Abstract
The C4′-oxidized abasic site is produced in DNA by a variety of oxidizing agents, including potent cytotoxic antitumor agents. Independent generation of this alkali-labile lesion at defined positions within nucleosome core particles reveals that the histone proteins increase strand scission between 130 and 550-fold. Strand scission proceeds via a Schiff base intermediate but the DNA protein cross-links are unstable. The oxidized abasic site is removed in its entirety from the DNA and transferred to the lysine rich tail region of the proximal histone protein in the form of a lactam. The modification is distributed over several residues within the amino terminal tail of the proximal histone. Transfer of DNA damage to histones could affect gene regulation.
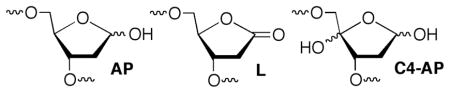
DNA lesion reactivity and their effects on biochemical processes may provide the chemical basis for therapeutics that damage nucleic acids. The C4′-oxidized abasic site (C4-AP) is a commonly formed DNA lesion.1,2 Its formation by diffusible and minor groove binding DNA damaging agents is attributed to the relatively low respective C-H bond dissociation energy and the C4′-hydrogen atom’s solvent accessibility. 3 C4-AP gives rise to interstrand cross-links (ICLs) that have been detected in cellular DNA and whose formation in naked DNA is catalyzed by the local nucleic acid sequence.4–6 These ICLs are converted to double strand breaks by the UvrABC bacterial nucleotide excision repair proteins.7 The oxidized abasic site also poses a challenge to base excision repair because the incision product formed by apurinic endonuclease I (Ape1) inactivates DNA polymerase β, the next essential protein in the repair pathway.8 C4-AP also exerts very unusual effects on replication in E. coli, resulting in 3 nucleotide deletions.9 C4-AP is an alkali-labile lesion and DNA containing it undergoes strand scission upon mild base treatment. Studies involving bleomycin, which produces C4-AP, indicate that DNA lability is enhanced when the lesion is produced within chromatin.10 Recent studies on related DNA lesions (AP, L) revealed that nucleosome core particles (NCPs) accelerate cleavage as much as 100-fold.11–13 We have now examined the effect of forming C4-AP in a NCP on the lesion’s reactivity. This was accomplished via independently generating the lesion from a stable photochemical precursor (eqn. 1) incorporated at specific locations within nucleosomal DNA.
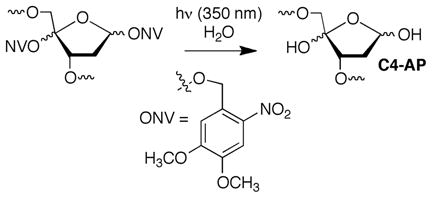 |
(1) |
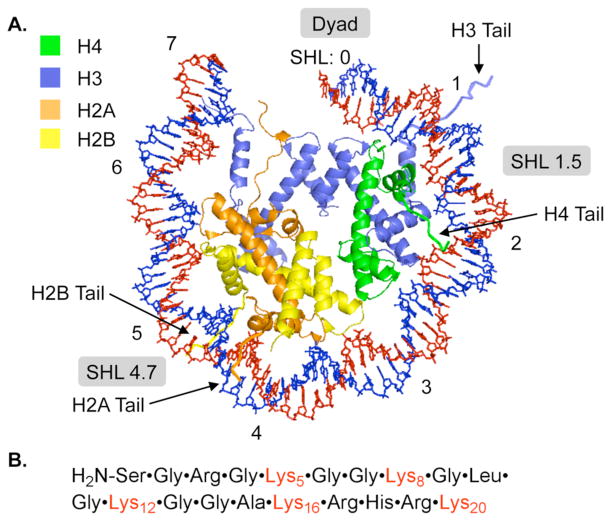
NCPs were reconstituted using FPLC purified octamers containing histone proteins that were expressed in E. coli.14 Polyacrylamide gel electrophoresis purified DNA was prepared by ligating chemically synthesized oligonucleotides.15 C4-AP reactivity was examined in 3 regions of the core particle comprised of two different DNA sequences. The DNA sequences were based on the α-satellite DNA employed in the original structural studies by Richmond, and the strong positioning 601 sequence discovered by Widom, which was also characterized by X-ray crystallography.16–18 The 3 regions within the core particle include superhelical location (SHL) 1.5, which is a hot spot for DNA damaging molecules and is in close proximity to the lysine rich amino terminus of histone H4 (Figure 1A).19 Reactivity was also examined at the dyad axis (SHL 0) where direct DNA-histone interaction is weak and lysine rich tails are more distal, and SHL 4.5 where the DNA is stretched and the lesion is in closer contact to H2A and H2B.16,20
Figure 1.
