Abstract
The dendrimer chemistry reported offers a route to synthetic target molecules with spherical shape, well-defined surface chemistries and dimensions that match the size of virus particles. The largest target, a generation 13 dendrimer comprising triazines linked by diamines, is stable across ranges of concentration, pH, temperature, solvent polarity and in the presence of additives. This dendrimer theoretically, presents 16384 surface groups and has a molecular weight exceeding 8.4 million Daltons. Transmission electron and atomic force microscopies, dynamic light scattering, and computation reveal a diameter of approximately 30 nm. The target is synthesized through an iterative divergent approach using a monochlorotriazine macromonomer providing two generations of growth per synthetic cycle. Fidelity in synthesis is supported by evidence from NMR spectroscopy, mass spectrometry, and high pressure liquid chromatography.
Viruses—with diameters ranging from 20 nm to 400 nm—occupy a length scale that represents the heart of all that is nano.1 The lower bounds of this length scale are a convergent point for both top-down and bottom-up synthesis. Existing organic building blocks providing access to this size regime derive from all disciplines including chemistry, molecular biology and virology and include micelles, liposomes, and viral capsids, respectively. These materials rely on self-assembly. Self-assembly reduces the burden of synthesis, but typically at the cost of stability as reflected in sensitivity to concentration, pH, temperature, solvents, and additives such as denaturants and detergents. Covalent synthesis offers an alternative to self-assembly and significant opportunities for compositional control. Virus-sized, covalent organic molecules have remained elusive targets.
First described by Tomalia,2 Newkome,3 and Vögtle4 in the late 1970s and early 1980s, dendrimers are highly branched, globular macromolecules that present multiple surface groups on the periphery. These architectures have been the focus of experimental and theoretical investigations in areas including materials science and medicinal chemistry.5 While a myriad of platforms have been described, a handful have been the focus of intense scrutiny based on the ease of synthesis and/or commercial availability. These materials include the polylysines introduced by Denkewalter6, the poly(propylene imine) (PPI) dendrimers of Vögtle4 and Meijer,7 the polyamidoamine (PAMAM) dendrimers of Tomalia,2 the polyaryl ethers of Fréchet8, the polyesters advanced by Frechet and others,9 and the phosphorous containing dendrimers of Majoral and Caminade.10 Still, the synthesis of large generation dendrimers is rare. Reports of large dendrimers include PAMAM11 and phosphorus containing dendrimers,10,12 both of which are commercially available up to generation 10. Phosphorus-based dendrimers have long occupied the benchmark for large molecules, but at G12 they encounter a solubility challenge that precludes additional pursuits.12b At these generations, however, both this platform and PAMAM dendrimers only reach diameters of ~14–15 nm. As such, these materials populate the length scale of proteins, but fall short of the length scales of viruses.
For many years, we have been interested in triazine dendrimers.13 Synthetically, these materials derive from cyanuric chloride and diamines that can be elaborated to dendrimer cores and monomers. As a platform, this class of dendrimers offers a number of benefits including i) low cost of reagents, ii) compositional diversity stemming from the wealth of available diamines and the step-wise substitution of the triazine nucleus, iii) ease of large scale synthesis, iv) stability to highly acidic (pH 0) or basic (pH 14) solutions, and v) long shelf-life. These materials can be prepared using either convergent or divergent approaches, although the only latter provides access to large molecules.
