Abstract
RNA interference (RNAi) caused by exogenous double-stranded RNA (dsRNA) has developed into a powerful technique in functional genomics, and to date it is widely used to down-regulate crucial physiology-related genes to control pest insects. A molt-regulating transcription factor gene, HaHR3, of cotton bollworm (Helicoverpa armigera) was selected as the target gene. Four different fragments covering the coding sequence (CDS) of HaHR3 were cloned into vector L4440 to express dsRNAs in Escherichia coli. The most effective silencing fragment was then cloned into a plant over-expression vector to express a hairpin RNA (hpRNA) in transgenic tobacco (Nicotiana tabacum). When H. armigera larvae were fed the E. coli or transgenic plants, the HaHR3 mRNA and protein levels dramatically decreased, resulting developmental deformity and larval lethality. The results demonstrate that both recombinant bacteria and transgenic plants could induce HaHR3 silence to disrupt H. armigera development, transgenic plant-mediated RNAi is emerging as a powerful approach for controlling insect pests.
Keywords: RNAi, Helicoverpa armigera, HaHR3, transgenic tobacco plants, systemic RNAi.
Introduction
RNA interference (RNAi) is an innovative and efficient molecular biology tool to down-regulate the expression of genes specifically. The first documentation of an RNA-silencing phenomenon was by Jorgensen and coworkers 1, who unexpectedly noted transgenic petunia-induced gene silencing when they tried to over-express the chalcone synthase gene to enrich flower pigmentation. The underlying mechanism of this phenomenon was unveiled by Andrew Fire and Craig Mello's work 2, for which they were awarded the Nobel Prize in 2006. They systematically clarified that double-stranded RNA (dsRNA) was more effective than either sense or antisense RNA in silencing genes, and they termed this dsRNA-induced silencing phenomenon RNAi. To date, many breakthroughs have been achieved in this area. Significant progress has been made in understanding the molecular mechanisms of RNAi and developing applications of it, and it is now clear that RNAi is a conserved mechanism in many eukaryotes 3,4,5.
Most eukaryotic organisms, including insects, possess common machinery for sequence-specific post-transcriptional gene silencing that is triggered by the presence of dsRNA, resulting in the silencing of expression of the targeted mRNA. RNAi has proven useful in functional genomic research on insects, and it also has been considered a potential tool for controlling agricultural pest insects because of its high specificity 6.
The cotton bollworm, Helicoverpa armigera, is one of the most serious herbivorous insect pests of cotton, corn, vegetables and other crops throughout the world. It can be controlled by insecticides and Bacillus thuringiensis (Bt) toxin-containing cotton, which is very effective in China 7. However, several important insect pests are not amenable to Bt-toxin control, and there is an imminent danger of at least some species, including H. armigera, developing Bt resistance 8,9,10. Field trials conducted over 10 years in northern China have shown that mirid bugs have progressively increased in population and have acquired major pest status because of a regional increase in Bt cotton adoption 11. At the same time, the increasing use of insecticides has resulted in many problems, such as environmental residual pollution, harm to natural enemies, insect resistance and secondary pest outbreaks 12,13,14.
With the progression of RNAi research, RNAi provides an alternative tool for controlling pests. Expression of dsRNA in transgenic plants directed against crucial insect target genes protects plants from insect pests, paving the way for a new generation of insect-resistant transgenic crops 15. Mao et al. silenced a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi, impairing larval tolerance in the gossypol 16,17. Baum et al. engineered transgenic corn plants expressing western corn rootworm (WCR) dsRNAs and observed a significant reduction in WCR feeding damage in a growth chamber assay 18. Zha et al. silenced the hemipteran insect Nilaparvata lugens midgut genes by dsRNA-transgenic plant-mediated RNAi 19. Pitino et al. developed plant-mediated RNAi technology for aphids to allow for gene silencing in the aphid natural environment 20. A tobacco rattle virus vector was used to produce dsRNA in the host plant Nicotiana attenuata to transiently silence midgut genes of the plant's lepidopteran specialist herbivore (Manduca sexta), providing a rapid and transient means of silencing insect pest genes by plant-mediated RNAi 21. Zhu et al. reported that pest resistance is improved in transgenic tobacco plants expressing dsRNA of EcR from the cotton bollworm 22. Thus, plant-mediated RNAi is a promising pest-control strategy for future plant protection, and transgene-encoded ingestible dsRNAs may one day stand alongside Bt transgenes in insect management programs 8.
Insects belong to the phylum Arthropoda, which undergo a well-known molting process. They are obliged to remove old shells or cuticles to grow and develop. Following a genetically specified number of molting cycles, they can enter a pupal stage, during which they undergo metamorphosis to their adult, reproductive form 23. 20-Hydroxyecdysone (20E) and juvenile hormone (JH) coordinately orchestrate insect growth and development 24. In recent decades, these two insect growth and developmental hormones have been mimicked by chemically insecticides in the field 25. Successful application of these chemical insecticides suggests that regulating molting or metamorphosis can be used as an effective means to control pest insects.
Because HR3 plays a key role during metamorphosis by regulating metamorphosis-related genes expression 26, 27, we selected it as a potential target gene for RNAi research to control H. armigera. In this context, a molt-regulating transcription factor gene, HaHR3, was cloned from cotton bollworm. Four dsRNA-HaHR3 fragments were expressed in E. coli, which was fed to the insects. The fragment that was most effective at silencing HaHR3 was then selected for transgenic tobacco plant generation. Feeding bioassay showed that, both recombinant bacteria and transgenic plants induced H. armigera developmental deformity and larval lethality.
Results and Discussion
Molecular cloning of HaHR3 transcription factor gene from H. armigera
A hormone receptor 3 (HR3) gene was selected as our target among insect hormone regulation network genes. A 1671 bp full CDS sequence was obtained by our early work 28. We named it HaHR3 (GenBank accession number: FJ009448). Multiple sequence alignment results showed that our PCR products shared 98.50% identity with HHR3 (GenBank accession number: AF337637) cloned by Zhao 29. There are also 2 amino acids differences between protein HaHR3 and HHR3. This difference can be explained by genetic isolation because insect samples are from different geographical regions.
Although the RNAi effect is specific, the underlying biological risk of plant-mediated RNAi still exists because of potential off-target silencing effects of homologous genes that share high identity with the target insect gene 22. We selected molting as the physiological target because it is one of the most important processes for insect development and it has been considered a target process to control pest insects for decades. Field applications show that diverse insecticides that target molting are safe to beneficial insects and are selectively toxic to lepidopteran pests 30. Therefore, we focused our attention on the insect-specific metamorphosis regulation network gene HR3, which regulates metamorphosis-related tissue-specific gene expression 29, to develop a transgenic plant that would induce RNAi.
The NCBI Conserved Domains prediction (http://www.ncbi.nlm.nih.gov/cdd) results showed that, like other members of the nuclear receptor (NR) superfamily of ligand-activated transcription factors, HaHR3 contained a predicted DNA-binding domain (DBD), a variable N-terminal domain, a non-conserved hinge and a ligand-binding domain (LBD) (Supplementary Material: Figure S1A). The DBD of retinoid-related orphan receptors is composed of two C4-type zinc fingers, and each zinc finger contains a group of four Cys residues that coordinate a single zinc atom.
