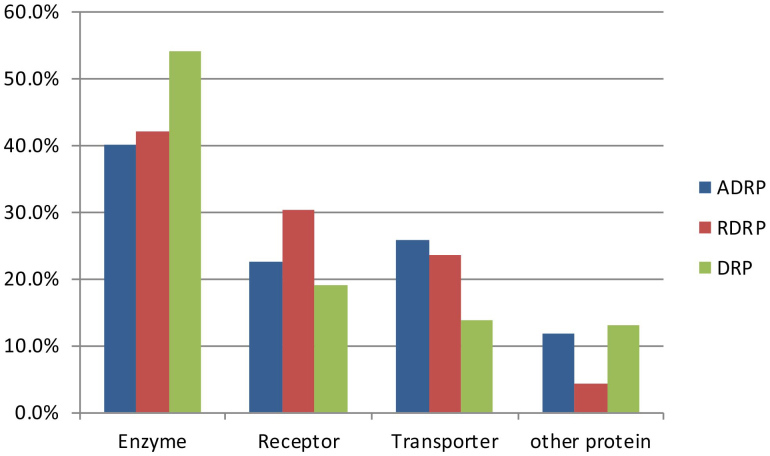
In reference to the categories of ADRPs in the DART database, we classified ADRPs and DRPs into four groups: enzymes, receptors, transporters, and other proteins (as shown in Fig. 3). In the case of ADRPs, 40% were enzymes, 25.6% were transporters, and 22.4% were receptors. For DRPs, 13.7% were transporters, while 54.1% were enzymes, which was much higher than the percentage found in ADRPs (chi-square test, p < 0.0001). It is clear that the biological processes involving enzymes play an important role in both therapeutic effects and adverse reactions. Furthermore, we analyzed the proportions of enzymes and transporters of RDRPs in all DRPs and found that 16.1% of drug-related enzymes directly induce ADRs, while 34.9% of drug-related transporters directly induce ADRs. Based on these findings, transporters are more likely to cause adverse reactions than enzymes, suggesting that the safety of both drug targets and transporters needs to be taken into account in rational drug design.
Figure 3. Compositional analysis of ADRPs, RDRPs, and DRPs.
Characteristics of tissue distribution
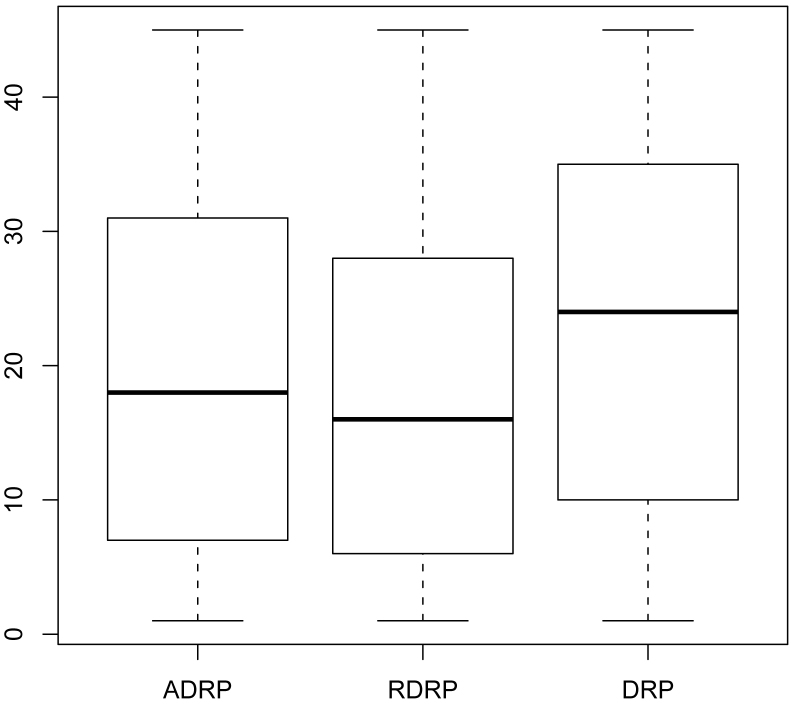
Because gene expression is tissue-specific, we analysed the tissue distributions of ADRPs, RDRPs, and DRPs (Fig. 4) using the Kruskal-Wallis rank-sum test. The results indicated that these three types of proteins were clearly distributed in different tissues (p = 4.61E-12). In addition, we applied a nonparametric multiple comparison test based on the result of Kruskal-Wallis rank-sum test, and these results showed that there was no significant difference between the distributions of ADRPs and RDRPs, although there was a significant difference between the distributions of ADRPs (RDRPs) and DRPs (Supplementary Fig. 6). On average, ADRPs were identified in 19.34 tissues, which is lower than the number of tissues in which DRPs presented (23.84), while the average tissue distribution of RDRPs was as low as 17.85. This result was not biased due to inequalities related to sample size, which is shown in Supplementary Fig. 7. Compared with DRPs, ADRPs tended to show more strict tissue specificity (Supplementary Fig. 8 and Fig. 9). Therefore, when a drug or metabolite interacts with an ADRP, fewer tissue types are affected, and this characteristic may explain why most ADRs affect specific parts of the body, such as headache, dizziness, liver damage, and kidney toxicity, etc.
