Abstract
Objective
Logical Observation Identifiers Names and Codes (LOINC) mapping of laboratory data is often a question of the effort of mapping compared with the benefits of the structure achieved. The new LOINC mapping assistant RELMA (version 2011) has the potential to reduce the effort required for semi-automated mapping. We examined quality, time effort, and sustainability of such mapping.
Methods
To verify the mapping quality, two samples of 100 laboratory terms were extracted from the laboratory system of a German university hospital and processed in a semi-automated fashion with RELMA V.5 and LOINC V.2.34 German translation DIMDI to obtain LOINC codes. These codes were reviewed by two experts from each of two laboratories. Then all 2148 terms used in these two laboratories were processed in the same way.
Results
In the initial samples, 93 terms from one laboratory system and 92 terms from the other were correctly mapped. Of the total 2148 terms, 1660 could be mapped. An average of 500 terms per day or 60 terms per hour could be mapped. Of the laboratory terms used in 2010, 99% could be mapped.
Discussion
Semi-automated LOINC mapping of non-English laboratory terms has become promising in terms of effort and mapping quality using the new version RELMA V.5. The effort is probably lower than for previous manual mapping. The mapping quality equals that of manual mapping and is far better than that reported with previous automated mapping activities.
Conclusion
RELMA V.5 and LOINC V.2.34 offer the opportunity to start thinking again about LOINC mapping even in non-English languages, since mapping effort is acceptable and mapping results equal those of previous manual mapping reports.
Keywords: LOINC, RELMA, single source, semantic interoperability, electronic health records, single source projects
Objective
The reuse of electronic health records for clinical research following the single-source paradigm is gaining acceptance.1–4 The implementation of clinical data warehouses has improved the accessibility and reuse of clinical documentation by integrating data sources throughout the enterprise.5–8 However, in many cases, this integration is restricted to a level of ‘syntactic interoperability’: data items are extracted from source systems and put into a common integration database suitable for querying. This approach is very effective with items for which specific coding schemes are mandated throughout the enterprise (eg, billing codes, ICD diagnosis codes, and procedure codes). However, other content of electronic medical records (EMRs), even if stored in a structured fashion, is often coded using disparate standards or locally developed schemes. Unless these individual interface terminologies are carefully inspected and mapped to a common set of concepts, querying these datasets remains cumbersome and error-prone. Thus the lack of ‘semantic interoperability’ remains a fundamental barrier to the full realization of the single-source paradigm.9–13
Laboratory analysis results offer an exceptional potential for reuse in a single-source scenario, as they are typically well-structured and well-defined data elements, often generated in an automated, quality-controlled fashion.14 As EMRs become more widespread, clinical laboratories are increasingly delivering reports electronically in a form that can be directly stored in the client's EMR.15–17 The choice of coding system for observation identifiers has, however, been left to the communicants, who still tend to use local and idiosyncratic codes, because no adequate coding scheme has yet gained universal acceptance.9 18–20
The Logical Observation Identifiers Names and Codes (LOINC) terminology18 21 22 has been reported to be a promising tool for unlocking the potential to reuse laboratory data. It provides a large, structured and multiaxial coding system for clinical observations focusing on, but not limited to, laboratory data. LOINC terminology covers at least 98% of the average laboratory's tests.18 During its development, researchers already used the logical structure of LOINC terms as a way to translate local terms into LOINC for the purpose of sharing patient data.23 24 Furthermore, LOINC can be used for automated information integration and decision support25 26 and also as a source of computable knowledge.27 28 Since LOINC was introduced in 1996, it has been cited in more than 130 peer-reviewed publications addressing its use for coding EMR data, mapping between terminologies and other topics.
Uptake of LOINC for routine use in clinical laboratories has, however, been limited so far. Published LOINC mapping projects indicate that the availability of adequate tools and the effort to initiate and maintain full coverage of laboratory observations are important factors influencing LOINC uptake.29 30
In this context, we present our experience with a semi-automated approach to mapping non-English local laboratory terms to LOINC with the Regenstrief LOINC Mapping Assistant (RELMA)22 and discuss its advantages and disadvantages in comparison with previously reported automatic and manual approaches. Our project focused on the following aspects.
Semantic integration of heterogeneous data in a clinical data warehouse by mapping the interface terminologies of two local clinical chemistry laboratories to LOINC.