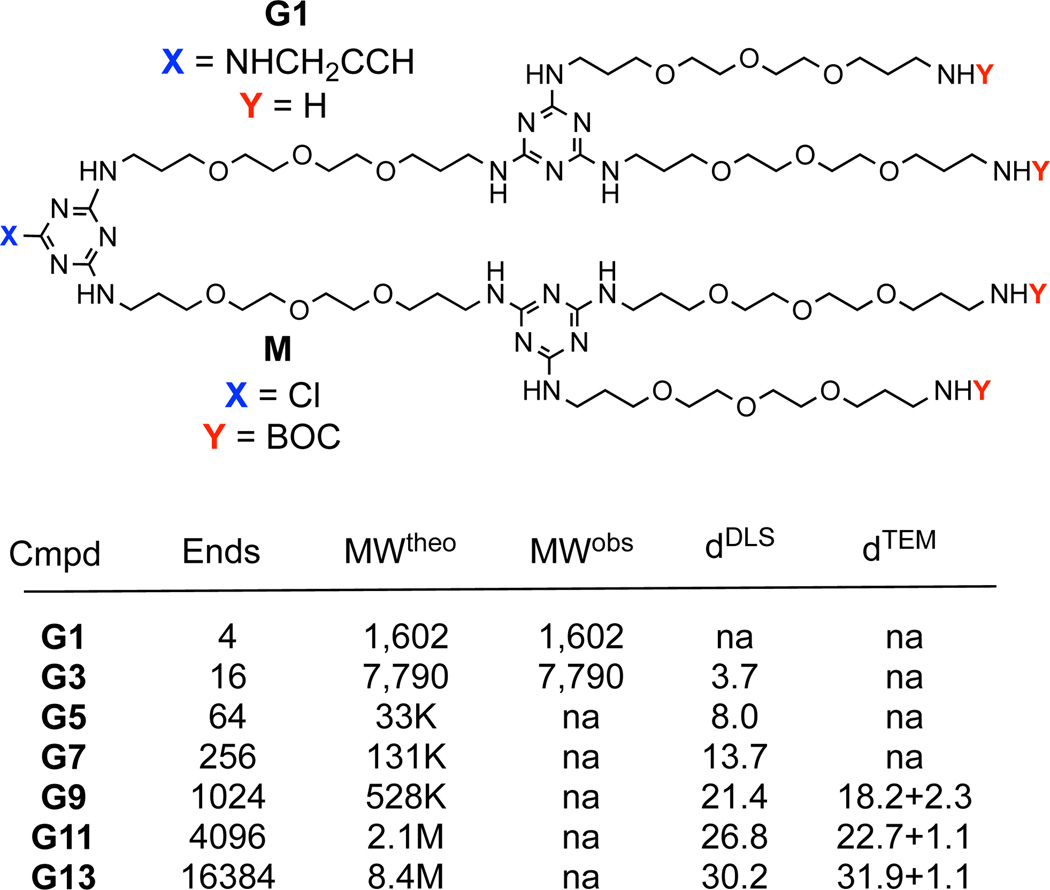
To reach the target generation 13 dendrimer, G13, the synthesis starts with a generation one amine core, G1, that is reacted with macromonomer M (Figure 1). We install an alkyne at the core in anticipation of future manipulation through click chemistry. Following deprotection of the resulting intermediate, the process is repeated. By design, macromonomer M has a single reactive site, a monochlorotriazine, that precludes formation of covalent dimers thereby limiting side products. Previously, in order to synthesize large dendrimers, we employed two different diamines as linkers; piperazine and an oligoether diamine.13c This synthetic approach was successful up to generation 9 before poor solubility was encountered at generation 11. The former diamine, piperazine, provides a more reactive, rigid linking group without hydrogen bond donors. The latter sacrifices reactivity for flexibility and length, and introduces hydrogen bond donors and acceptors. Here, only the latter flexible diamine is adopted. Dendrimers comprising this diamine show excellent solubility (> 100 mg/mL) at all generations explored. The protected dendrimers are readily soluble in organic solvents. Amine-terminated dendrimers show good solubility in both water and organic solvents.
Figure 1.
Structures of the G1 dendrimer with alkyne core and the monomer, M, that is reacted to install two additional generations per iteration. “Cmpd” lists the intermediate dendrimers compounds accessed in route to G13. The number of end groups, “Ends”, the theoretical and observed molecular weights (determined by ESI-TOF) in Daltons, “MWtheo” and “MWobs”, and the diameters in nanometers recorded with dynamic light scattering and TEM, “dDLS” and “dTEM” are indicated. The dTEM data includes the stain in the measurement and provide an upper limit on size.