Figure 4. Tissue distributions of ADRPs, RDRPs, and DRPs.
The y-axis represents the tissue number.
Characteristics of subcellular location
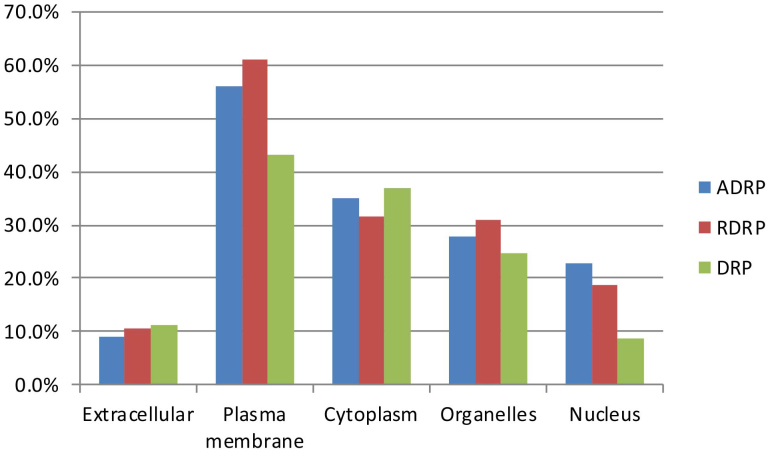
Typically, proteins in different subcellular locations have different biological functions. In accordance with the locations of proteins in the cell, subcellular locations are classified as extracellular, plasma membrane, cytoplasm, organelles, or nucleus. A ‘multiple location protein' is defined as a protein that is present in more than one subcellular location, and 36.9% of ADRPs are multiple location proteins. Figure 5 shows the subcellular location distribution of ADRPs and DRPs. Both groups were found to be highly expressed in the plasma membrane, followed by the cytoplasm, organelles, nucleus, and extracellular compartment. Plasma membrane proteins accounted for 56.1% and 43.2% of ADRPs and DRPs, respectively (chi-square test, p < 0.0001). Most plasma membrane ADRPs were receptors and transporters, while few of them were enzymes. Moreover, membrane proteins accounted for as many as 61.1% of all RDRPs. Clearly, the subcellular location of ADRPs is not random, and these proteins are more likely to be located on the cell membrane. Due to the complications of transporting drugs across membranes, membrane proteins are easier to target11. While DRPs tend to be distributed on the membrane, RDRPs are the main source of ADRPs. Therefore, DRPs in the plasma membrane should be utilised prudently in drug research and development.
Figure 5. Subcellular location analysis of ADRPs, RDRPs, and DRPs.
Characteristics of network properties
To examine the global relationships between ADRPs and DRPs, we analysed the topological properties of these two groups of proteins in the human PPI network, and the results are displayed in Table 1. Compared to DRPs, ADRPs tended to form hub nodes (10.97) and play an important role in the PPI network. Essential genes are defined as genes that are indispensable to cellular life, and these genes tend to coordinate the activity of diverse biological processes or ‘modules' and also tend to form hub nodes in the PPI network12. We found that only 5.8% of ADRPs and 10% of DRPs were essential genes, which shows that ADRPs tend not to code essential genes (chi-square test, p = 0.0026). As we know, drugs with serious ADRs cannot be permitted to enter the market, and most ADRs do not show lethal effects. Furthermore, Wu et al. demonstrated varying functions between ADRPs and essential proteins13, which may explain why ADRPs tend not to be essential genes.