Evaluation of the semi-automated mapping approach and comparing it with methods of previously reported LOINC mapping projects.
Establishing the conditions for sustainable use of LOINC once integrated into our system.
Background
Existing LOINC mapping approaches can be categorized into two groups: manual mapping by human experts and automated mapping using tools such as RELMA.22 The Regenstrief Project,31 32 for example, demonstrated the success of manual LOINC mapping in three large institutions. We cite the results in table 1 for later reference. Up to 79% of laboratory terms could be mapped. The low coverage at ARUP resulted from data such as ‘Doctor review’ or ‘final diagnosis’, which could not be mapped to LOINC.31 32 Quality control at all three sites in a sample of 884 manually mapped terms demonstrated four completely false mappings and 36 mappings containing at least one error in one of the six axes of LOINC.31 32 We could not find exact data in the publications quoting the effort required for manual LOINC mapping, but ‘high effort’ was reported in four studies.31 32 35 36
Table 1.
Results of manual LOINC mapping31 32 compared with the automated mapping results in the Map to LOINC Project33 34
| Manual mapping | ||
| Institute | Source terms | Manual mapping (%) |
| ARUP | 4321 | 44 |
| Intermountain | 1667 | 78 |
| Regenstrief | 7387 | 79 |
| Total | 13 375 | 67 |
| Automated mapping | ||||
| Site | Source terms | Automated mapping (%) | Manual mapping (%) | Uncodeable tests (%) |
| 1 | 1050 | 76 | 11 | 13 |
| 2 | 1098 | 63 | 19 | 19 |
| 3 | 1315 | 66 | 27 | 6 |
| 4 | 1213 | 63 | 20 | 17 |
| 5 | 291 | 70 | 12 | 17 |
| Total | 4967 | 67 | 19 | 14 |
Automated LOINC mapping with a second stage manual mapping process is reported in the ‘Map to LOINC’ Project.33 34 Again we cite the results in table 1 for later reference. As expected, results are worse than with the manual approach, with 63–76% successfully mapped terms. There, mapping results could be improved within the following manual mapping step. However, no quality control for mapped terms is described, and the authors reported problems with naming choices and unspecified measurement units.33 34 None of these publications specified the effort needed to perform the mapping.
Other projects have tried automated LOINC mapping—for example, with natural language processing tools37 38—but no data on mapping quality are available.
Although use of LOINC has been reported in 140 countries,39 40 according to a presentation from the German LOINC User Group in 2008, its use in Germany is still restricted to only a few institutions,41 possibly because of the mapping effort required. However, in January 2011, a new version of RELMA was released, which supports semi-automated mapping for those foreign languages supported in the RELMA program. To our knowledge, no evaluation of these RELMA cross-language mapping features has yet been published.
Methods
Erlangen University Hospital (EUH) in Germany is a 1316 bed maximum care facility spread over a campus of about 13 km2 with more than 50 buildings. Laboratory services are performed in 13 different laboratories, with some overlap in the diagnostic spectrum. All laboratories use the Swisslab commercial laboratory information system, LIS, with disparate but non-unified codes for all observations. Laboratory data are transferred to the clinical workstation (Soarian; Siemens Inc in normal wards and for intensive care units) via HL7 using proprietary so-called interface terms. In addition, all laboratory results are transferred and loaded into the EUH clinical data warehouse (COGNOS), but, owing to the different coding systems used by the 13 laboratories, the implementation of semantically exact analyses and reports is very cumbersome and time consuming. Thus the goal was to establish LOINC as an additional coding system for analyses and query purposes in a single-source context, not to replace the existing interface terms of the routine systems.
To establish semi-automated LOINC mapping, we generated a table containing all German language interface terms from the Swisslab LIS, including available additional information such as specimen type, measurement scale (eg, narrative, quantitative), time aspect (eg, 30 min after 100 g glucose by mouth), and property (eg, substance concentration) when available. For most analyses, no test method was coded in the Swisslab LIS. Test methods were available in the standard operating procedures of the laboratory technicians—mostly written documents in pdf or word format. The respective test method for each laboratory value was extracted manually and added to the table mentioned above.