In the process of RNAi, the siRNAs serve as guides for endonucleolytic cleavage of homologous mRNA in association with the RNA-induced silencing complex (RISC) 31. When RISC combines with the target mRNA, large variations in degradation efficiency can be observed, because different siRNAs can bind to different sites on the same target mRNA 32,33. In present research, different dsRNAs were produced in bacteria, and every dsRNA was assumed to be processed into a group of siRNAs for RISC after insect ingestion. Different mortalities were observed with different dsRNAs. Several hypotheses have also been proposed to explain this RNAi efficiency phenomenon, such as local protein factor positional effects, sequence-dependent mRNA phosphorylation and the local structure of the target mRNA 34. The target mRNA secondary structure and accessibility may be the most important factor determining silencing efficiency 35 because different mRNA secondary structures have different binding or recognition capacities for guide siRNAs. Similarly to our study, Zhang et al. 36 selected two interference fragments of CiHR3 for feeding bioassays, and their results showed that feeding with CiHR3-I2 dsRNA was more effective than CiHR3-I1. Our prediction results of siRNA binding with Sfold also indicated the possibility of various RNAi efficiencies.
The HaHR3 mRNA secondary structure was predicted by the Sfold web server 37(http://sfold.wadsworth.org) to design interference fragments. A higher probability means a fragment was more likely to silence HaHR3 (Supplementary Material: Figure S1B). Because there was no obvious concentrated region of high probability, four fragments (1, 2, 3 and 4) covering the full CDS, with lengths between 400 and 600 bp, were selected for further research (Supplementary Material: Figure S1A).
H. armigera shows developmental deformities after feeding on bacteria expressing dsRNAs-HaHR3
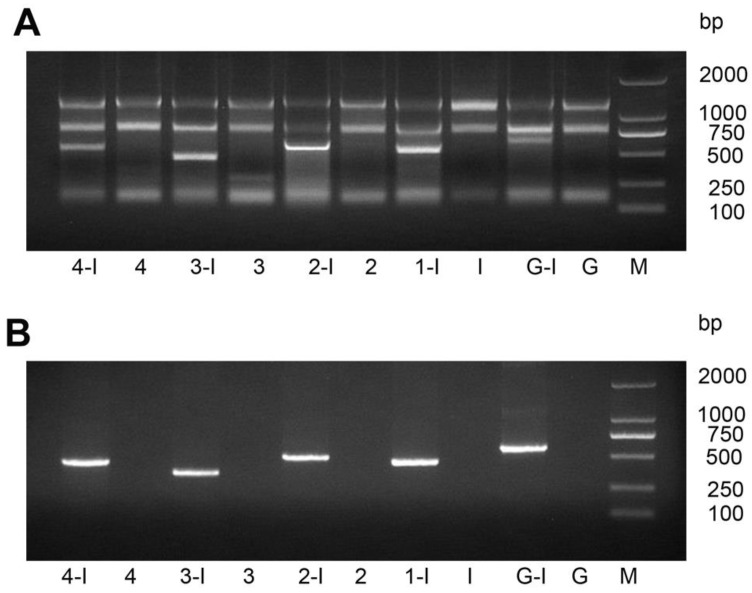
After we transformed the constructed vector L4440-HaHR3 into HT115 competent cells, total RNA and dsRNA were extracted by different methods. Gel separation showed that dsRNAs were extracted effectively by both methods and that every dsRNA fragment, including dsRNA-EGFP, was expressed successfully in HT115 cells (Figure 1).
Figure 1.
Identification of dsRNA-HaHR3 and dsRNA-EGFP fragments produced in bacteria. (A) Total RNA extracted from bacteria using a bacterial RNA-specific extraction kit. (B) dsRNAs extracted from bacteria using the modified LiCl method. M: DL2000 Marker; Lanes 1-I, 2-I, 3-I, 4-I and G-I mean IPTG was added into the bacterial samples L4440-frag.1, L4440-frag.2, L4440-frag.3, L4440-frag.4 and L4440-frag.EGFP, respectively; lanes 1, 2, 3, and 4 and G mean no IPTG was added (negative controls).
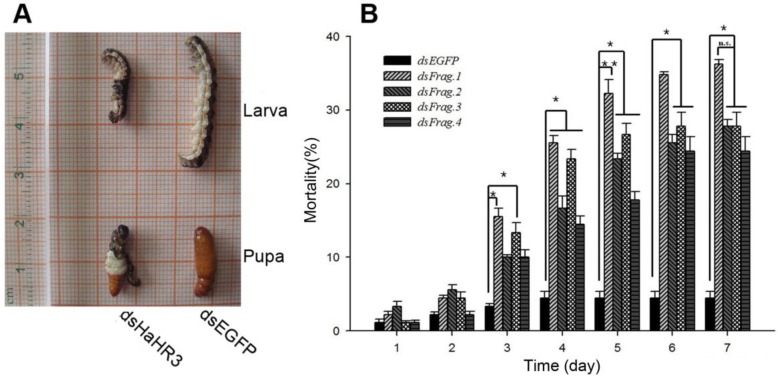
After feeding H. armigera the artificial diet containing dsRNAs for 7 days, the bioassay results showed that the four different dsRNA-HaHR3 fragments increased H. armigera mortality. The mortality increased rapidly on day 3-4 after feeding dsRNA-HaHR3; on day 3, the mortality of feeding with dsRNA-HaHR3-fragment 1 (dsFrag.1) and dsRNA-HaHR3-fragment 3 (dsFrag.3) showed significant differences with dsRNA-EGFP. On day 4, the mortality caused by all four dsRNA-HaHR3-fragments showed significant differences compared to dsRNA-EGFP (P<0.05). The mortality plateaued on day 6-7. dsFrag.1, which caused the highest mortality, showed a significant difference (P<0.01) with dsEGFP starting on day 5. On day 7 after feeding with dsFrag.1, the mortality went up to 36.22% (Figure 2B).
Figure 2.
Phenotype and mortality of H. armigera feeding with different bacterial-expressed dsRNAs. (A) H. armigera molting and pupation failure led to mortality. (B) Ingestion of artificial diet containing different dsRNA fragments on the indicated days (1-7) caused lethality. dsFrag.1, -2, -3, and -4 and dsEGFP mean dsRNA-HaHR3-fragment 1, 2, 3, and 4 and dsRNA-EGFP, respectively. Mortality values consist of means ± standard deviation. Asterisk indicates significant difference from control group, P<0.05; two asterisks indicate P<0.01.
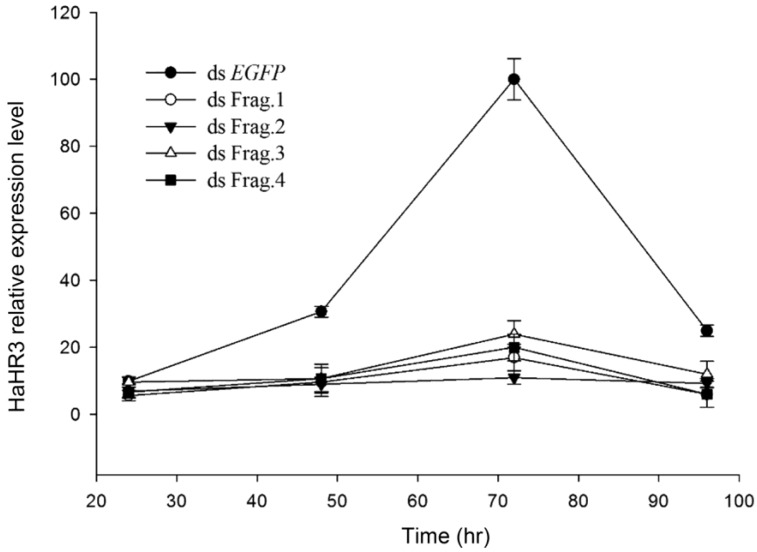
Quantitative RT-PCR showed HaHR3 mRNA had a periodic expression profile of 72 h, coincident with the H. armigera molting progress. HaHR3 mRNA increased ~35-fold, then decreased significantly following this peak. Zhao also proved that HHR3 expression exhibited developmental variations in various tissues 29. This periodic expression profile of HaHR3 in whole larvae was coincident with the previous results in some extend. After feeding H. armigera with four different dsRNA-HaHR3 fragments for 48 hours, the HaHR3 gene was silenced successfully (Figure 3). All four fragments showed steady suppression compared to the dsEGFP (P<0.05), and there was no significant difference among four different fragments (Figure 3).