Table 1. Topological properties of ADRPs and DRPs in the PPI network.
| Metrics | ADRPs | DPRs |
|---|---|---|
| Average degree | 10.97 | 7.80 |
| Average clustering coefficient | 0.057 | 0.059 |
| Average betweenness centrality | 7.06*E-4 | 4.20*E-4 |
The average degree of ADRPs in the PPI network was 10.97, the average connecting number of ADRPs in the ADR-protein network was 8.48, and 56% of all ADRPs were identified as membrane proteins. We hypothesize that a protein associated with many ADRs would also show a high degree in the PPI. Namely, supposing the reason of a protein associated with more ADRs is due to its high protein interactions. We therefore analysed the relationship between the degrees of ADRPs in the ADR-protein network and the PPI network and the membrane protein proportion of ADRPs (Table 2). We found an inverse relationship between the number of ADRs and the degree of protein interactions in the PPI network, while the proportion of membrane proteins showed an upward trend with increasing ADRs. When proteins were associated with more than 40 ADRs, the average degree of protein interactions of these proteins was as low as 3.97, while the membrane protein proportion was as high as 78.4%. It is therefore not the case that proteins with more protein interactions relate to more ADRs. On the other hand, it was clear that proteins with multiple ADRs tend to be located on the plasma membrane. In the PPI network, the degree of protein interactions is correlated with the essentiality of the protein14; if a protein with a high number of protein interactions showed an association with more ADRs, this protein might affect organismal survival, which would prohibit the development of rational drugs. Furthermore, membrane proteins may have multiple biological functions; if a membrane protein involved in a certain biological process is targeted by a drug, then other intracellular proteins involved in different biological processes will also be affected, and this characteristic of membrane proteins may explain why proteins with multiple ADRs tend to be located on the membrane.
Table 2. Analysis of association between the degree in the ADR-protein network and the PPI and the membrane protein proportion of ADRPs.
| Degree in ADR-protein network | Average degree in PPI | Membrane protein proportion |
|---|---|---|
| Proteins with more than 1 ADR | 9.36 | 0.64 |
| Proteins with more than 2 ADRs | 9.45 | 0.68 |
| Proteins with more than 3 ADRs | 10.16 | 0.70 |
| Proteins with more than 4 ADRs | 8.00 | 0.71 |
| Proteins with more than 5 ADRs | 7.84 | 0.66 |
| Proteins with more than 6 ADRs | 6.95 | 0.67 |
| Proteins with more than 7 ADRs | 7.30 | 0.67 |
| Proteins with more than 8 ADRs | 7.74 | 0.68 |
| Proteins with more than 10 ADRs | 7.10 | 0.72 |
| Proteins with more than 20 ADRs | 7.53 | 0.75 |
| Proteins with more than 30 ADRs | 4.29 | 0.76 |
| Proteins with more than 40 ADRs | 3.97 | 0.78 |
| Proteins with more than 50 ADRs | 4.32 | 0.76 |
Discussion
Based on the integration of information from DART, CTD, DrugBank, and other public databases, we constructed an ADR-protein network to study system level associations of ADR functions, identify ADR-ADR interactions, and predict new ADRPs. Using such an ADR-protein network, it is possible to comprehensively understand the relationship between an ADR and its related proteins, which will facilitate the study of ADR-related pathogenesis. Furthermore, network module analysis can help detect associations between ADRs as well as find new ADRPs.
ADRs are caused by interactions between drugs or their metabolites and specific proteins. Research on the characteristics of ADRPs will not only promote research of the mechanisms underlying ADRs but will also promote rational drug design and safety evaluation. We therefore analysed the composition, subcellular location, tissue distribution, and PPI network properties of ADRPs.