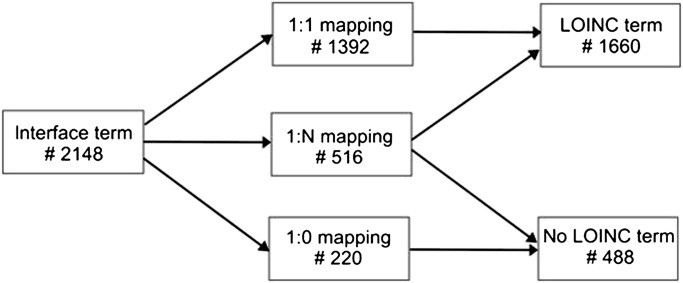
Then the resulting table was imported into RELMA's local term file to start the concept extraction. We used RELMA V.5 and LOINC V.2.34 (German Translation by DIMDI, β version42). A linguistic variant index for the German language had to be added in order to correctly process the German terms. RELMA tries to find matching LOINCs for each analyte resulting in three distinctive cases (figure 1):
No appropriate LOINC code found (1:0 mapping)
Exactly one matching LOINC code found (1:1 mapping)
Several potential LOINC codes found (1:N mapping)
Figure 1.
Results of concept mapping and manual review.
Therefore the semi-automated mapping process required an additional task when author CZ reviewed all 1:N mappings to select the matching LOINC code. Furthermore, when no matching code was found in the German β version, we screened the original English LOINC V.2.34 for matching codes as well.
For the project, we decided to map all the codes from the two laboratories with the highest throughput. In our case, these were the central laboratory (called laboratory A hereafter) and the pediatric laboratory (called laboratory B hereafter). For quality assessment, we first selected 100 interface terms from each laboratory by stratified randomization. We randomly extracted 50 of the most commonly and 50 of the most rarely used terms from each laboratory. For these 200 terms, we performed the described semi-automated mapping process and forwarded the results to two laboratory physicians in each of the laboratories for independent manual expert review. The result of this expert review was used to calculate precision, recall, and F-measure (table 2). In addition, we used Cohen's kappa to check the inter-rater reliability between the two physicians from each laboratory.
Table 2.
Mapping quality according to expert validation of 100 term samples from each laboratory
| Laboratory | ∑ | Mapped | Unmapped | Mistakes | Cohen's kappa | Precision | Recall | F-measure |
| A | 100 | 93 | 7 | 2 | 0.96 | 0.98 | 0.91 | 0.95 |
| B | 100 | 92 | 8 | 2 | 1 | 0.98 | 0.90 | 0.95 |
Next we performed semi-automated mapping of the remaining 1948 terms from laboratory A and laboratory B. Based on all 2148 terms, we calculated the coverage—that is, the fraction of terms that could be mapped—and classified the terms that could not be mapped (table 3).
Table 3.
Mapping results and categories of terms that could not be mapped
| Laboratory | Source terms | Mapped terms | Unmapped terms | ||||
| Comment | Service | Observation | Other | Total | |||
| A | 787 | 642 (82%) | 72 | 0 | 70 | 3 | 145 |
| B | 1361 | 1018 (75%) | 146 | 2 | 186 | 9 | 343 |
Finally we used the mappings for a data warehouse analysis to assess the sustainability of the mapping process when laboratory working methods change. First, we calculated the percentage of mapped observation results imported in 2010 into the data warehouse (table 4). Then we counted the new laboratory tests per month that were added to the Swisslab LIS for these two laboratories for each of the last 8 years to determine the required yearly and monthly mapping effort (figure 2).
Table 4.
Amount of terms used in 2010 that could be mapped successfully
| Laboratory | ∑ terms in use in 2010 | Terms mapped |
| A | 578 | 571 (99%) |
| B | 958 | 943 (99%) |
Figure 2.
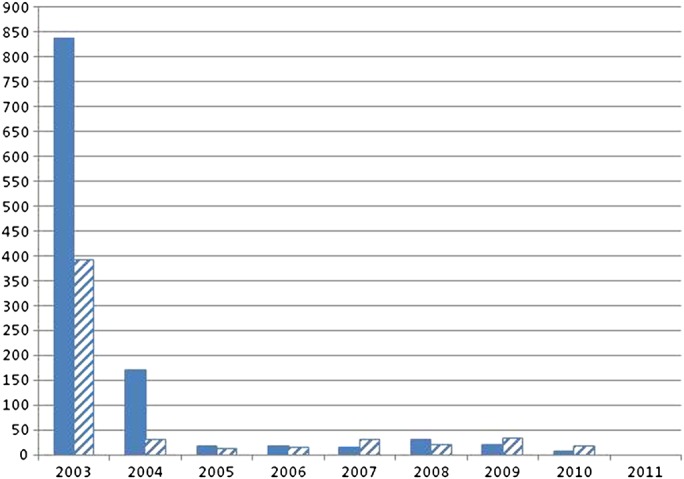
New interface terms introduced at laboratory A (hatched) and B (solid) per year.
