ABSTRACT
Aeromonas hydrophila has increasingly been implicated as a virulent and antibiotic-resistant etiologic agent in various human diseases. In a previously published case report, we described a subject with a polymicrobial wound infection that included a persistent and aggressive strain of A. hydrophila (E1), as well as a more antibiotic-resistant strain of A. hydrophila (E2). To better understand the differences between pathogenic and environmental strains of A. hydrophila, we conducted comparative genomic and functional analyses of virulence-associated genes of these two wound isolates (E1 and E2), the environmental type strain A. hydrophila ATCC 7966T, and four other isolates belonging to A. aquariorum, A. veronii, A. salmonicida, and A. caviae. Full-genome sequencing of strains E1 and E2 revealed extensive differences between the two and strain ATCC 7966T. The more persistent wound infection strain, E1, harbored coding sequences for a cytotoxic enterotoxin (Act), a type 3 secretion system (T3SS), flagella, hemolysins, and a homolog of exotoxin A found in Pseudomonas aeruginosa. Corresponding phenotypic analyses with A. hydrophila ATCC 7966T and SSU as reference strains demonstrated the functionality of these virulence genes, with strain E1 displaying enhanced swimming and swarming motility, lateral flagella on electron microscopy, the presence of T3SS effector AexU, and enhanced lethality in a mouse model of Aeromonas infection. By combining sequence-based analysis and functional assays, we characterized an A. hydrophila pathotype, exemplified by strain E1, that exhibited increased virulence in a mouse model of infection, likely because of encapsulation, enhanced motility, toxin secretion, and cellular toxicity.
IMPORTANCE
Aeromonas hydrophila is a common aquatic bacterium that has increasingly been implicated in serious human infections. While many determinants of virulence have been identified in Aeromonas, rapid identification of pathogenic versus nonpathogenic strains remains a challenge for this genus, as it is for other opportunistic pathogens. This paper demonstrates, by using whole-genome sequencing of clinical Aeromonas strains, followed by corresponding virulence assays, that comparative genomics can be used to identify a virulent subtype of A. hydrophila that is aggressive during human infection and more lethal in a mouse model of infection. This aggressive pathotype contained genes for toxin production, toxin secretion, and bacterial motility that likely enabled its pathogenicity. Our results highlight the potential of whole-genome sequencing to transform microbial diagnostics; with further advances in rapid sequencing and annotation, genomic analysis will be able to provide timely information on the identities and virulence potential of clinically isolated microorganisms.
Introduction
The Gram-negative bacterium Aeromonas hydrophila, a ubiquitous inhabitant of fresh and estuarine waters (1), has increasingly been implicated as an etiologic agent in a variety of human diseases (2). The spectrum of disease severity is broad, ranging from mild diarrhea to life-threatening necrotizing fasciitis, septicemia, meningitis, cholera-like illness, and hemolytic-uremic syndrome (3). The mere presence of A. hydrophila in an infected wound is an independent predictor of death among patients with necrotizing fasciitis (4). While traditionally regarded as a pathogen of immunocompromised humans, there have been several recently reported Aeromonas infections of immunocompetent individuals (5–8).
Recently, a case report by Shak et al. described a human wound infection involving a mixture of Gram-positive and Gram-negative bacteria, including two distinct strains of A. hydrophila, Aero 1 and Aero 2 (8), which we refer to here as strains E1 and E2, respectively. Both of these strains were recovered when the patient was initially admitted to the hospital and were identified as A. hydrophila on the basis of cell wall fatty acid analysis and biochemical characterization (8). While strain E1 was resistant to ampicillin and tetracycline, strain E2 exhibited additional antimicrobial resistance, specifically, to aminoglycosides and several expanded-spectrum cephalosporin β-lactams (8). Despite aggressive antibiotic treatment and surgical debridement, A. hydrophila strain E1 continued to be cultured from advancing cellulitis. With repeated surgical debridement and treatment with vancomycin and piperacillin-tazobactam, the infection cleared and the patient made a full recovery.