Figure 3.
The relative expression of HaHR3 in H. armigera. qRT-PCR analysis of HaHR3 expression. HaHR3 relative expression level in H. armigera that fed on dsRNA fragments; ds Frag.1,2,3,4 and EGFP: dsRNA of HaHR3 frag.1,2,3,4 and EGFP fragment.
From basic to applied research, RNAi induction approaches in insects have mainly gone through injection, feeding and transgenic plants mediation. RNAi has been used as a tool in reverse genetics to study insect gene function. Most of these experiments have been conducted by injecting dsRNA into the insects directly 38,39, which is able to operate under experimental conditions but not feasible to control pest insects in the field. The RNAi feeding protocol has several distinct advantages over injection 40. For instance, it is extremely convenient for performing RNAi experiments in a large number of insects or with different genes, and it has potential for applied research. Since the first two successful experiments on transgenic plant-mediated RNAi to control insects were published 16,18, this approach has emerged as an effective tool for protection of plants against insect pests 19,20,22.
In our study, bacteria expressing dsRNAs were used to screen the most effective target fragment. This is a quicker, cheaper and more convenient approach to produce large amounts of dsRNA compared to synthesizing dsRNA with kits. Feeding experiments were then used to induce RNAi in H. armigera, which is more convenient than injection 41. Thus, we screened for the most effective fragment with an efficient protocol. Based on the dsRNA feeding bioassay results, transgenic plant-mediated, which is more applicable for field pest control than other methods, was used for our further experiments with that selected fragment.
Generation of transgenic tobacco plants
Because the dsRNA-HaHR3-fragment 1 expressed by bacteria caused the highest mortality, only fragment 1 was selected for expression in transgenic plants. The hairpin-EGFP was expressed as a control at the same time. Sequencing results of single colonies indicated that both sense-HaHR3-fragment 1 and antisense-HaHR3-fragment 1 were cloned into the plant over-expression vector in the right orientation.
After transforming pCAMBIA-RNAi-HaHR3 to tobacco calli and one month of tissue culture, 106 regenerated plants were obtained (Supplementary Material: Figure S2). PCR amplification analysis of the sense fragment and antisense fragment showed that 56 of those regenerated plants were positive (Figure 4).
Figure 4.
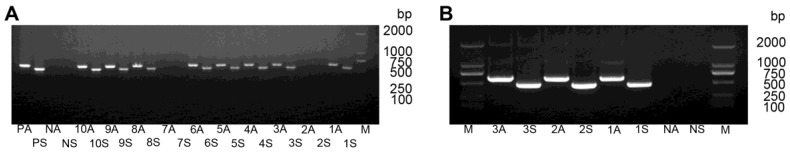
PCR analysis of T0-generation transgenic tobacco lines. (A) PCR analysis of plants expressing hpRNA-HaHR3 with sense fragment primers and anti-sense fragment primers. Lanes 1S-10S denote PCR analysis of line 1-line 10 with sense fragment primers. Lanes 1A-10A denote PCR analysis of line 1-line 10 with anti-sense primers. Lanes PA and PS denote antisense and sense positive controls, respectively. NA and NS are antisense and sense negative controls, respectively. (B) PCR analysis of hpRNA-EGFP expressed plants with two pairs of primers. M: DL2000 Marker. 1S-3S, 1A-3A, NA and NS have the same meanings as in (A).
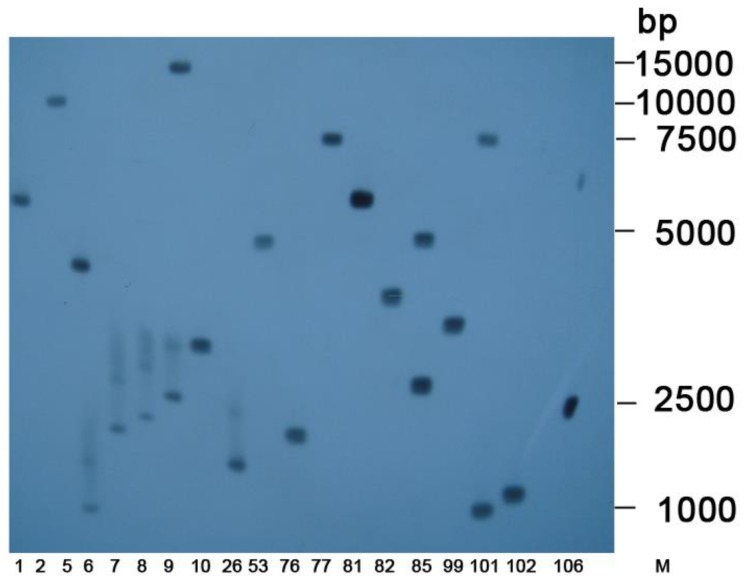
Southern blots indicated that exogenous gene fragments were integrated into the genomes of the tobacco plants at different locations (Figure 5). In total, 38 single-copy lines were identified.
Figure 5.
Southern blot of transgenic tobacco. Southern blot analysis of T0-generation positive lines expressing hpRNA-HaHR3. M: DL15000 Marker; 1, 2, 5, 6, …, 106 denote different transgenic tobacco lines.
Seeds of three self-fertilized single-copy lines from T0-generation positive transgenic plants (line 1, line 5 and line 99) were planted in the greenhouse to obtain T1-generation lines for further research. The PCR results in T1-generation plants showed that the ratio of positive plants was in accordance with Mendelian law (data not shown).
Silencing H. armigera HaHR3 gene expression by ingestion of transgenic tobacco plants expressing hairpin-RNA
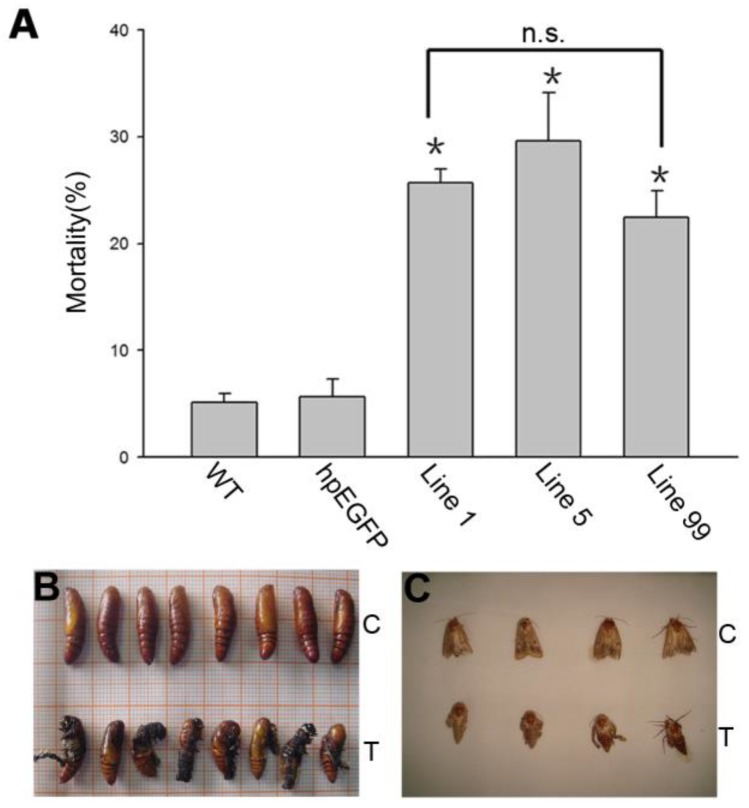
We calculated the mortality in each H. armigera feeding group. After 5 days of feeding, the mortality was 25.71%, 29.65% and 22.5%, respectively, in groups hpRNA-HaHR3 1, hpRNA-HaHR3 5, and hpRNA-HaHR3 99. In the wild-type and hpEGFP control groups, mortality was only 5.16% and 5.69%, respectively (Figure 7A). The three treatment groups were similar to each other in mortality but significantly different (P<0.05) from the two control groups (Figure 7A).