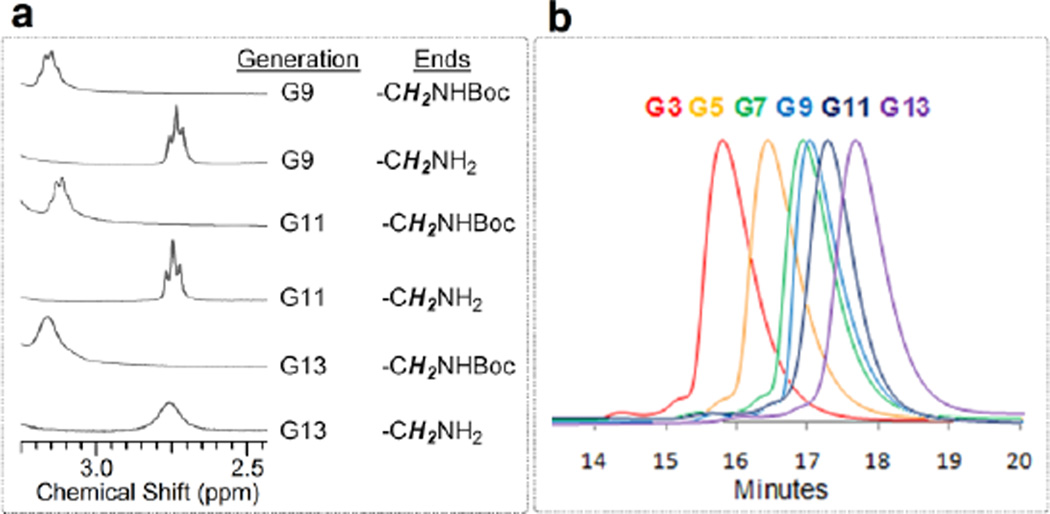
Target G13 is the largest dendrimer reported to date, with 13 branching points (generations) between the core and periphery (Figure 1). This target, theoretically, presents 16,384 amine groups on the surface and has a molecular weight exceeding 8.4 million Daltons. However, due to the size of the G13, the extent to which the actual target(s) of this synthesis represent the idealized structure is unknown. Evidence for success derives from multiple techniques including the behavior of lower generation intermediates as well as the characterization of the final product. That is, the iterative synthesis reveals trends that match expectations with regard to both differences in polarity of the protected and deprotected materials, as well as the appearance and disappearance of characteristic lines derived from the peripheral groups in the NMR spectra (Figure 2a). Specifically, removing the t-butoxycarbonyl (BOC) protecting group leads to a change in chemical shift of the vicinal —CH2— group from 3.2 ppm to 2.75 ppm. While the spectra suggest a clean reaction, confidence in this interpretation is compromised by the limits of detection afforded by 1H NMR. Given the large number of protons in the final product—737,240 protons theoretically—1H NMR spectroscopy cannot provide evidence of complete success in conversion at the final product stage, or even beyond the generation 3 intermediates due to inherent limits in signal-noise.
Figure 2.
(a) The 1H NMR spectra of the large generation dendrimers (G9–G13) have a finger print region for monitoring the reiterative addition of monomer M and deprotection: The vicinal proton signals of NHBoc groups appear around at 3.2 ppm, while the vicinal proton signals of NH2 groups appear around at 2.75 ppm. (b) HPLC traces of the dendrimers G3–G13.
High pressure liquid chromatography (HPLC) shows that larger generations elute later than smaller generation materials (Figure 2b). Based on the amount of readily discerned impurities in the traces, we can assign the purity of the target G13 and other lower generation intermediates to be ~95%. This purity label reflects purity in size, and not atomic composition of the theoretical structure depicted. We infer that these impurities are lower molecular weight species that may arise due to incomplete deprotection or incomplete reactions of M with the deprotected dendrimer.