According to traditional pharmacology, ADRs are mediated by the off-target proteins. Our findings, however, indicated that ADRs are mainly caused by risk drug-related proteins, whereas drug off-target effects represented the secondary reason. The composition analysis of ADRPs indicated that biological processes involving enzymes are the main causes of ADRs occurrence, but the risk of inducing an ADR is greater for drug-related transporters than for drug-related enzymes. These findings suggest that the safety of both drug targets and transporters needs to be taken into account for rational drug design. Compared to DRPs, ADRPs more tend to be expressed in the plasma membrane; moreover, ADRPs located in the cell membrane tend to induce multiple ADRs. Therefore, DRPs in the plasma membrane should be utilised prudently in drug development. Topological feature analysis results in the human PPI network suggested that ADRPs tend to be located in hub nodes, but not to be essential genes, which reflects the fact that the majority of approved drug ADRs are not lethal and module analysis results suggested that we should more strictly prevent potential accompanying ADRs to avoid serious complications.
Experimental validation of these findings will be required to further estimate their potential clinical value. Although our results may be constrained by data incompleteness, they are based on high-quality ADR and ADRP interaction annotations.
Furthermore, our investigation supports the notion of applying system-based approaches to elucidate ADR and ADRP characteristics. The prevailing approach based on descriptive views of approved ADRs and ADRP interactions cannot adequately characterize the complexity of the clinical value, which results in reduced opportunities to anticipate potential ADR-ADR associations and other findings. Thus, we expect new research directions to further address the network-driven complexity and clinical value of concrete ADRs, especially serious ADRs.
Methods
Definitions
‘Drug-related proteins' (DRPs) was the generic term for pharmacokinetics and pharmacodynamics proteins that were related to the therapeutic actions of drugs. Here, based on the protein taxonomy of the DART database, we divided DRPs into 4 classes: transporters, enzymes, receptors, and other proteins. DRPs were further separated into drug targets and non-drug targets. ‘ADR-related proteins' (ADRPs) are proteins that mediate ADRs or toxicities by binding to drugs or their reactive metabolites. The intersection of ADRPs and DRPs was defined as ‘risk drug-related proteins' (RDRPs) because these DRPs located at the intersection were related not only to therapeutic action but also to the risk of inducing ADRs occurrence.
Datasets
The data on ADRs and ADRPs were sourced from DART5 (Version March/03/2009) and CTD6 (Version Oct/18/2012). All ADRs were uniformly named according to FDA MedWatch, and the ADRP names were converted to gene symbols. PPI networks were obtained from the HPRD. DRPs that were drug targets, drug-related enzymes, or drug-related transporters were collected from DrugBank15 (Version Jan/14/2011). Essential genes, which are indispensable for cellular life, were taken from the literature published by Tu et al16. Protein-tissue distribution data were obtained from the EST databases of the NCBI17. Protein subcellular location data were obtained from UniProt Knowledgebase (UniProtKB18, Version, Oct/3/2012), and proteins were classified as extracellular, plasma membrane, cytoplasm, organelles, or nucleus. By integrating these data, we obtained 622 ADRs, 547 ADRPs, 1,445 approved DRPs, and 298 RDRPs.
ADR-protein network
To generate the ADR-protein network, we first incorporated relationships between ADR and ADRP sourced from DART and CTD. An ADR and an ADRP were connected to each other if the ADRP was a known related protein of the ADR, giving rise to a bipartite graph of ADR-ADRP interactions. Next, to better understand ADR mechanisms, we added the directed interactions between ADRPs from human PPIs to the bipartite network of ADR-ADRP interactions. The union of the ADR-ADRP and ADRP-ADRP interactions resulted in our ADR-protein network. In this network, nodes represented either ADRs or ADRPs and edges encoded either ADR-ADRP or ADRP-ADRP interactions. The ADR-protein network was constructed by connecting the relationships between ADRs and ADRPs and the interactions between ADRPs from PPI. We then analysed the topological characteristics, mined modules using MINE19, a plugin of Cytoscape20, and identified associations between modules and GO biological processes using BinGO21. Default parameters were used for both plugins, with the exception of selecting Homo sapiens as the species. Random networks were generated by applying RandomNetworks while keeping the size of nodes and edges the same as in the ADR-gene network (Azuaje, F.J et al., 2011)22. First, we chose the parameter ‘Compare against generated random networks'. In the ‘Generate Random Network' section, we chose the Erdos Renyi Model to generate a flat random network with 1,000 permutations. In the Erdos Renyi Model, we chose G (n, m), which uniformly generates a random undirected-edge graph with 1,169 nodes and 9,613 edges. We then compared the two networks using the following metrics: clustering coefficient, average degree, degree distribution, and mean shortest path.