Figure 7.
H. armigera mortality and phenotype of developmental deformity (A) Mortality after feeding on different lines of tobacco leaves for 5 d. WT: wild-type tobacco plants; hpEGFP: transgenic tobacco plants expressing hairpin-EGFP; Line 1, Line 5, Line 99: line 1, line 5, and line 99 of transgenic tobacco expressing hp-HaHR3. Asterisk indicates P<0.05 compared to either of the two control groups; n.s. means no significant difference among line 1, line 5 and line 99. (B) The inhibition of pupation after feeding with hpRNA-HaHR3 tobacco. (C) The deformity of adult emergence. C: control group fed with hpRNA-EGFP or wild-type tobacco; T: treatment groups fed with hpRNA-HaHR3 tobacco.
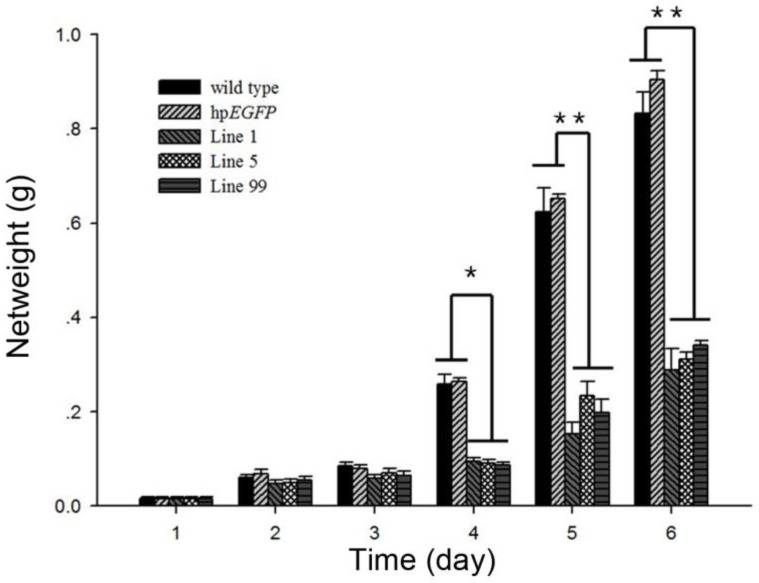
The average net weight of survival H. armigera was also suppressed after feeding with hpRNA-HaHR3-expressing tobacco leaves. On day 4 of feeding, the three treatment groups (feeding on hpHaHR3 line 1, 5 and 99 leaves, respectively) had an average net weight of ~0.0900 g, while the two control groups (wild-type and hpEGFP tobacco) weighed ~0.2600 g. On day 6, the control groups increased to ~1.000 g, while the three treatment groups remained at ~0.300 g. The three hpHaHR3 lines weighed significantly less than the wild-type and hpEGFP control groups on day 4 (P<0.05) and on days 5 and 6 (P<0.01) (Figure 6).
Figure 6.
Net weight of H. armigera after feeding on hpRNA-tobacco leaves. After feeding on hpRNA-EGFP, hpRNA-HaHR3 line 1, hpRNA-HaHR3 line 5 or hpRNA-HaHR3 line 99, the net weight of each insect was recorded daily (days 1, 2, 3, 4, 5, and 6). Error bars represent ± SD of means. Essentially identical results were obtained in three independent experiments. WT: wild-type tobacco plants; hpEGFP: transgenic tobacco plants expressing hairpin-EGFP; Line 1, Line 5, Line 99: line 1, line 5, and line 99 of transgenic tobacco expressing hp-HaHR3. One asterisk indicates P<0.05 compared with wild-type and hpEGFP control groups; two asterisks indicate P<0.01.
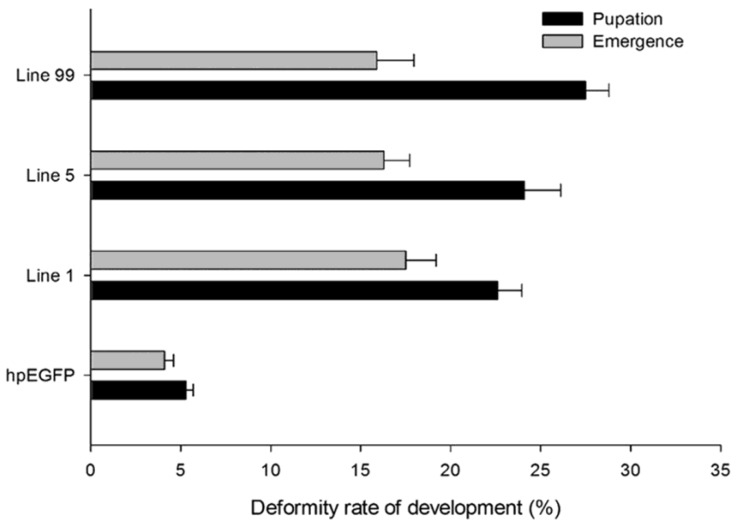
In addition to higher mortality, developmental deformity at metamorphosis was observed in the hpRNA-HaHR3 treatment groups. In the first two days that the cotton bollworms fed on hpRNA-HaHR3, larvae exhibited a normal appearance. However, abnormal phenotypes were observed 4-5 days later. In the larval growth stage, some bollworms moved slowly or not at all, and some failed to complete the molting progress because the older cuticle could not detach from the insect body or the new cuticle was too weak (Figure 2A). In pupation, the hpRNA-HaHR3 treatment groups deformity rate was ~25.50% (Figure 8), showed significant difference with hpEGFP (P<0.05); some insects could not detach the larval cuticle from the last instar of the larval body, and some new puparia were not integrated into the pupal body (Figure 7B). Adult emergence was also impaired, the treatment groups emergence deformity rate was ~16.23% (Figure 8) showed significant difference from control group (P<0.05); in the form of deficient wing length and width, smaller body size, and lower weight (Figure 7C).
Figure 8.
The deformity rate of H. armigera development. Deformity rate after feeding on different lines of tobacco leaves. hpEGFP: transgenic tobacco plants expressing hairpin-EGFP; Line 1, Line 5, Line 99: line 1, line 5, and line 99 of transgenic tobacco expressing hp-HaHR3.
Functionally, HR3 plays a key role during metamorphosis by regulating gene expression 26,27, and it directly induces the fushi tarazu (ftz) gene that encodes the prepupal regulator FTZ-F1 (NR5A3) 42,43. HR3 orthologs have been identified in various insect species 44. Mutation of HR3 in D. melanogaster reveals that it is required for metamorphosis 45, especially for the correct prepupal-pupal transition and for the differentiation of adult structures 26. When HaHR3 was silenced in this context, larval lethality and metamorphosis deformities indicated that HaHR3 is a vital factor for cotton bollworm molting and metamorphosis. The HaHR3 expression profile suggested that it has a periodic expression coincident with the cotton bollworm molting process. Cruz et al. also found that, at each Blattella germanica nymphal instar, the expression of three isoforms of BgHR3 coincides with the peak of circulating ecdysteroid concentrations 46. The timing of the HaHR3 expression profile suggests HaHR3 regulates insect metamorphosis directly or indirectly. Outside of insects, CHR3 is a critical transcriptional regulator in C. elegans: disrupted CHR3 expression leads to developmental changes, including incomplete molting 47.