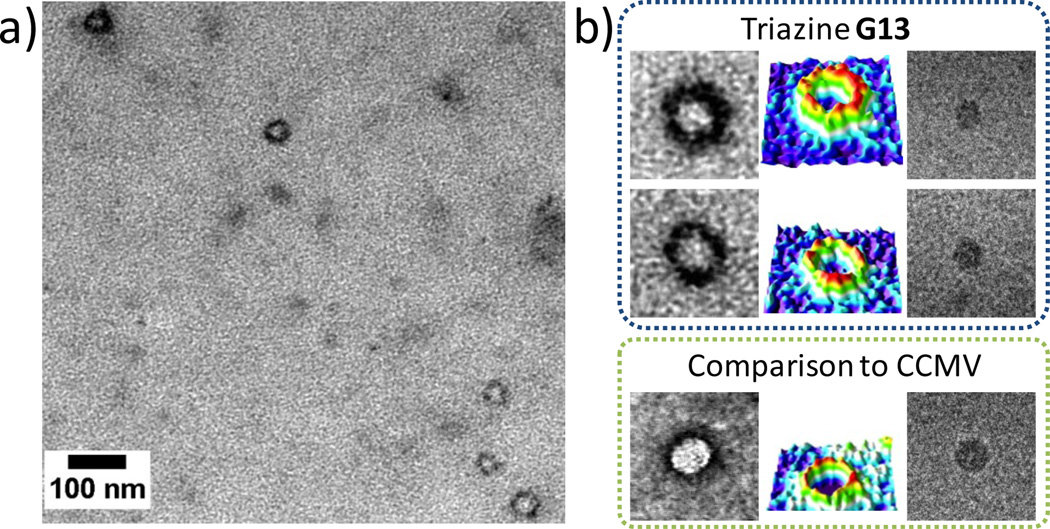
The most compelling evidence for success in the preparation of virus-sized particles comes from imaging G13 using cryogenic transmission electron microscopy (cryo-TEM). Figure 3 shows cryo-TEM images of both G13 and the cowpea chlorotic mottle virus (CCMV), an icosahedral RNA plant virus. X-ray crystallography and transmission electron microscopy reveal that the virus has a diameter of ~28 nm. The microscopy images show similarities in size and shape between these the virus and G13. Both the virus and the G13 appear spherical when imaged as either dried samples stained to provide high contrast, or vitrified aqueous solutions. Because the staining is imperfect, size values derived yield a range rather than a single number. If the stain does not penetrate the dendrimer significantly (the standard assumption), the diameter for G13 is 24.4 ± 2.3 nm. If the dye layer is included with the belief that partial penetration occurs due to the nature of the periphery, the dimension increases to 31.9 ± 1.1 nm. Unstained samples derived from vitrified solutions provide a diameter of 25.2 ± 2.3 nm, although this may underrepresent the size of the particle slightly because the image contrast at the edges of the G13 particle is low.
Figure 3.
Transmission electron microscopy (TEM) of G13 triazine dendrimer and comparison to CCMV virus, (a) G13 imaged with TEM (sample dried and negatively stained with uranyl acetate), (b) Close view of the G13 dendrimer and comparison to CCMV with dry samples prepared by negative staining (left columns), colored 3D intensity profiles (middle columns), and samples derived from vitrified aqueous solutions (right columns). Image sizes 100 × 100 nm.