Network topology characteristics analysis in PPI
The human PPI network was constructed based on PPI data from the HPRD. We compared the topological characteristics of ADRPs, DRPs, and essential genes in the PPI network using the Network Analysis Cytoscape plugin20.
Author Contributions
X.J.C. designed the study and revised the manuscript. X.Q.L., X.D.J., F.J.T. and S. C. collected data, constructed the network, analysed the topology properties, and wrote the manuscript and revised the manuscript. R.Z.Y. implemented the enrichment analysis and revised the manuscript. L.L., Y.F.W. and Y.L.C. implemented the characteristic analysis of composition, subcellular location, and tissue distribution of ADRPs. All authors reviewed the manuscript.
Supplementary Material
Supplementary Material List
Supplementary Table S3-S5
Acknowledgments
The authors would like to acknowledge the support to this work by the Graduate Innovation Foundation of Heilongjiang province, China (Grant No. YJSCX2012-223HLJ, YJSCX2012-205HLJ, YJSCX2012-341HLJ), the Provincial Education Department Project of Heilongjiang, China (Grant No. 11541121), and the Innovation Manpower Fund of Harbin Science and Technology Bureau, China (Grant No. 2010RFXXS053).
References
- Wishart D. S. et al. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36, D901–906 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews J. Stategic trends in the drug industry. Drug Discov Today. 8, 411–420 (2003). [DOI] [PubMed] [Google Scholar]
- Schmid E. F., Smith D. A. & Ryder S. W. Communicating the risks and benefits of medicines. Drug Discov Today. 12, 355–364 (2007). [DOI] [PubMed] [Google Scholar]
- Keshava Prasad T. S. et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 37, D767–772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z. L. et al. Drug Adverse Reaction Target Database (DART) : proteins related to adverse drug reactions. Drug Saf. 26, 685–690 (2003). [DOI] [PubMed] [Google Scholar]
- Watts D. J. & Strogatz S. H. Collective dynamics of small-world networks. Natrure. 393, 440 (1988). [DOI] [PubMed] [Google Scholar]
- Davis A. P. et al. Comparative Toxicogenomics Database: a knowledgebase and discovery tool for chemical-gene-disease networks. Nucleic Acids Res. 37, D786–792 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G. & Agarwal P. Human disease-drug network based on genomic expression profiles. PLoS One. 4, e6536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillos M., Kuhn M., Gavin A. C., Jensen L. J. & Bork P. Drug target identification using side-effect similarity. Science. 321, 263–266 (2008). [DOI] [PubMed] [Google Scholar]
- Ashburner M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim M. A., Goh K. I., Cusick M. E., Barabasi A. L. & Vidal M. Drug-target network. Nat Biotechnol. 25, 1119–1126 (2007). [DOI] [PubMed] [Google Scholar]
- Goh K. I. et al. The human disease network. Proc Natl Acad Sci U S A. 104, 8685–8690 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. A. New Method to Identify Repositioned Drugs for Prostate Cancer. IEEE. (2012). [Google Scholar]
- Jeong H., Mason S. P., Barabasi A. L. & Oltvai Z. N. Lethality and centrality in protein networks. Nature. 411, 41–42 (2001). [DOI] [PubMed] [Google Scholar]
- Knox C. et al. DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 39, D1035–1041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z. et al. Further understanding human disease genes by comparing with housekeeping genes and other genes. BMC Genomics. 7, 31 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K. D., Tatusova T. & Maglott D. R. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutet E., Lieberherr D., Tognolli M., Schneider M. & Bairoch A. UniProtKB/Swiss-Prot. Methods Mol Biol. 406, 89–112 (2007). [DOI] [PubMed] [Google Scholar]
- Rhrissorrakrai K. & Gunsalus K. C. MINE: Module Identification in Networks. BMC Bioinformatics. 12, 192 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., Heymans K. & Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 21, 3448–3449 (2005). [DOI] [PubMed] [Google Scholar]
- Azuaje F. J., Zhang L., Devaux Y. & Wagner D. R. Drug-target network in myocardial infarction reveals multiple side effects of unrelated drugs. Sci Rep. 1, 52 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material List
Supplementary Table S3-S5