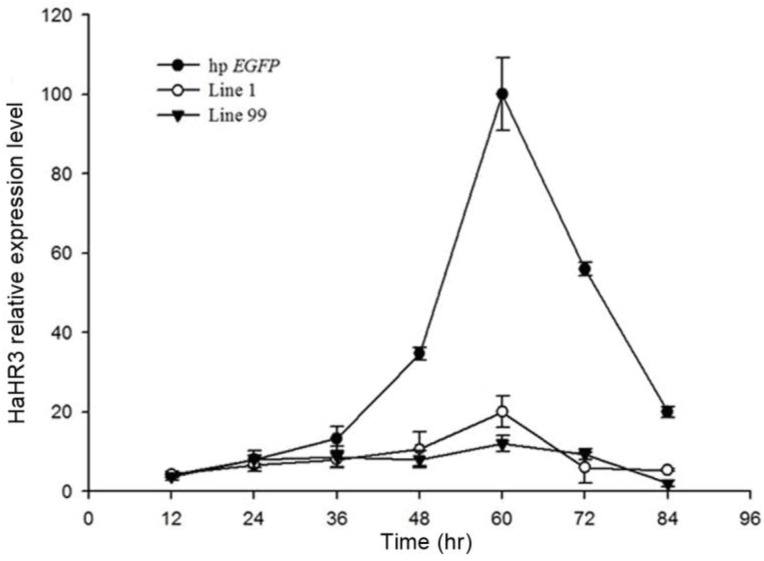
Quantitative RT-PCR showed that, after feeding H. armigera with transgenic tobacco leaf plates for several days, the HaHR3 gene was silenced successfully, especially after 48 h. Two independent single-copy lines of transgenic tobacco, line 1 and line 99, were used to test the gene-silencing efficiency of the dsRNA. Both of them showed steady suppression compared to the hp-EGFP-expressing tobacco (P<0.05), and there was no significant difference in gene expression between line 1 and line 99 (Figure 9). After feeding with hp-HaHR3 for 60 h, the relative expression level of HaHR3 under both treatments decreased by ~85% (Figure 9).
Figure 9.
The relative expression of HaHR3 in H. armigera. qRT-PCR analysis of HaHR3 expression. HaHR3 relative expression level in H. armigera that fed on transgenic plants expressing hp-HaHR3 or hp-EGFP; Line 1, Line 99: line 1 and line 99 of transgenic tobacco expressing hp-HaHR3.
The bioassay with bacteria or transgenic tobacco expressing this dsRNA indicated that feeding on either food source could induce gene silencing and kill H. armigera. Transgenic tobacco lines 1, 5, and 99 all caused similar mortality rates, indicating that the silencing effect was not an accident of random insertion in the plant genome. Importantly, although the larval mortality caused by line 5 was 29.65%, which was not as high as typical chemical pesticides, but the larval deformity rate caused later was ~30% (data not shown), and deformity was also observed in subsequent development. This indicates that HaHR3 gene silencing interferes with the development of insect pests in the latter metamorphosis stage if it does not kill the larvae, suggesting RNAi would be effective to keep the insect population density under an economic threshold. qRT-PCR and western blot analyses indicated that HaHR3 mRNA and protein expression were decreased by 48 h after ingestion. Compared to the transgenic plants, the bacteria expressing dsRNA-fragment 1 caused slightly higher mortality. The exact dose of dsRNA or hpRNA taken up by the tested insects could not be calculated, and there is no report about the dsRNA and hpRNA silencing efficiency in insects, so we can not draw any conclusion about the RNAi efficiency between the two delivery methods. However, transgenic plant-mediated RNAi is obviously much more applicable than bacterially expressed dsRNA in field applications, and it does possess more commercial value in plant protection.
More importantly, the current investigation confirmed the possibility of RNAi as a tool for specific and strong silencing of a gene in adult hemipteran pests that are not sensitive to Bt transgenic plants 48. This approach provides a new mode of controlling insect pests that might complement the currently used transgenic plants expressing Bt toxin 8. On the other hand, a non-midgut gene of the lepidopteran pest Spodoptera exigua can be silenced by ingestion of dsRNA 49, suggesting that dsRNA introduced into the midgut of larvae may be transported to other insect cells or tissues. This fact broadens the target gene spectrum from gut-specific genes to the whole insect. Transgenic plant-mediated gene silencing therefore presents many possibilities that were unavailable only recently.
HaHR3 protein expression in H. armigera after feeding with transgenic tobacco plants
When the recombinant vector pET-28a-HaHR3, pET-30a-HaHR3, pET-32a-HaHR3 or pGEX-6p-1-HaHR3 was transformed into Rosetta(DE3) competent cells, SDS-PAGE analysis showed that the HaHR3 protein was expressed successfully in the first three groups (as determined by the formation of insoluble inclusion bodies), but unfortunately, it was not expressed with the pGEX-6p-1-HaHR3 vector, for unknown reasons (Supplementary Material: Figure S3). The native PAGE and electroelution methods were used to separate and purify the HaHR3 protein for antibody preparation.
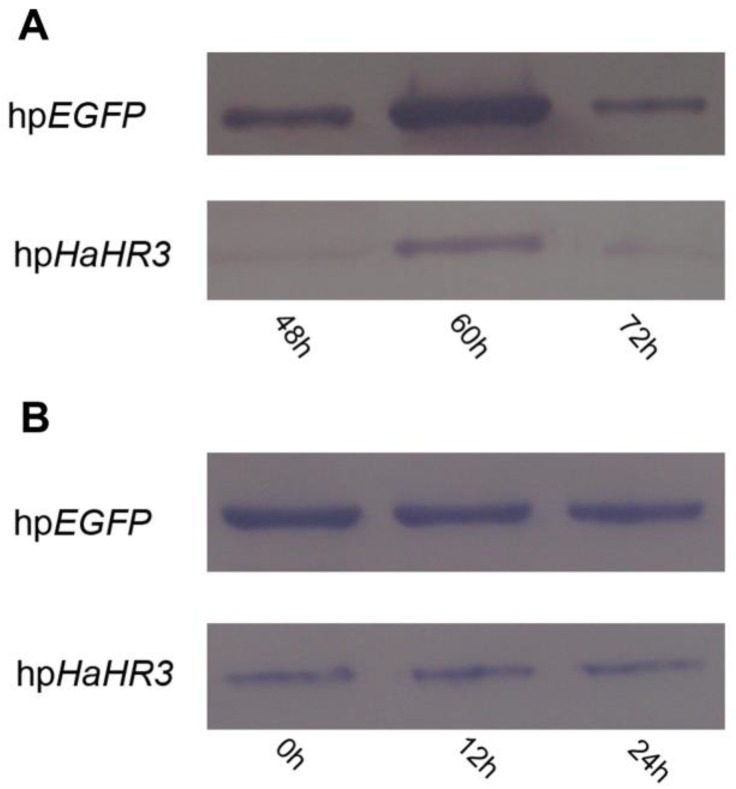
Western blot analysis revealed that expression of HaHR3 protein was suppressed in the high-expression period of this protein after feeding with transgenic tobacco plants expressing hpRNA-HaHR3. After feeding for 48 h, the HaHR3 expression of larvae was suppressed successfully; 60 h later, the HaHR3 expression had decreased dramatically compared to the hpEGFP group, in which HaHR3 expression increased and reached a peak at 60 h (Figure 10A). Not only in the total protein from the whole larval body of H. armigera (Figure 10A) but also in the protein of the female adult ovary (Figure 10B), HaHR3 protein expression was repressed at the same time. Our western blot results suggests that siRNAs may be transported from the midgut to the ovaries or other tissues in cotton bollworm, suggesting that systemic RNAi might exists in this insect.
Figure 10.