General structural features of a nucleosome core particle (A.) Partial X-ray crystal structure of NCP containing α-satellite DNA (B.) Amino acid sequence of histone H4 tail. Data taken from PDB: 1aoi. (See supporting information for complete DNA sequences.)
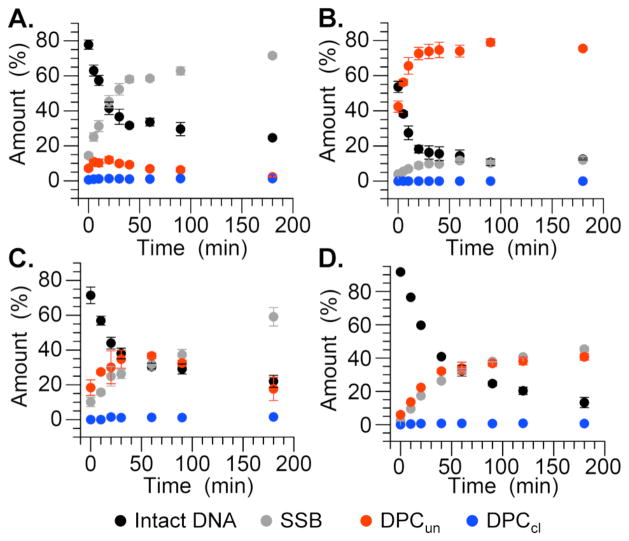
When produced at defined sites within NCPs C4-AP decomposition yields DNA-protein cross-links (DPCs) and single strand breaks (SSBs) (Figure 2). The overall disappearance of intact DNA containing C4-AP in nucleosome core particles fit well to first order kinetics (kDec, Table 1).15 The half-life of the DNA ranged from 14 to 120 min, depending upon its position within the nucleosome and DNA sequence. C4-AP half-life was longest in both DNA sequences at the dyad axis where direct interactions with the octameric core are weakest.16 In contrast, the half-life for C4-AP in naked DNA within identical sequences ranged from 98 h at position 202 to 268 h at position 89. Hence, depending upon the position within the nucleosome, C4-AP decomposition (kDec) is accelerated at least 130-fold (positions 73 and 202) compared to that in naked DNA, and as much as 550-fold when incorporated at position 89 of the 601 DNA. This acceleration in the nucleosome core particle is greater than that experienced by either AP or L and the absolute half-life of C4-AP is also considerably shorter.11,13 The rate constants for ICL formation on naked DNA are uncompetitive with kDec, and cross-linked DNA was not observed in the NCPs.4–6 These data suggest that the C4-AP derived ICLs observed in cells are formed in yields below the detection limits of these experiments (~1%) and/or in the DNA linker regions between NCPs in chromatin.
Figure 2.
Time dependent product distribution from C4-AP89 in a nucleosome core particle composed of 601 DNA and (A.) wild type H4 in the absence, (B.) presence of NaBH3CN, (C.) H4 Lys16,20Ala variant, (D.) H4 Del.1–20.
Table 1.
Decomposition kinetics of C4-AP as a function of position within nucleosome core particles at 37 °C.
| Position (SHL) | DNA Seq. | kDec (×10−4 s−1)a | t1/2 (min) |
|---|---|---|---|
| 73 (0) | 601 | 1.0 ± 0.1 | 120 ± 13 |
| 74 (0) | α-Satellite | 1.5 ± 0.3 | 83 ± 23 |
| 89 (1.5) | 601 | 4.0 ±0.8 | 29 ± 4 |
| 89 (1.5) | α-Satellite | 2.6 ± 0.5 | 46 ± 13 |
| 202 (1.5) | 601 | 2.7 ± 0.8 | 45 ± 12 |
| 119 (4.5) | 601 | 2.8 ± 0.7 | 43 ± 10 |
| 124 (4.7) | α-Satellite | 8.7 ± 1.4 | 14 ± 3 |
Rate constants are averages ± std. dev. of at least 2 experiments, each consisting of 3 independent reactions.