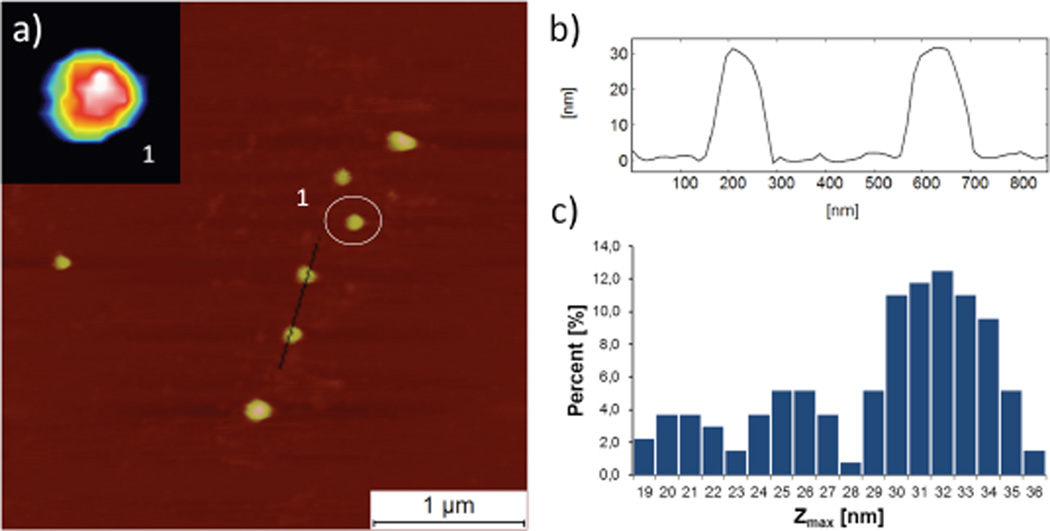
Atomic force microscopy (AFM) corroborates the measurements of size and shape and suggests a hydration-state dependent size. Figure 4 shows the AFM analysis of G13 on atomically flat hydrophilic mica in water. The average height of G13 (Zmax parameter) which corresponds to diameter of dendrimer assuming the ideal spherical shape is Zmax = 31.5 ± 1.9 nm. AFM analysis of dry samples suggests that the den-drimers collapse significantly in Z-scale (Zmax = 9.8 ± 1.9, Supporting Information). This collapse suggests a high degree of hydration of internal void space of dendrimer, or at least, a higher sensitivity to the force at the AFM tip when dendrimers are dehydrated. The results are similar to those observed by Haag with hydrogels that collapsed from ~100 nm to ~20 nm on drying.14 The size distribution function from AFM also provides insights into the dispersity of the samples. The histogram in Figure 4c shows evidence for two smaller materials present in the sample of G13 with diameters of ~20 and 25 nm. Populated at almost 35% of the sample, these entities appear to be discrete populations of macromolecules and may correspond to materials with sizes similar to G7 and G9/G11 as suggested by the HPLC traces. Their appearance is consistent with failures of purification using conventional chromatographic methods and incomplete reactions throughout the synthesis. Clearly, room for improvement exists, but this has not proven trivial to date. Filtration during sample preparation and aggregation of G13 appears to enrich the populations of smaller particles. Accordingly the extent of smaller particles is likely to be closer to the HPLC-based estimates of ~5% rather than the AFM-based estimates of 35%. The HPLC traces suggest that these impurities are elaborated throughout the synthesis—perhaps due to a failed deprotection or macromonomer addition early in the process. Their existence, however, suggests opportunities for finer control over the size of particles by manipulating the valency of the dendrimer core.
Figure 4.
Atomic force microscopy (AFM) analysis of G13 in liquid environment, (a) G13 imaged in water on mica surface. Inset: close view of G13 dendrimer marked by circle (1) with image size 230 × 230 nm. (b) Cross section profiles (average of 3 lines) of two dendrimers. (c) Histogram showing heights of two subpopulations of particles in addition to G13.