Western blot detection of HaHR3 protein expression in H. armigera after feeding with transgenic tobacco plants. hpEGFP denotes protein extracted from cotton bollworm fed with plants expressing hpRNA-EGFP; hpHaHR3 denotes protein extracted from cotton bollworm fed with plants expressing hpRNA-HaHR3. Loaded protein of each lane were equated (40 μg protein per lane) and separated on 15% SDS-PAGE gel. (A) Protein isolated from the whole insect body. Total protein of H. armigera larvae was extracted and separated by SDS-PAGE for western blot detection of HaHR3; 48 h, 60 h and 72 h mean protein expression was detected after feeding with hpEGFP or hpHaHR3 plants 48 hours, 60 hours and 72 hours. (B) Protein isolated from female adult ovary. After ovaries were dissected from adults, the ovary total protein was extracted at 0 h, 12 h or 24 h after insect emergence for western blot analysis.
After 24 h of emergence, the HaHR3 expression level of each treatment group, which fed on hpHaHR3 during the larval stage, was still lower than the control (Figure 10B). In the control group (hpEGFP), HaHR3 showed consistent expression in female adult ovary (Figure 10B). During the first 24 h after emergence, HaHR3 expression in the treatment group was still lower than the control group. We can conclude that the RNAi effect can persist in H. armigera for at least 24 h.
Materials and methods
Insect and plant culture
Cotton bollworm eggs were obtained from cotton pest group of the Institute of Plant Protection, Chinese Academy of Agricultural Sciences. Larvae were fed an artificial diet as described 50 and reared in a versatile environmental test chamber (SANYO, MLR-350HT) at 28°C and 70% relative humidity under a 14-h-light/10-h-dark photoperiod. Larvae were fed individually in 24-well plates.
Tobacco plants (Nicotiana tabacum) were grown in sterilized soil at 30°C and 60% relative humidity on a 16-h-day/8-h-night cycle.
dsRNA-HaHR3 expression in bacteria
According to the mRNA secondary structure online prediction tool Sfold (http://sfold.wadsworth.org), four fragments (400-600 base pairs each) that covered the full CDS of HaHR3 were selected to investigate their RNAi effects. Four pairs of specific primers were designed to amplify these four RNAi-HaHR3-fragments to express dsRNAs in bacteria (Table 1). An EGFP fragment of similar length was amplified as a dsRNA negative control. The XhoI restriction site CTCGAG and the BglII restriction site AGATCT were added to the primer pairs so that these restriction sites would appear at opposite ends of every fragment. Amplification reactions comprised 35 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s, with a final extension step of 72°C for 5 min. The PCR products were ligated into pMD18-T. The pMD18-T-HaHR3-fragments were then cloned into plasmid L4440. The constructed plasmids in single colonies were verified by PCR and sequencing.
Table 1.
Primers used in this study.
| Target gene | Annotation | Accession NO. | Primer sequence | PCR product size(bp) |
|---|---|---|---|---|
| HaHR3-Frag.1 | Molt-regulating transcription factor fragment one | FJ009448 | RNAi-F1: 5'-ccCTCGAGATGAACAACAACCAGTTCCACGAT-3' RNAi-R1: 5'-gaAGATCTCACGCAGGCTTTATTCCGTGGACA-3' |
450 (1-450)a |
| HaHR3-Frag.2 | Molt-regulating transcription factor fragment two | FJ009448 | RNAi-F2: 5'-ccCTCGAGACCAGTGTCCACGGAATAA-3' RNAi-R2: 5'-gaAGATCTTTGCTGATGTCACCCTCC -3' |
492 (422-914)a |
| HaHR3-Frag.3 | Molt-regulating transcription factor fragment three | FJ009448 | RNAi-F3: 5'-ccCTCGAGGGGTGACATCAGCAAAGT -3' RNAi-R3: 5'-gaAGATCTCTCTGATGGACAGCACCG -3' |
358 (900-1258)a |
| HaHR3-Frag.4 | Molt-regulating transcription factor fragment four | FJ009448 | RNAi-F4: 5'-ccCTCGAGCAAGTGCTATACGGTGATG -3' RNAi-R4: 5'-gaAGATCTTTAACCGTGGGTGTAGTC -3' |
449 (1222-1671)a |
| EGFP-Frag. | Enhanced green fluorescent protein | JQ733047 | RNAi-EGFP-F: 5'-ccCTCGAGAGCTGACCCTGAAGTTCATCTGCACCAC-3' RNAi-EGFP-R: 5'-gaAGATCTGTCACGAACTCCAGCAGGACCATGTGAT-3' |
552 (125-677)a |
| Sense-Frag. | Molt-regulating transcription factor sense fragment for plant transformation | FJ009448 | RNAi-450s-F: 5'-ccCTCGAGATGAACAACAACCAGTTCCACGAT-3' RNAi-450s-F: 5'-gaAGATCTCACGCAGGCTTTATTCCGTGGACA-3' |
450 (1-450)a |
| Anti-sense Frag. | Molt-regulating transcription factor anti-sense fragment for plant transformation | FJ009448 | RNAi-450a-F: 5'-gcGTCGACATGAACAACAACCAGTTCCACGAT -3' RNAi-450a-R: 5'-cgGGATCCCACGCAGGCTTTATTCCGTGGACA -3' |
450 |
| Sense frag. in plants | Molt-regulating transcription factor sense fragment with partial intron | FJ009448 | Detection sense fragment F: 5'-ATGAACAACAACCAGTTC -3' Detection sense fragment R: 5'-TGTTTATCTGGTTTTGAGT -3' |
571 |
| Anti-sense frag. in plants | Molt-regulating transcription factor anti-sense fragment with partial intron | FJ009448 | Detection anti-sense fragment F: 5'-CGGACCGTACTACTCTATTC-3' Detection anti-sense fragment R: 5'-CAACCAGTTCCACGATTT-3' |
640 |
| qRT-HaHR3 fragment | Molt-regulating transcription factor fragment qRT-PCR product 1 | FJ009448 | qRT- HaHR3 F: 5'-GCCACAGGATGTCTCCAAGC-3' qRT- HaHR3 R: 5'-GAGCCAGATTTCAAGAGCAAAA-3' |
177 (993-1169)a |
| qRT-actA3b fragment | actin-A3b fragment | X97615 | qRT- actA3b F: 5'-CCCCGTCCACAATGAAGATC-3' qRT- actA3b R: 5'-GGCCAGACTCGTCGTACTCCT-3' |
140 |
| Protein expression full length CDS | Molt-regulating transcription factor | FJ009448 | protein expression primer F: 5'-CGGAATTCATGAACAACAACCAGTTCCACGATT-3' protein expression primer R: 5'-CCGCTCGAGTTAACCGTGGGTGTAGTCCAGGAC-3' |
1688 |
a Positions correspond to the gene sequence.
HT115 competent cells (a mutant E. coli strain lacking RNaseIII) were prepared by the CaCl2 precipitation method and transformed with verified recombinant plasmids. Single colonies of HT115 were cultured in LB at 37°C with shaking at 220 rpm overnight. The bacterial culture was diluted 100-fold in 1000 ml LB medium supplemented with 100 μg/ml ampicillin plus 12.5 μg/ml tetracycline and cultured at 37°C to OD600 = 0.4. The bacteria were incubated with shaking for overnight at 37°C after adding 0.5 mM IPTG to induce dsRNA expression.
Confirmation of dsRNA expression in HT115 cells
Total RNA was extracted from HT115 cells using a bacterial RNA-specific extraction kit (Sangon, China). dsRNA was extracted with a modified LiCl protocol 51. The extracted total RNAs or dsRNAs were loaded onto a 1.2% agarose gel, stained with GoldView Dye and then photographed.
Plant expression vector construction and tobacco transformation
XhoI and BglII restriction sites were added to the sense fragment 1 ends with primer RNAi-450s-F/R. SalI and BamHI restriction sites were added to the anti-sense ends with primer RNAi-450a-F/R at the same time. We ligated the sense fragment followed by the anti-sense fragment into the pRNAi1017 vector in sequence. PCR detection and double enzyme digestion were used to verify the correct construction of the intermediate vector pRNAi1017-HaHR3sa.