Results
Our first goal was semantic integration of our heterogeneous laboratory data by mapping the local interface terms to LOINC. A total of 2148 interface terms were processed, and 1660 could be mapped to LOINC (figure 1). The time required to complete mapping was 4 working days, which equals 500 interface terms per day or 60 terms per hour on average. The time required per term, however, varied widely, ranging from mapping more than 200 ‘simple’ terms (eg, serum creatinine) per hour to <10 terms per hour; further investigation was required (eg, allergen mixtures for Prick tests with sometimes more than 10 different allergens).
The 1660 successfully mapped interface terms related to only 1208 individual LOINC codes because several interface terms mapped to the same LOINC concept (eg, because of the same laboratory test being offered by multiple laboratories).
Table 3 details the amount of terms that could be mapped from the central laboratory (laboratory A) and the pediatric laboratory (laboratory B). The terms that could not be mapped were classified into the four categories ‘comment’ (eg, ‘sample was hemolytic’ or ‘sample was clotted’), ‘service’ (eg, blood sampling performed by laboratory member), ‘observation’ (the term codes a laboratory value that is not yet available in LOINC), and ‘other’ (codes for internal use, eg, diagnosis imports from Intensive Care Manager (ICM); Draeger Inc, Industrial Solution Hospital (ISH); SAP Inc or LAURIS; Roche Inc).
Our second goal was validation of the mappings. As described, a random sample of 100 terms from each laboratory was selected and independently manually reviewed by two physicians in each laboratory. The quality parameters precision, recall and F-measure for each laboratory are presented in table 2. Cohen's kappa was calculated to check the inter-rater agreement between the two physicians reviewing the same sample. The physicians from laboratory A showed disagreement in the case of two terms, which was subsequently resolved by discussion. No disagreement was found in laboratory B. The physicians reported that it took 1–2 h to validate the 100 terms.
The third goal was to check the conditions for sustainable use of LOINC at our hospital. Sustainable use implies that the effort to cover the currently used terms is minimized. Using the data warehouse content, we checked how many of the terms used in the year 2010 could be mapped. Results are given in table 4. We showed a coverage of 99% for the most recently used terms.
Next we examined the turnover rate—that is, the number of new interface terms introduced per year. Results are given in figure 2. The large number of new terms in 2003 results from the initial import of the laboratory repository into the clinical data warehouse. Thereafter, the uptake of additional laboratory terms per year is much lower with <50 new terms per year since 2005 in both laboratories.
Discussion
We have been able to map the majority of more than 2000 non-English local laboratory interface terms from two laboratories to LOINC with a semi-automated approach using RELMA V.5. The total mapping effort of 4 working days was acceptable. Mapping quality in two samples of 100 terms was good; discussion is underway on validation of the semi-automated mapping results of the remaining terms. The LOINC mapping at EUH has the potential for semantic integration of laboratory values from the 11 other different laboratories and enables comprehensive analysis of laboratory values in our clinical data warehouse. Furthermore, LOINC mapping can be leveraged for routine care—for example, to structure the display format of laboratory results in the EMR or to facilitate communication between clinical systems. Use in routine clinical care, however, is subject to full validation of all mappings, which has not yet been carried out.
Mapping methods
The routine laboratory system (Swisslab) did not provide all data elements needed to perform the mapping in every case. Even though many items could be mapped on the basis of the existing structured metadata, additional specifications (eg, analysis method of a laboratory test) were required to resolve some cases. The missing information, mostly the analysis method, was available in standard operating procedures provided by the laboratories and could be manually extracted and added to the tabular source data.