Measurements from dynamic light scattering were performed on high-generation dendrimers in aqueous salt solutions at neutral pH. Results for both G9 and G11 were obtained in phosphate buffered saline (PBS) at pH 7.0. Aggregation of G13 in PBS required the use of 0.1 M NaCl (aq) at pH=7. In all cases, distributions of light-scattering intensities were found to be bimodal. The fast-diffusion mode was related to the size of dendrimer using the Stokes-Einstein equation. The extracted hydrodynamic diameters, which are shown in Figure 1, increase with dendrimer generation going from G9 to G13. The slow-diffusion mode can be related to the presence of large dendrimer aggregates with diameters ranging from 50 to 150 nm. Considering that hydration contributes to the particle hydrodynamic diameter in solution, the DLS values are consistent with ranges provided by TEM and AFM analyses. DLS measurements were also performed on G13 in the absence of salts yielding dendrimer diffusion coefficients that were about 10% higher. This effect can be related to dendrimer charge and the absence of electrostatic screening.15
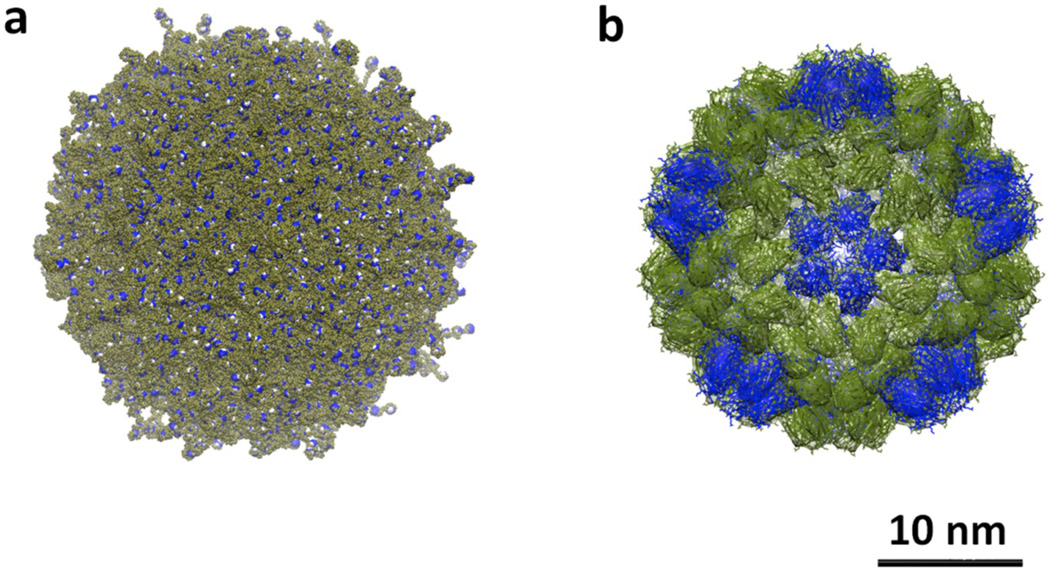
Computational models provide additional insights into structure and anchor our intuitive picture of these molecules. A fully atomistic simulation in explicit solvent was not possible for G13, as explicit molecules of water exceeded the computational infrastructure necessary for this 1.3 million atom molecule: G13 is theoretically C376812H737240N114682O98298. Moreover, the use of coarse-grained simplified models for G13 was incompatible with the linear and extremely flexible character of the monomers that constitute the dendritic scaffold. The construction of the G13 atomistic model was extremely challenging due to high structural complexity emerging from the large number of monomer connections. MD simulations reveal that G13 reaches a “hard sphere” limit (Figure 5). Peripheral amine groups both extend outward and inward by backfolding into the dendrimer resulting in high structural density (see RDF plots in Supporting Information). The measured radius of gyration of this molecule is Rg= 11.0–11.4 nm. This value is in good agreement with the diameters measured using dynamic light scattering, as Rg and the radius of hydration, Rh, are related by the equation, Rh ≈ Rg * 1.29 for spherical molecules of uniform density.16 Accordingly, these diameters of 28.6–29.4 nm match estimates derived from TEM and AFM.
Figure 5.
(a) Computational model of G13 derived from molecular dynamics simulations. Terminal amine groups are shown in blue, (b) CCMV particle colored olive and blue to illustrate the icosahedral structure.
The success in generating virus-sized materials observed here and not yet seen in PAMAM or phosphorous platforms may derive from more than just the solubility of the linking diamine. Clearly, the generation unit of the triazine dendrimers is approximately twice as long as the other platforms. PAMAM relies on –N(R)CH2CH2C(O)NHCH2CH2—with 7 atoms. Phosphorous dendrimers rely on –P(R)(S)O-(p-C6H4)-C=NNH—with 9 atoms. This triazine platform utilizes 18 atoms per generation. Relying on the number of flexible atoms, P, de Gennes predicts the maximum size a dendrimer with perfect branching before defects are necessitated by steric crowding of the periphery (eq 1).17 Using P=18, the maximum generation achieved before defects is 12.6.
| eq 1 |
The nature of the branching unit may be less relevant. While both PAMAM and phosphorous dendrimers branch (shown as R) from a single atom that is shown in bold above—either a tertiary amine nitrogen or pentavalent phosphorus atom, respectively—both triazines and cyclophosphazene dendrimers branch from a larger rigid ring.