We then double-digested the plant over-expression vector pCAMBIA2300-35s-OCS and intermediate vector pRNAi1017-HaHR3sa with SalI and PstI. We ligated the pCAMBIA2300-35s-OCS and target gene fragments with introns using TOYOBO Ligation high Ver. 2 (Japan, lot 94190H6, code LGK-101). The vector for hpRNA-EGFP was similarly constructed. pCAMBIA-RNAi-HaHR3 and pCAMBIA-RNAi-EGFP were transformed into competent Agrobacterium tumefaciens strain LBA4404 by electroporation. The presence of the constructed plasmids in single colonies were verified by sequencing.
The leaf disc cocultivation method was followed using the standard procedure for tobacco transformation 52. After one month of tissue culture, the rooted plants were transferred into sterilized soil under the greenhouse conditions described above. T0-generation seeds (T1 progeny) were germinated on 1/2 MS medium plus 100 mg/l kanamycin for one week to screen heterologous or homologous plants. Transgenic tobacco plants expressing hpRNA-EGFP were used as controls in subsequent experiments.
Transgenic tobacco molecular analysis
Transgenic tobacco plant lines were identified by PCR, southern blot. Genomic DNA was extracted from regenerated tobacco leaves with the cetyltrimethylammonium bromide (CTAB) method 53. Transgenic plants were confirmed by PCR with two pairs of primers that covered an intron (Detection sense fragment F/R, Detection anti-sense fragment F/R). For Southern blot hybridizations, 30 μg of the genomic DNA was completely digested with SalI overnight. The DNAs were separated in 1.5% agarose gel and transferred to a Hybond-N+ filter membrane (Amersham Pharmacia Biotech) with a Vacuum Blotter (Bio-Rad, Model 785). After electrophoresis, they were electroblotted onto a Hybond-N+ filter membrane. The membranes were UV-cross-linked and hybridized with expressHyb solution (Clontech). PCR-positive plants were confirmed by Southern blot according to the DIG High Prime DNA labeling and Detection Starter kit II instruction manual (Roche, USA, Cat. No. 1585614).
Feeding bioassays with bacteria expressing different dsRNAs or plants expressing hairpin RNAs
Bacteria expressing four different dsRNA fragments were prepared with the bacterial expression system. We centrifuged the bacterial solution at 5,000 g for 15 min, resuspended bacteria in ddH2O, and mixed the artificial diet with condensed bacteria expressing dsRNA-HaHR3 or dsRNA-EGFP in the mass: volume ratio of 100:1. The 3rd-instar synchronous larvae were selected and reared individually with 1.0 g of an artificial diet in 6-well plates after 24 h starvation. Thirty biological repeats were used in each treatment, and there were 3 replicates of each treatment. The diet was replaced daily. After feeding on artificial diets supplemented with different dsRNAs, larvae were weighed and lethality was recorded for further analysis.
To investigate the transgenic tobacco activity, three independent single-copy lines were chosen for the hpHaHR3 feeding bioassay. Wild-type tobacco and hpEGFP-expressing tobacco lines were used as controls. Thirty synchronous 3rd-instar larvae starved for 24 h were fed leaf discs of the same size individually in 6-well plates. There were 3 replicates of each transgenic, control and wild-type line. Wet filter paper of the same size was placed in every chamber to maintain humidity. The leaf disc was replaced every 12 hours. After feeding on different diets for the indicated times, each larva was weighed individually, and RNA and protein were extracted for further analysis. The survival larvae (10-20) of each treatment were kept and recorded for further pupation or emergence deformity rate calculation.
Statistical analysis was performed with Student's t-test in the SPSS 13.0 program.
qRT-PCR
Total RNA was extracted from the experimental and control cotton bollworms using Trizol reagent according to the manufacturer's instructions. First-strand cDNA was synthesized using reverse TranscriptaseII (Transgen, China) under the following reaction conditions: 37°C for 10 min, 42°C for 1 h, and 95°C for 5 min. Specific primerswere designed to amplify the partial CDS of the HaHR3 (qRT-HaHR3 F and R, Table 1) and Actin genes. All of the qRT-PCR experiments were performed with a Bio-Rad iQ5 Real-Time PCR Detection System (Bio-Rad, USA) in a reaction volume of 20 μl using SYBR Green Master Mix (TaKaRa). The PCR program was 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 15 s. All the results were obtained from 4 independent biological replicates. Threshold cycle (CT) values were calculated using Bio-Rad CFX ManagerTM software. The relative gene expression data were analyzed using the 2-△△CT method as described previously 54. The results were analyzed for significant differences with Student's t-test.
Protein detection
We amplified the HaHR3 gene with a pair of protein expression primers. The amplified PCR product was double-digested with EcoRI and XhoI. We cloned this fragment into pET-28a, pET-30a, pET-32a and pGEX-6p-1 protein expression vectors for screening out the best soluble protein expression. Each recombinant vector was then transformed into Rosetta(DE3) competent cells. The DE3 cell cultures were incubated in LB liquid medium and induced for 20 h with 0.1 mM isopropyl β-D-thiogalactopyranoside (IPTG) at 20°C. The bacteria were collected by centrifugation, resuspended in PBS and sonicated. Unfortunately, recombinant protein HaHR3 was expressed in insoluble inclusion bodies. We isolated total proteins and separated them by SDS-PAGE. The pET-28a constructed expression vector was selected for the following antibody preparation experiments, because it contained the shortest protein expression tag. The HaHR3 protein was extracted from the gel and then purified with electrical washing for antibody preparation. Polyclonal rabbit antiserum against HaHR3 was raised in New Zealand white rabbits following standard protocols. Rabbits were immunized with HaHR3 weekly, and the antiserum was harvested one month later. We then purified the antibody with a HiTrap Protein A HP column.
Total proteins of H. armigera were extracted for western blot analysis. We determined the concentration of total protein by bicinchoninic acid (BCA) method (Pierce Chemical). Total protein of each lane were equated (40 μg protein per lane) and separated by 15% SDS-PAGE, and transferred to a polyvinylidene fluoride (PVDF) membrane (Bio-Rad) with a Trans-Blot SD semi-dry electrophoretic transfer cell (Bio-Rad). The membrane was blocked overnight with 5% non-fat milk in Tris-buffered saline containing 0.1% Tween 20 (TBST) and then incubated with HaHR3 antiserum (1:6000) overnight at 4°C. We washed the membrane 4 times with TBST every 5 minutes, then incubated the membrane with alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma) at a dilution of 1:2000 for 1 h. The membrane was washed 3 times with TBST and visualized with 1 mL BCIP/NBT solution (Amresco, USA).
Supplementary Material
Fig.S1 - S3.
Acknowledgments
We wish to express our gratitude to Professor Gemei Liang (Institute of Plant Protection, Chinese Academy of Agricultural Sciences) for kindly providing eggs of the cotton bollworm, Helicoverpa armigera. This work was supported by the National Natural Science Foundation of China (31171914).