Unlike AP, C4-AP does not yield persistent DPCs.11,12 Maximum DNA-protein cross-link yields vary depending upon the DNA sequence and the location of C4-AP. For instance, DPC containing uncleaved DNA (DPCun, Scheme 1) reach only 12% at position 89 in the 601 DNA (Figure 2A). Furthermore, the DPCs containing cleaved DNA (DPCcl) typically form in <1% yield and in some cases are undetectable. SSBs are the final end product in the core particles. Enzymatic analysis of the fragments’ end groups produced upon cleavage revealed that they consist solely of phosphates at the 5′-termini of the 3′-fragments, as well as at the 3′-termini of 5′-fragments using shrimp alkaline phosphatase and polynucleotide T4 kinase, respectively.15
Scheme 1.
The intermediacy of DPCs from decomposition of C4-AP containing NCPs (Scheme 1) was probed with NaBH3CN, which reduces Schiff bases to stable alkylamines. DPCun are formed in >90% yield at the expense of SSBs when the core particle is incubated in the presence of NaBH3CN (Figure 2B). In addition, NaBH3CN trapping of DPCun decreases the half-life for C4-AP89 decomposition from 29 to 17 minutes by preventing reversion of DNA-protein cross-links to starting material. Furthermore, DPCun trapping by NaBH3CN, in conjunction with internally 32P-labeled C4-AP89 containing DNA also revealed that histone H4 was solely responsible for DPC formation at this position (Figure 3A).12
Figure 3.
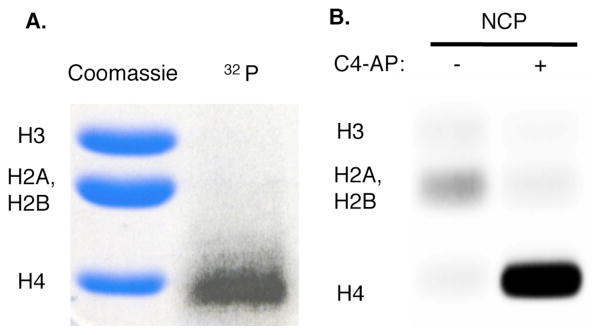
Identification of histone protein(s) reacting with C4-AP89. (A.) Determination using internal 32P-labeling and NCP incubation with NaBH3CN (B.) Histone protein tagging by 2.
The lysine rich histone H4 tail exits the octameric core in close proximity to C4-AP89.16 The role of these amino acids in lesion decomposition was examined using histone H4 variants in which one or more lysines in the protein tail (Figure 1B) were mutated to alanine. Substituting alanines for lysines in the H4 tail decreased the rate constant for C4-AP89 decomposition by less than 50%, and removing the entire tail had only an ~2-fold effect on kDec.15 In contrast, kDec for C4-AP89 decreased ~6-fold when the 5 lysines in the H4 tail were replaced with arginines. The polyarginine variant retained the H4 tail’s overall charge, but reduced its basicity and nucleophilicity. The smaller effect on kDec of deleting the entire H4 tail is attributed to the proximity of the truncated protein’s N-terminal amine, which is more nucleophilic than the lysine ε-amino group, to the lesion.21
The lysine to alanine mutations did significantly affect the product distribution (Figures 2C and 2D). Larger bursts of DPCs containing uncleaved DNA (DPCun) were formed and these products persisted longer than in NCPs composed of wild type H4. This was particularly true when Lys16 and/or Lys20, which were expected to be proximal to C4-AP89, were replaced by Ala. In addition, DPCun were formed in comparable yields to SSBs when the entire H4 tail was removed. These observations are reminiscent of the effects that H4 variants had on AP decomposition in NCPs, and suggest histone protein contribution to strand scission is not limited to Schiff base formation.11 The amino acid side chains may also contribute to β-elimination from DPCun to DPCcl.
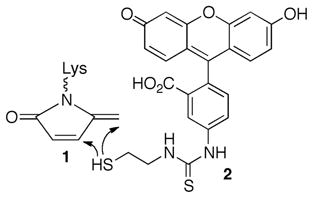 |
(2) |
The above experiments do not describe the fate of the C4-AP lesion. Previous work led us to examine whether the oxidized deoxyribose is transferred to the histone protein that is involved in Schiff base formation, resulting in the formation of an unsaturated lactam (1, Scheme 1).22,23 We anticipated detecting 1 via fluorescence by taking advantage of its electrophilicity in reaction with 2 (eqn. 2).24 This possibility was validated by examining the latter’s reactivity with a small molecule model.15 NCP with and without C4-AP89 were incubated as above, and then reacted with 2. The histones were separated by SDS gel and detected via fluorescence (Figure 3B). The presence of all histones in the sample was verified by Coomassie staining. 15 Although a weak false positive was detected in the absence of C4-AP89, fluorescence was detected only in histone H4 in lesion containing NCPs, consistent with the internal 32P-labeling experiment (Figure 3A).