The pathogenic potential of A. hydrophila has been related to several virulence factors, including the cytotoxic enterotoxin Act (9), which has hemolytic, cytotoxic, and enterotoxic activities; a variety of proteases (10, 11); cytotonic enterotoxins Ast and Alt (12); type 3 secretion systems (T3SSs) (13); and motility factors such as lateral and polar flagella (14). As in previous case reports of Aeromonas-associated human wound infections (15, 16), the report by Shak et al. (8) did not describe the genotypic or mechanistic determinants of virulence and antibiotic resistance. We hypothesized that the persistence of strain E1 in the wound of this patient could be attributed to known Aeromonas virulence factors identifiable at the genotypic and phenotypic levels.
To examine this hypothesis and further develop the earlier findings of Joseph et al. (15) and Shak et al. (8), the whole genome of each clinical strain was sequenced and the resulting draft genomes were compared to other Aeromonas genomes, including the closed genome of the type strain, A. hydrophila ATCC 7966T (17). Comparative genomics revealed several differences in the genomes of A. hydrophila strains E1 and E2 that suggested that strain E1 was more virulent because of the presence of several virulence factor-encoding genes. To complement the genomic findings and investigate the functionality of these virulence traits, bioassays of strains E1 and E2 were conducted alongside environmental isolate A. hydrophila ATCC 7966T (17) and virulent diarrhea isolate SSU (18) to assess motility, cytotoxicity, protease activity, secretion system functionality, the ability to form biofilms, and serum resistance. Finally, a septicemic-mouse model of infection was used to investigate the virulence potential of both strains E1 and E2 in comparison with those of other well-studied clinical and environmental Aeromonas isolates. Altogether, we demonstrated that genotypic differences correlated with functional virulence factor assays, strongly suggesting the existence of an identifiable virulent pathotype of A. hydrophila that leads to wound infections in humans.
RESULTS
Genomic characteristics of A. hydrophila strains E1 and E2.
Pyrosequencing of A. hydrophila strains E1 and E2 resulted in draft genomes with calculated G+C contents of 61.3 and 61.5%, respectively, similar to those of A. hydrophila ATCC 7966T and the closely related species A. aquariorum and A. caviae (Table 1). The genomes of strains E1 and E2 contained an estimated 10 rRNA operons, equal to the genomes of A. hydrophila ATCC 7966T, A. aquariorum AAK1, and A. veronii B565 (Table 1). No plasmids were harbored in the genome of either strain E1 or E2 at the time of sequencing. Average nucleotide identity (ANI) analysis by BLAST confirmed that both strains E1 and E2 belong to A. hydrophila (Table 2). Importantly, E1 and E2 were distinct strains, with 96.88 and 96.96% ANIs, and both strains E1 and E2 were equally dissimilar to A. hydrophila ATCC 7966T, with ANIs of approximately 97% (Table 2). Bidirectional BLAST analyses of annotated coding DNA sequences (CDSs) of A. hydrophila strains ATCC 7966T, E1, and E2, as well as A. aquariorum AAK1, A. caviae Ae398, A. salmonicida A449, and A. veronii B565, confirmed the results of the ANI genomic comparison (see Table S1 and Fig. S1 in the supplemental material).
TABLE 1.