The covalent, single-molecule nanomaterial described contributes a new platform for nanosynthesis. Its viral dimensions and spherical shape are complemented by a well-defined (although not uniquely perfect) composition. Extending opportunities for functional group variation both within and on the periphery of these building materials is precedent in smaller generation dendrimers of all classes. Polymerization chemistry offers another route to such targets. Indeed, linear and hyperbranched polymers of similar molecular weight can be achieved, although control over shape in linear systems is not as readily guaranteed as it is with dendrimers, nor is control over composition in hyperbranched systems.14 Self-assembly18 has achieved these dimensions including Percec’s dendrisomes19 and other systems that can be subsequently cross-linked,20 although such materials are usually more sensitive to factors including temperature, detergents, and concentration. The primary disadvantage of this system, the burden of synthesis, is reduced significantly by using an iterative route with a common macromonomer. Accordingly, we believe that these materials offer a complementary avenue to emulating some of the properties of viruses, and further expand the nanoperiodic table.21 These properties could include i) attachment to biological surfaces including bacteria, cells, or bone, ii) delivery of cargo including nucleic acids or small molecules, iii) or in the current pursuit as use as building blocks for hybrid materials.22
Supplementary Material
ACKNOWLEDGMENT
The authors acknowledge the following: NIH (NIGMS R01 65460, EES), the Robert A. Welch Foundation (A-0008, EES), the Academy of Finland (13137582, MAK), the Emil Aaltonen Foundation and Aalto starting grant (MAK), the Czech national COST project (OC10053, JM), the Czech Science Foundation (13-06609S, JM), and the ACS Petroleum Research Fund (47244-G4, OA). This work made use of the Aalto University Nanomicroscopy Center (Aalto-NMC) with assistance on the cryo-TEM from J. Seitsonen.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Details of synthesis, characterization, and computation. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Levine A. Viruses: A Scientific American Book. New York: W.H. Freeman and Co.; 1992. p. 240. [Google Scholar]
- 2.Tomalia DA, Baker H, Dewald J, Hall M, Kallos G, Martin S, Roeck J, Ryder J, Smith P. Polym. J. 1985;17:117. [Google Scholar]
- 3.Newkome GR, Yao ZQ, Baker GR, Gupta VK. J. Org. Chem. 1985;50:2003. [Google Scholar]
- 4.Buhleier E, Wehner W, Vögtle F. Synthesis. 1978;2:155. [Google Scholar]
- 5.(a) Helms B, Meijer EW. Science. 2006;313:929. doi: 10.1126/science.1130639. [DOI] [PubMed] [Google Scholar]; (b) Hu J, Xu T, Cheng Y. Chem. Rev. 2012;112:3856. doi: 10.1021/cr200333h. [DOI] [PubMed] [Google Scholar]; (c) Percec V, Peterca M, Dulcey AE, Imam MR, Hudson SD, Nummelin S, Adelman P, Heiney PA. J. Am. Chem. Soc. 2008;130:13079. doi: 10.1021/ja8034703. [DOI] [PubMed] [Google Scholar]