A high throughput was achieved by implementing a semi-automated mapping approach with RELMA V.5, followed by manual resolution of items with either no direct hits or multiple candidates. Other published mapping projects have neither disclosed the actual time required nor the throughput for manual31 32 35 36 or automated33 34 37 38 mapping, but we believe that the mapping rate of 500 terms per day or 60 terms per hour on average in this project shows that it is feasible to convert even large repositories of heterogeneous laboratory findings into LOINC in a reasonable amount of time. Our approach was limited by the German LOINC translation, which covered only 11 00043 of the most common terms from a total of 61 255 terms in LOINC V.2.34. We believe that in an English language environment, or by having a complete translation available, a higher rate of direct matches and thus an even higher throughput could have been achieved. Also, the qualification of the person carrying out the manual stage of the mapping has to be taken into account, which may affect either the mapping rate (because of delegating queries to the laboratory team) or the mapping quality (because of mismappings). In this project, the manual mapping stage was carried out by a third-year medical student (CZ).
Validation was carried out by submitting a stratified sample of 100 interface terms to two members of the respective laboratory team for review. Depending on the planned use of the LOINC codes, different validation approaches may be appropriate. A partial validation may be sufficient for use in single-source scenarios—for example, supporting patient recruitment for clinical studies with subsequent mandatory manual supervision of query results. If mapped terms are intended for use in routine clinical care, full validation of all mappings should be mandatory. While the actual time needed to perform the validation was limited (2 h for 100 terms per laboratory), it should be noted that it is subject to availability of qualified routine laboratory personnel.
Mapping results
We were able to map 82% (laboratory A) and 75% (laboratory B) of our interface terms. Lin et al 31 32 reported a coverage of 44%, 78% and 79% of their terms at three different sites, whereas Khan et al 33 34 produced a coverage of 67% with their fully automated mapping tool. Their manual review33 34 showed that, after automated mapping, there were many unmapped terms remaining for which a precise LOINC existed, but the tool did not find it. The coverage of our semi-automated mapping using RELMA V.5, however, was just as good as the coverage of the manual mapping of Lin et al,31 32 whereas the full automatic mapping tool of Khan et al 33 34 missed 11–27% of interface terms (compare with table 1).
Our mapping precision was 0.98, whereas Lin et al 31 32 reported a precision of 0.95, with 40 mistakes in a sample of 884 terms. Khan et al 33 34 did not disclose their mapping errors. Mapping errors noted during our validation never concerned more than one LOINC axis (eg, mistaking only the specimen ‘platelet depleted plasma’ with ‘platelet rich plasma’). These results show that the quality of mapping performed with RELMA is comparable to the quality of manual mapping.
Of 488 terms that we could not map to LOINC, only 256 concerned actual laboratory findings, with the others being mostly service codes and comments (table 3). A total of 149 of these laboratory values were in use in 2010. Discussion about submitting these interface terms to the Regenstrief Institute for inclusion in future releases of the LOINC is ongoing with both laboratory teams.
Considerations regarding sustainability
Once the initial mapping has been completed, it appears that only a limited number of new laboratory tests have to be added per year to keep the mapping up to date. With approximately two new terms per laboratory and month, the time required to carry out these additional mappings should be less than an hour per month.
Conclusion
The results of our project have shown that, using a semi-automated approach based on RELMA V.5, full mapping of non-English routine clinical laboratory interface terms to LOINC can be achieved in a reasonable amount of time. We established LOINC as a method of coding in addition to the existing interface terms, thus clinical routine and laboratory infrastructure have not been impeded, but additional use cases in a single-source context are facilitated. The extra effort required to maintain the mapping in future appears to be limited.
Acknowledgments
We would like to thank Drs Seefried, Schmid and Parsch from the Central Laboratory and Dr Rauh and Mrs Porzelius from the Pediatric Laboratory of Erlangen University Hospital for their support and collaboration during the mapping and validation of their respective laboratory interface terms. We would also like to thank Dr Vreeman and Mr Hook from the Regenstrief Institute for providing a linguistic variant index used to make the German LOINC translation available in RELMA V.5.