Genome characteristics of three A. hydrophila strains and four closely related Aeromonas species as determined by the RAST annotation pipeline
| Organism | Source | No. of contigs |
No. of bp |
No. of CDSs |
No. of RNAs |
No. of tRNAs |
No. of rRNAs |
No. of rRNA operonsb |
G+C content (%) |
|---|---|---|---|---|---|---|---|---|---|
| A. hydrophila E1 | Wound infection | 249 | 4,754,562 | 4,373 | 76 | 70 | 6 | 10 | 61.3 |
| A. hydrophila E2 | Wound infection | 426 | 4,564,644 | 4,241 | 60 | 56 | 4 | 10 | 61.5 |
| A. hydrophila ATCC 7966T | Fishy milk | 1 | 4,744,448 | 4,279 | 158 | 127 | 31 | 10 | 61.6 |
| A. aquariorum AAK1 | Septicemia, necrotizing fasciitis |
36 | 4,763,532 | 4,275 | 100 | 88 | 12 | 10 | 61.8 |
| A. veronii B565 | Pond | 1 | 4,551,783 | 3,936 | 133 | 102 | 31 | 10 | 58.7 |
| A. salmonicida A449 | Furunculosis, brown trout | 6a | 5,040,536 | 4,306 | 137 | 109 | 28 | 9 | 58.5 |
| A. caviae Ae398 | Diarrhea, child | 149 | 4,439,218 | 3,912 | 78b | 72 | 6 | ? | 61.4 |
Includes one chromosome and five plasmids.
rRNA operon numbers for draft genomes were estimated on the basis of the presence of full and partial 16S and 23S rRNA genes and comparison to A. hydrophila ATCC 7966T.
TABLE 2.
Pairwise ANIs, by BLAST, of three strains of A. hydrophila and four closely related Aeromonas species
| Organism |
A. hydrophila E1 |
A. hydrophila E2 |
A. hydrophila ATCC 7966T |
A. aquariorum AAK1 |
A. veronii B565 |
A. caviae Ae398 |
A. salmonicida A449 |
|---|---|---|---|---|---|---|---|
| A. hydrophila E1 | 96.88a | 96.84 | 92.8 | 85.73 | 86.6 | 86.49 | |
| A. hydrophila E2 | 96.96 | 97.16 | 92.95 | 85.79 | 86.79 | 86.82 | |
| A. hydrophila ATCC 7966T | 96.81 | 97.06 | 92.92 | 85.5 | 86.45 | 86.47 | |
| A. aquariorum AAK1 | 92.93 | 93.04 | 93.00 | 85.24 | 86.57 | 85.92 | |
| A. veronii B565 | 85.8 | 85.77 | 85.55 | 85.18 | 83.74 | 84.10 | |
| A. caviae Ae398 | 86.63 | 86.68 | 86.48 | 86.54 | 83.63 | 83.97 | |
| A. salmonicida A449 | 86.58 | 86.87 | 86.54 | 86.03 | 84.15 | 84.22 |
Values in bold indicate strains that belong to the same species (i.e., ANI of >95).
Comparative genomics of A. hydrophila.
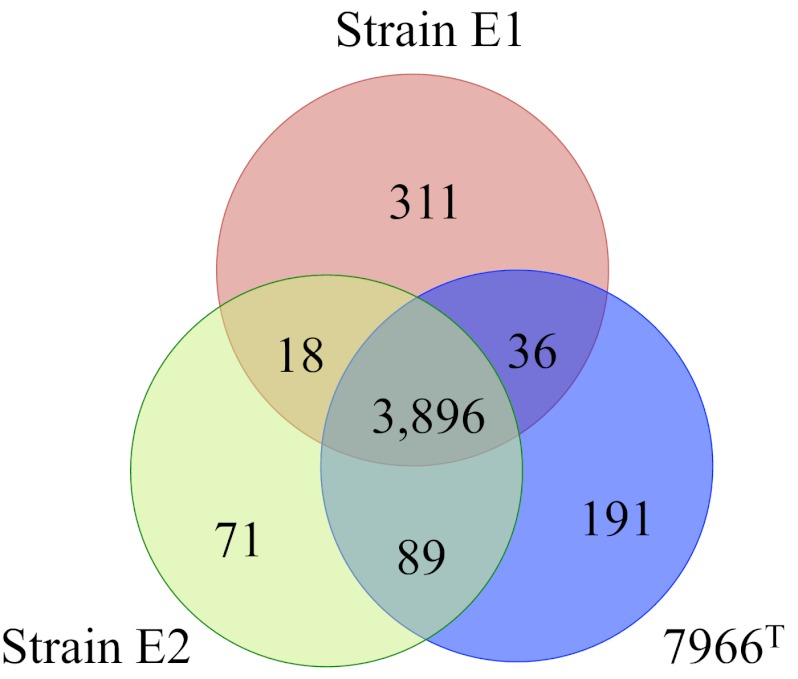
The genome sequences of strains E1, E2, and ATCC 7966T contained 4,373, 4,241, and 4,279 CDSs, respectively, upon initial annotation by the RAST server (Table 1). Manual curation was performed to reconcile genes that were fragmented because of sequence quality or disrupted by gaps in contigs and a number of small hypothetical protein-encoding genes. Following curation, we found that all three A. hydrophila strains shared a core genome of 3,896 genes, which was, on average, 93% of the total CDSs in each genome (see Fig. 1 and Table S1 in the supplemental material).