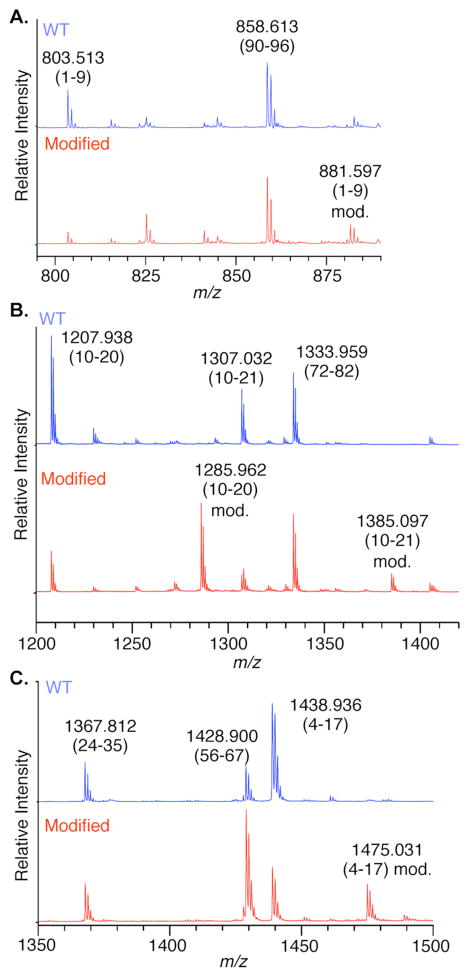
Mass spectrometric analysis confirmed that C4-AP89 was in essence transferred to histone H4. In order to maximize the amount of H4 modified, a NCP containing C4-AP at position 89 and its equivalent site on the second gyre (234) was prepared.15 The MALDI-TOF MS of H4 consisted of a broad peak that was 78 Da greater than the unmodified protein.15 Higher resolution analysis of the positions modified was achieved following a series of protease digestions in which the masses of peptide fragments obtained from the C4-AP containing NCP were compared to those from unmodified H4 protein (Figure 4). Combining GluC and AspN, which cleave proteins at the carboxyl group of glutamic acid and the amino group of aspartic acid respectively, yielded a peptide fragment consisting of amino acids 1–23 whose mass increased by 78 Da when NCP containing C4-AP89 and C4-AP234 was analyzed.15 Thermolysin, which generally cleaves proteins at the amino termini of hydrophobic residues, yielded a greater number of peptides whose masses increased by 78 Da in the doubly modified NCP, including amino acids 1–9, 10–20, and 10–21 (Figures 4A and 4B). Finally, in-gel acetylation of lysine residues, followed by trypsin digestion (which cleaves at the carboxyl group of unmodified lysines and arginines) yielded peptide 4–17 whose mass was 36 Da greater than the respective product obtained from unmodified H4 (Figure 4C). The mass changes correspond to the difference in masses between a lysine modified by C4-AP to produce 1 and an acetylated one. In each of the digests all modified peptides detected were derived from the N-amino terminal tail region of histone H4. Consistent with the effects of H4 mutants on kinetics, these data indicate that more than one lysine in the histone tail is capable of inducing cleavage at C4-AP.
Figure 4.
MALDI-TOF MS of proteolyzed histone H4 fragments (A & B) Thermolysin digestion. (C) Trypsin digestion. Top (blue): unmodified H4; Bottom (red): H4 from NCP containing C4-AP89 and C4-AP234.
The mass spectral, product, and kinetic data are consistent with a mechanism wherein reversible Schiff base formation leads to a double elimination reaction (Scheme 1). During this process one or more other amino acids contributes to strand scission by catalyzing β-elimination from the Schiff base. C4-AP lifetime in the NCP is significantly shorter than that of AP and L, and DNA interstrand cross-linking does not compete with the reactivity described herein.4 Although C4-AP’s lifetime in cells is unknown, the intracellular half-life of an AP site is believed to be on the order of 1 h.25 Given that DNA containing C4-AP is processed more slowly by the primary enzymes responsible for abasic site repair, Ape1 and DNA polymerase β, its lifetime in cells should be at least as long as that of AP.8 This suggests that formation of the histone modification by C4-AP described above is efficient enough to occur in cells, just as N-formylation resulting from oxidative DNA damage has been observed.26 The ability of DNA lesions to alter chromatin structure raises the specter that damaging agents indirectly affect cell signaling, providing additional sources for biological effects.