References
- 1.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fire A, Xu SQ, Montgomery MK. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Belles X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annu Rev Entomol. 2010;55:111–128. doi: 10.1146/annurev-ento-112408-085301. [DOI] [PubMed] [Google Scholar]
- 4.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 5.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol. 2010;56:227–235. doi: 10.1016/j.jinsphys.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Wu KM, Lu YH, Feng HQ. et al. Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science. 2008;321:1676–1678. doi: 10.1126/science.1160550. [DOI] [PubMed] [Google Scholar]
- 8.Gordon KH, Waterhouse PM. RNAi for insect-proof plants. Nat Biotechnol. 2007;25:1231–1232. doi: 10.1038/nbt1107-1231. [DOI] [PubMed] [Google Scholar]
- 9.Bravo A, Soberón M. How to cope with insect resistance to Bt toxins? Trends Biotechnol. 2008;26:573–579. doi: 10.1016/j.tibtech.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Tabashnik BE, Gassmann AJ, Crowder DW. et al. Insect resistance to Bt crops: evidence versus theory. Nat Biotechnol. 2008;26:199–202. doi: 10.1038/nbt1382. [DOI] [PubMed] [Google Scholar]
- 11.Lu Y, Wu K, Jiang Y. et al. Mirid bug outbreaks in multiple crops correlated with wide-scale adoption of Bt cotton in China. Science. 2010;328:1151–1154. doi: 10.1126/science.1187881. [DOI] [PubMed] [Google Scholar]
- 12.Chadwick D, Marsh J. Crop protection and sustainable agriculture. John Wiley & Sons. 1993.
- 13.Pimentel D. Encyclopedia of pest management. CRC. 2007.
- 14.Kortenhoff A. Developments in the use of pesticides. Modern crop protection: development and perspectives; 1993. pp. 3–10. [Google Scholar]
- 15.Price DR, Gatehouse JA. RNAi-mediated crop protection against insects. Trends Biotechnol. 2008;26:393–400. doi: 10.1016/j.tibtech.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Mao YB, Cai WJ, Wang JW. et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- 17.Mao YB, Tao XY, Xue XY. et al. Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Res. 2011;20:665–673. doi: 10.1007/s11248-010-9450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baum JA, Bogaert T, Clinton W. et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- 19.Zha W, Peng X, Chen R. et al. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS One. 2011;6:e20504. doi: 10.1371/journal.pone.0020504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitino M, Coleman AD, Maffei ME. et al. Silencing of aphid genes by dsRNA feeding from plants. PLoS One. 2011;6:e25709. doi: 10.1371/journal.pone.0025709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar P, Pandit SS, Baldwin IT. Tobacco rattle virus vector: A rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS One. 2012;7:e31347. doi: 10.1371/journal.pone.0031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu JQ, Liu S, Ma Y. et al. Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PLoS One. 2012;7:e38572. doi: 10.1371/journal.pone.0038572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar BA. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7:907–916. doi: 10.1038/nrg1989. [DOI] [PubMed] [Google Scholar]
- 24.Dubrovsky EB. Hormonal cross talk in insect development. Trends Endocrinol Metab. 2005;16:6–11. doi: 10.1016/j.tem.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Dhadialla TS, Carlson GR, Le DP. New insecticides with ecdysteroidal and juvenile hormone activity. Annu Rev Entomol. 1998;43:545–569. doi: 10.1146/annurev.ento.43.1.545. [DOI] [PubMed] [Google Scholar]
- 26.Lam G, Hall BL, Bender M. et al. DHR3 is required for the prepupal-pupal transition and differentiation of adult structures during Drosophila metamorphosis. Dev Biol. 1999;212:204–216. doi: 10.1006/dbio.1999.9343. [DOI] [PubMed] [Google Scholar]
- 27.Koelle MR, Segraves WA, Hogness DS. DHR3: a Drosophila steroid receptor homolog. P Natl Acad Sci USA. 1992;89:6167. doi: 10.1073/pnas.89.13.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin FS, Zeng HM, Zhang YL. et al. Cloning of Helicoverpa armigera gene HaHR3 and construction of its RNA interference vector. Cotton Science. 2010;22:157–162. [Google Scholar]
- 29.Zhao XF, Wang JX, Xu XL. et al. Molecular cloning and expression patterns of the molt-regulating transcription factor HHR3 from Helicoverpa armigera. Insect Mol Bio. 2004;13:407–412. doi: 10.1111/j.0962-1075.2004.00499.x. [DOI] [PubMed] [Google Scholar]
- 30.Dhadialla TS, Carlson GR, Le DP. New insecticides with ecdysteroidal and juvenile hormone activity. Annu Rev Entomol. 1998;43:545–569. doi: 10.1146/annurev.ento.43.1.545. [DOI] [PubMed] [Google Scholar]
- 31.Matzke M, Matzke A, Kooter J. RNA: guiding gene silencing. Science. 2001;293:1080–1083. doi: 10.1126/science.1063051. [DOI] [PubMed] [Google Scholar]
- 32.Holen T, Amarzguioui M, Wiiger MT. et al. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds A, Leake D, Boese Q. et al. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 34.Luo KQ, Chang DC. The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem Biophys Res Commun. 2004;318:303–310. doi: 10.1016/j.bbrc.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 35.Shao Y, Chan CY, Maliyekkel A. et al. Effect of target secondary structure on RNAi efficiency. Rna. 2007;13:1631–1640. doi: 10.1261/rna.546207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Zhang S, Kulye M. et al. Silencing of molt-regulating transcription factor gene, CiHR3, affects growth and development of sugarcane stem borer, Chilo infuscatellus. J insect science. 2012;12:9. doi: 10.1673/031.012.9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding Y, Chan CY, Lawrence CE. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terenius O, Papanicolaou A, Garbutt JS. et al. RNA interference in lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231–245. doi: 10.1016/j.jinsphys.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 39.Burand JP, Hunter WB. RNAi: Future in insect management. J Invertebr Pathol. 2012. [DOI] [PubMed]
- 40.Kamath RS, Martinez-Campos M, Zipperlen P. et al. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:0002.0001–0002.0010. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Timmons L, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 42.White KP, Hurban P, Watanabe T. et al. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science. 1997;276:114–117. doi: 10.1126/science.276.5309.114. [DOI] [PubMed] [Google Scholar]
- 43.Lam GT, Jiang C, Thummel CS. Coordination of larval and prepupal gene expression by the DHR3 orphan receptor during Drosophila metamorphosis. Development. 1997;124:1757–1769. doi: 10.1242/dev.124.9.1757. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa Y, Henrich VC. Arthropod nuclear receptors and their role in molting. FEBS Journal. 2009;276:6128–6157. doi: 10.1111/j.1742-4658.2009.07347.x. [DOI] [PubMed] [Google Scholar]
- 45.Carney GE, Wade AA, Sapra R. et al. DHR3, an ecdysone-inducible early-late gene encoding a Drosophila nuclear receptor, is required for embryogenesis. P Natl Acad Sci USA. 1997;94:12024–12029. doi: 10.1073/pnas.94.22.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cruz J, Martin D, Bell¨¦s X. Redundant ecdysis regulatory functions of three nuclear receptor HR3 isoforms in the direct-developing insect Blattella germanica. Mech Dev. 2007;124:180–189. doi: 10.1016/j.mod.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Kostrouchova M, Krause M, Kostrouch Z. et al. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. P Natl Acad Sci USA. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borgio JF. RNAi mediated gene knockdown in sucking and chewing insect pests. J Biopesticides. 2010;3:386–393. [Google Scholar]
- 49.Tian H, Peng H, Yao Q. et al. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One. 2009;4:340–347. doi: 10.1371/journal.pone.0006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou L, Fang Y, Yang J. Investigation on artificial diet in Heliothis armigera. Acta Entomol Sinica. 1981;24:108–110. [Google Scholar]
- 51.Morris T, Dodds J. Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology. 1979;69:854–858. [Google Scholar]
- 52.Gallois P, Marinho P. Leaf disk transformation using Agrobacterium tumefaciens-expression of heterologous genes in tobacco. Methods Mol. Biol. 1995;49:39–48. doi: 10.1385/0-89603-321-X:39. [DOI] [PubMed] [Google Scholar]
- 53.Stewart Jr CN, Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques. 1993;14:748–750. [PubMed] [Google Scholar]
- 54.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig.S1 - S3.