FIG 1 .
Venn diagram of the distribution of protein CDSs inferred from the genomes of A. hydrophila ATCC 7966T, E1, and E2. Numbers of genes unique to and common to ATCC 7966T, E1, and E2 are indicated within the Venn diagram. The values are gene counts following manual curation and differ from the gene counts in the automated RAST pipeline results presented in Table 1.
Several of the core A. hydrophila genes were shared with the other Aeromonas species analyzed in this study. For example, all seven genomes contained the following notable operons or features: T1SS (tolC); Sec and Tat secretion systems; a mannose-sensitive hemagglutinin (MSHA) bundle-forming type IV pilus (BFP; Table 3); polar flagella encoded in four gene clusters; the cytotonic enterotoxin gene alt; glycerophospholipid:cholesterol acyltransferase (GCAT); enolase; the vibriolysin/pseudolysin-like extracellular zinc protease/elastase (Table 3); the cephalosporinase β-lactamase gene cepS; the adenylate kinase β-lactamase gene ampS (or ampH); the carbapenemase gene cphA; multiple RND and ABC multidrug resistance (MDR) efflux pump genes; an ABC antimicrobial peptide transporter, the N-acetylglucosamine (NAG) utilization operon; an N,N′-diacetylchitobiose [(GlcNAc)2] utilization operon; the phenolate siderophore amonabactin gene cluster; the fbp gene for a ferric iron ABC transporter that transports iron from the periplasmic space into the cytosol; a hemin utilization locus, hut; ferric iron transporter genes; the ferrous iron transporter-encoding gene feo cluster; the luxS gene; the QS pair qseBC; and the QS regulator of virulence gene hapR (data not shown). Interestingly, the Tap type IV pilus, encoded in four clusters, was present in all seven genomes, but the level of amino acid identity between the species (~20 to 60%) was relatively low.
TABLE 3.
Presence or absence of protein CDSs in the genomes of three strains of A. hydrophila and four closely related Aeromonas species
| Putative protein CDS |
A. hydrophila ATCC 7966T |
A. hydrophila E1 |
A. hydrophila E2 |
A. aquariorum AAK1 |
A. veronii B565 |
A. salmonicida A449 |
A. caviae Ae398 |
|---|---|---|---|---|---|---|---|
| Metabolic and related | |||||||
| pgt utilization operon | + | + | + | + | − | − | − |
| Maltose homolog PTS, glucosidase | + | − | + | + | + | + | − |
| N-Acetylgalactosamine, aga | + | + | + | + | + | − | − |
| N-Acetylmuramic acid transporter | + | − | + | − | − | − | + |
| Xanthosine | + | + | + | − | − | − | − |
| Uncharacterized hexose-P PTS, uhpABC | + | + | + | + | + | + | − |
| Arabinose utilization | + | + | + | − | − | + | − |
| 2-Aminoethylphosphonate | + | + | + | + | + | − | − |
| Xanthine, yge, yqe cluster | + | + | + | + | + | − | − |
| Mannose transporter | + | + | + | − | − | − | − |
| l-Lactate utilization | + | + | + | + | − | − | + |
| l-Cystine transporter | + | + | + | − | − | − | + |
| Glutamate-aspartate transporter | − | + | − | + | − | − | + |
| Taurine transporter | + | − | + | + | − | − | − |