- 6.Denkewalter RG, Kolc J, Lukasavage WJ. Macromolecular highly branched homogeneous compound based on lysine units. 4,289,872. U.S. Patent. 1981
- 7.Brabander-van den Berg EMM, Meijer WW. Angew. Chem. Int. Ed. Engl. 1993;32:1308. [Google Scholar]
- 8.Grayson SM, Fréchet JMJ. Chem. Rev. 2001;101:3819. doi: 10.1021/cr990116h. [DOI] [PubMed] [Google Scholar]
- 9.Gillies ER, Fréchet JMJ. J. Am. Chem. Soc. 2002;124:14137. doi: 10.1021/ja028100n. [DOI] [PubMed] [Google Scholar]
- 10.Caminade AM, Majoral JP. Acc. Chem. Res. 2004;37:341. doi: 10.1021/ar020077n. [DOI] [PubMed] [Google Scholar]
- 11.(a) Maiti PK, Çağin T, Wang G, Goddard WA., III Macromolecules. 2004;37:6236. [Google Scholar]; (b) Li J, Piehler LT, Qin D, Baker JR, Jr, Tomalia DA. Langmuir. 2000;16:5613. [Google Scholar]
- 12.(a) Caminade AM, Laurent R, Majoral JP. Adv. Drug Deliv. Rev. 2005;57:2130. doi: 10.1016/j.addr.2005.09.011. [DOI] [PubMed] [Google Scholar]; (b) Majoral JP, Caminade AM. Top. Curr. Chem. 1998;197:79. [Google Scholar]
- 13.(a) Simanek EE, Abdou H, Lalwani S, Lim J, Mintzer MA, Venditto VJ, Vittur B. Proc. R. Soc. A. 2010;466:1445. [Google Scholar]; (b) Lim J, Simanek EE. Adv. Drug Deliv. Rev. 2012;64:826. doi: 10.1016/j.addr.2012.03.008. [DOI] [PubMed] [Google Scholar]; (c) Lim J, Pavan GM, Annunziata O, Simanek EE. J. Am. Chem. Soc. 2012;134:1942. doi: 10.1021/ja210122z. [DOI] [PubMed] [Google Scholar]
- 14.Zhou H, Steinhilber D, Schlaad H, Sisson AL, Haag R. Reactive Functional Polym. 2011;71:356. [Google Scholar]
- 15.Schmitz KS. An introduction to Dynamic Light Scattering by Macromolecules. San Diego: Academic Press; 1990. pp. 205–214. [Google Scholar]
- 16.Jensen LB, Mortensen K, Pavan GM, Kasimova MR, Jensen DK, Gadzhyeva V, Nielsen HM, Foged C. Biomacromolecules. 2010;11:3571. doi: 10.1021/bm101033g. [DOI] [PubMed] [Google Scholar]
- 17.de Gennes PG, Hervet H. J. Physique – LETTRES. 1983;44:L351. [Google Scholar]
- 18.Whitesides GM, Mathias JP, Seto CT. Science. 1991;254:1312. doi: 10.1126/science.1962191. [DOI] [PubMed] [Google Scholar]
- 19.(a) Peterca M, Percec V, Leowanawat P, Bertin A. J. Am. Chem. Soc. 2011;133:20507. doi: 10.1021/ja208762u. [DOI] [PubMed] [Google Scholar]; (b) Percec V, Wilson DA, Leowanawat P, Wilson CJ, Hughes AD, Kaucher MS, Hammer DA, Levine DH, Kim AJ, Bates FS, Davis KP, Lodge TP, Klein ML, DeVane RH, Aqad E, Rosen BM, Argintaru AO, Sienkowska MJ, Rissanen K, Nummelin S, Ropponen J. Science. 2010;328:1009. doi: 10.1126/science.1185547. [DOI] [PubMed] [Google Scholar]
- 20.O’Reilly RK, Hawker CJ, Wooley KL. Chem. Soc. Rev. 2006;35:1068. doi: 10.1039/b514858h. [DOI] [PubMed] [Google Scholar]
- 21.(a) Tomalia DA. Soft Matter. 2010;6:456. [Google Scholar]; (b) Tomalia DA. New J. Chem. 2012;36:264. [Google Scholar]; (c) Tomalia DA. In: Dendrimers Dendrons and Dendritic Polymers. Tomalia DA, Christensen JB, Boas U, editors. Cambridge UK: Cambridge University Press; 2012. 420 pp. [Google Scholar]
- 22.Kostiainen MA, Hiekkataipale P, Laiho A, Lemieux V, Seitsonen J, Ruokolainen J, Ceci P. Nature Nanotech. 2013;8:52. doi: 10.1038/nnano.2012.220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.