Footnotes
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Herzberg S, Dugas M. Single source information systems can improve data completeness in clinical studies: an example from nuclear medicine. Stud Health Technol Inform 2011;169:872–6 [PubMed] [Google Scholar]
- 2. El Fadly A, Rance B, Lucas N, et al. Integrating clinical research with the healthcare enterprise: from the RE-USE project to the EHR4CR platform. J Biomed Inform 2011;44(Suppl 1):S94–102 [DOI] [PubMed] [Google Scholar]
- 3. Fritz F, Ständer S, Breil B, et al. CIS-based registration of quality of life in a single source approach. BMC Med Inform Decis Mak 2011;11:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fritz F, Balhorn S, Riek M, et al. Qualitative and quantitative evaluation of EHR-integrated mobile patient questionnaires regarding usability andcost-efficiency. J Biomed Inform 2012;81:303–13 [DOI] [PubMed] [Google Scholar]
- 5. Ganslandt T, Prokosch HU, Beyer A, et al. Single-source-projekte am universitätsklinikum Erlangen. Forum der Medizin_Dokumentation und Medizin_Informatik 2010;2:62–6 [Google Scholar]
- 6. Prokosch HU, Ries M, Beyer A, et al. IT infrastructure components to support clinical care and translational research projects in a comprehensive cancer center. Stud Health Technol Inform 2011;169:892–6 [PubMed] [Google Scholar]
- 7. Hu H, Correll M, Kvecher L, et al. DW4TR: a data warehouse for translational research. J Biomed Inform 2011;44:1004–19 [DOI] [PubMed] [Google Scholar]
- 8. Cuggia M, Garcelon N, Campillo-Gimenez B, et al. Roogle: an information retrieval engine for clinical data warehouse. Stud Health Technol Inform 2011;169:584–8 [PubMed] [Google Scholar]
- 9. McDonald CJ. The barriers to electronic medical record systems and how to overcome them. J Am Med Inform Assoc 1997;4:213–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med 1998;37:394–403 [PMC free article] [PubMed] [Google Scholar]
- 11. Nagy M, Hanzlicek P, Precková P, et al. Semantic interoperability in Czech healthcare environment supported by HL7 version 3. Methods Inf Med 2010;49:186–95 [DOI] [PubMed] [Google Scholar]
- 12. Scott P, Worden R. Semantic mapping to simplify deployment of HL7 v3 Clinical document Architecture. J Biomed Inform. Published Online First: 24 March 2012. doi:10.1016/j.jbi.2012.02.006 [DOI] [PubMed] [Google Scholar]
- 13. Berges I, Bermudez J, Illarramendi A. Towards semantic interoperability of electronic health records. IEEE Trans Inf Technol Biomed. Published Online First: 30 December 2011. doi:10.1109/TITB.2011.2180917 [DOI] [PubMed] [Google Scholar]
- 14. Emons MF. Integrated patient data for optimal patient management: the value of laboratory data in quality improvement. Clin Chem 2001;47:1516–20 [PubMed] [Google Scholar]
- 15. McDonald CJ, Overhage JM, Tierney WM, et al. The regenstrief medical record system: a quarter century experience. Int J Med Inform 1999;54:225–53 [DOI] [PubMed] [Google Scholar]
- 16. Reichert JC, Glasgow M, Naru SP, et al. Using LOINC to link an EMR to the pertinent paragraph in a structured reference knowledge base. Proc AMIA Symp 2002;2002:652–6 [PMC free article] [PubMed] [Google Scholar]
- 17. Wurtz R, Cameron BJ. Electronic laboratory reporting for the infectious diseases physician and clinical microbiologist. Clin Infect Dis 2005;40:1638–43 [DOI] [PubMed] [Google Scholar]
- 18. Forrey AW, McDonald CJ, DeMoor G, et al. Logical observation identifier names and codes (LOINC) database: a public use set of codes and names for electronic reporting of clinical laboratory test results. Clin Chem 1996;42:81–90 [PubMed] [Google Scholar]
- 19. Kroth PJ, Daneshvari S, Harris EF, et al. Using LOINC to link 10 terminology standards to one unified standard in a specialized domain. J Biomed Inform. Published Online First: 19 October 2011. doi:10.1016/j.jbi.2011.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vreeman DJ, Chiaravalloti MT, Hook J, et al. Enabling international adoption of LOINC through translation. J Biomed Inform. Published Online First: 21 January 2012. doi:10.1016/j.jbi.2012.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huff SM, Rocha RA, McDonald CJ, et al. Development of the logical observation identifier names and codes (LOINC) vocabulary. J Am Med Inform Assoc 1998;5:276–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem 2003;49:624–33 [DOI] [PubMed] [Google Scholar]