Supplementary Material
Acknowledgments
We are grateful for generous financial support from the National Institute of General Medical Science (GM-063028). We thank Ms. Elaine To for preparing the polyarginine histone H4 variant.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supporting Information. Experimental procedures for all experiments. Complete sequences of all DNAs used to prepare nucleosome core particles. Representative autoradiograms and kinetic plots. Mass spectra of modified oligonucleotides, histone H4 variants, and GluC/AspN digest of modified H4. Enzyme end group analysis This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pitié M, Pratviel G. Chem Rev. 2010;110:1018–1059. doi: 10.1021/cr900247m. [DOI] [PubMed] [Google Scholar]
- 2.Pogozelski WK, Tullius TD. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian B, Pogozelski WK, Tullius TD. Proc Nat Acad Sci USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Regulus P, Duroux B, Bayle PA, Favier A, Cadet J, Ravanat JL. Proc Nat Acad Sci USA. 2007;104:14032–14037. doi: 10.1073/pnas.0706044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sczepanski JT, Jacobs AC, Majumdar A, Greenberg MM. J Am Chem Soc. 2009;131:11132–11139. doi: 10.1021/ja903404v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sczepanski JT, Jacobs AC, Greenberg MM. J Am Chem Soc. 2008;130:9646–9647. doi: 10.1021/ja8030642. [DOI] [PubMed] [Google Scholar]
- 7.Sczepanski JT, Jacobs AC, Van Houten B, Greenberg MM. Biochemistry. 2009;48:7565–7567. doi: 10.1021/bi901006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobs AC, Kreller CR, Greenberg MM. Biochemistry. 2011;50:136–143. doi: 10.1021/bi1017667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroeger KM, Kim J, Goodman MF, Greenberg MM. Biochemistry. 2004;43:13621–13627. doi: 10.1021/bi048337r. [DOI] [PubMed] [Google Scholar]
- 10.Bennett RAO, Swerdlow PS, Povirk LF. Biochemistry. 1993;32:3188–3195. doi: 10.1021/bi00063a034. [DOI] [PubMed] [Google Scholar]
- 11.Zhou C, Sczepanski JT, Greenberg MM. J Am Chem Soc. 2012;134:16734–16741. doi: 10.1021/ja306858m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Proc Natl Acad Sci U S A. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou C, Greenberg MM. J Am Chem Soc. 2012;134:8090–8093. doi: 10.1021/ja302993h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyer PN, Edayathumangalam RS, White CLB, Yunhe, Chakravarthy S, Muthurajan UM, Luger K. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 15.See Supporting Information.
- 16.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 17.Lowary PT, Widom J. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 18.Vasudevan D, Chua EYD, Davey CA. J Mol Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 19.Kuduvalli PN, Townsend CA, Tullius TD. Biochemistry. 1995;34:3899–3906. doi: 10.1021/bi00012a005. [DOI] [PubMed] [Google Scholar]
- 20.Davey G, Wu B, Dong Y, Surana U, Davey CA. Nucleic Acids Res. 2010;38:2081–2088. doi: 10.1093/nar/gkp1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raindlova V, Pohl R, Hocek M. Chem Eur J. 2012;18:4080–4087. doi: 10.1002/chem.201103270. [DOI] [PubMed] [Google Scholar]
- 22.Aso M, Usui K, Fukuda M, Kakihara Y, Goromaru T, Suemune H. Org Lett. 2006;8:3183–3186. doi: 10.1021/ol060987v. [DOI] [PubMed] [Google Scholar]
- 23.Aso M, Kondo M, Suemune H, Hecht SM. J Am Chem Soc. 1999;121:9023–9033. [Google Scholar]
- 24.Baumann S, Schoof S, Bolten M, Haering C, Takagi M, Shin-ya K, Arndt H-D. J Am Chem Soc. 2010;132:6973–6981. doi: 10.1021/ja909317n. [DOI] [PubMed] [Google Scholar]
- 25.Atamna H, Cheung I, Ames BN. Proc Nat Acad Sci USA. 2000;97:686–691. doi: 10.1073/pnas.97.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang T, Zhou X, Taghizadeh K, Dong M, Dedon PC. Proc Nat Acad Sci USA. 2007;104:60–65. doi: 10.1073/pnas.0606775103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.