- 23. Cimino JJ, Zhu X. The practical impact of ontologies on biomedical informatics. Yearb Med Inform 2006;2006:124–35 [PubMed] [Google Scholar]
- 24. Abhyankar S, Lloyd-Puryear MA, Goodwin R, et al. Standardizing newborn screening results for health information exchange. AMIA Annu Symp Proc 2010;2010:1–5 [PMC free article] [PubMed] [Google Scholar]
- 25. Ahmadian L, Engen-Verheul M, Bakhshi-Raiez F, et al. The role of standardized data and terminological systems in computerized clinical decision support systems: literature review and survey. Int J Med Inform 2011;80:81–93 [DOI] [PubMed] [Google Scholar]
- 26. Bonney W. Is it appropriate, or ethical, to use health data collected for the purpose of direct patient care to develop computerized predictive decision support tools? Stud Health Technol Inform 2009;143:115–21 [PubMed] [Google Scholar]
- 27. Srinivasan A, Kunapareddy N, Mirhaji P, et al. Semantic web representation of LOINC: an ontological perspective. AMIA Annu Symp Proc 2006;2006:1107. [PMC free article] [PubMed] [Google Scholar]
- 28. Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform 2008;2008:67–79 [PMC free article] [PubMed] [Google Scholar]
- 29. Lin MC, Vreeman DJ, Huff SM, et al. Investigating the semantic interoperability of laboratory data exchanged using LOINC codes in three large institutions. AMIA Annu Symp Proc 2011;2011:805–14 [PMC free article] [PubMed] [Google Scholar]
- 30. Lin MC, Vreeman DJ, McDonald CJ, et al. Auditing consistency and usefulness of LOINC use among three large institutions –Using version spaces for grouping LOINC codes. J Biomed Inform. Published Online First: 28 January 2012. doi:10.1016/j.jbi.2012.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin MC, Vreeman DJ, McDonald CJ, et al. A characterization of local LOINC mapping for laboratory tests in three large institutions. Methods Inf Med 2011;50:105–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin MC, Vreeman DJ, McDonald CJ, et al. Correctness of voluntary LOINC mapping for laboratory tests in three large institutions. AMIA Annu Symp Proc 2011;2011:447–51 [PMC free article] [PubMed] [Google Scholar]
- 33. Khan AN, Russel D, Moore C, et al. The map to LOINC project. AMIA Annu Symp Proc 2003;2003:890. [PMC free article] [PubMed] [Google Scholar]
- 34. Khan AN, Griffith SP, Moore C, et al. Standardizing laboratory data by mapping to LOINC. J Am Med Inform Assoc 2006;13:353–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Baorto DM, Cimino JJ, Parvin CA, et al. Using logical observation identifier names and codes (LOINC) toelaboratory data among three academic hospitals. Proc AMIA Annu Fall Symp 1997;1997:96–100 [PMC free article] [PubMed] [Google Scholar]
- 36. Baorto DM, Cimino JJ, Parvin CA, et al. Combining laboratory data sets from multiple institutions using the logical observation identifier names and codes (LOINC). Int J Med Inform 1998;51:29–37 [DOI] [PubMed] [Google Scholar]
- 37. Lau LM, Johnson K, Monson K, et al. A method for the automated mapping of laboratory results to LOINC. Proc AMIA Symp 2003;2003:472–6 [PMC free article] [PubMed] [Google Scholar]
- 38. Lau LM, Johnson K, Monson K, et al. Mapping department of defense laboratory results to logical observation identifiers names and codes (LOINC). AMIA Annu Symp Proc 2005;2005:430–4 [PMC free article] [PubMed] [Google Scholar]
- 39. http://loinc.org/international (accessed 16 Apr 2012).
- 40. McDonald CJ, Huff SM, Mercer K, et al. LOINC User's Guide. Indianapolis, IN: Regenstrief Institute, 2011 [Google Scholar]
- 41. http://www.tmf-ev.de/DesktopModules/Bring2mind/DMX/Download.aspx?Method=attachment&Command=Core_Download&EntryId=3270&PortalId=0 (accessed 16 Apr 2012)
- 42. http://www.dimdi.de/static/de/klassi/loinc/index.htm (accessed 16 Apr 2012).
- 43. http://www.dimdi.de/static/de/klassi/aktuelles/news_0259.htm_319159480.htm (accessed 16 Apr 2012).