| Cytochrome o | + | + | + | + | − | + | + |
| Methylamine homolog utilization | + | + | + | + | + | − | − |
| Benzoate, N-acetylglucosamine | + | + | + | + | + | − | + |
| Phosphonate transporter | + | − | − | + | − | + | − |
| Decaheme cytochrome c, nrf | + | + | + | + | − | − | − |
| Cysteine operon, cysPTWA | + | + | + | + | − | + | + |
| Glutamine/glutamate transporter | + | + | + | + | − | + | + |
| Chemotaxis cluster, AHA_2527-2538 | + | + | + | − | − | − | − |
| N-Ribosylnicotinamide transporter | + | + | + | + | + | + | − |
| NO reductase | + | + | + | + | − | + | + |
| Dienelactone hydrolase | + | − | + | − | − | − | − |
| Quaternary ammonium compound resistance, propanediol, ethanolamine |
+ | − | + | + | − | − | − |
| Anaerobic sulfite reductase | + | + | + | − | + | − | − |
| Tungstate transporter | + | + | + | + | + | + | − |
| Tetrathionate reductase | + | + | + | + | − | − | + |
| Nitrate-nitrite reductase | + | + | + | − | − | − | + |
| Appendages | |||||||
| CFA/I (α C/U) fimbriae, AHA_0060 | + | + | + | + | − | + | + |
| CFA/I (α C/U) fimbriae, AHA_1021 | + | − | + | − | + | + | − |
| P (π C/U) fimbriae, AHA_0521 | + | + | + | + | + | + | − |
| Tap type IV pilus | + | + | + | + | + | + | + |
| TAD Flp pilus | + | − | − | − | + | + | − |
| MSHA BFP type IV pilus | + | + | + | + | + | + | + |
| Polar flagellum | + | + | + | + | + | + | + |
| Lateral flagellum | − | + | − | − | − | + | − |
| Toxins and exoenzymes | |||||||
| T3SS, aexU | − | + | − | − | − | + | − |
| T6SS, hcp | + | + | + | + | − | + | − |
| Cytotonic enterotoxin/lipase, alt | + | + | + | + | + | + | + |
| Cytotoxic enterotoxin/hemolysin, act | + | + | − | + | + | + | − |
| Cytotonic enterotoxin, ast | + | + | + | − | − | − | − |
| Enolase | + | + | + | + | + | + | + |
| Elastase | + | + | + | + | + | + | + |
| RTX toxin, AHA_1359, and transporter cluster | + | − | + | + | − | − | − |
| FHA family, RTX toxin | − | + | − | − | − | − | − |
| Pore-forming cytolysin/hemolysin, hlyA | + | + | + | + | − | + | − |
| Phospholipase/lecithinase/hemolysin-GCAT | + | + | + | + | + | + | + |
| Capsule | − | + | + | + | + | − | − |
| Antibiotic and multidrug resistance | |||||||
| Macrolide-specific ABC efflux pump | + | + | + | + | − | + | + |
| Polymyxin B resistance (arn) | + | + | + | + | − | + | − |
| Acr family RND efflux pump, AHA_2959-60 | + | + | + | + | + | + | − |
| ABC-type multidrug transport system, AHA_0484-6 |
+ | + | + | + | − | + | − |
| MATE efflux pump | + | + | − | + | + | + | + |
| OmpK-AmpG | + | + | + | + | + | + | + |
| NodT family RND efflux pump | − | + | − | − | − | − | + |
| Acriflavin RND transporter | + | − | − | + | + | − | + |
A certain number of A. hydrophila core genes were not shared with all other Aeromonas species genomes (i.e., absent from one or more non-A. hydrophila species; Table 3). Interestingly, this group contained genes that encode an alpha, or class 5, chaperone/usher (CU) fimbrial operon (AHA_0060, usher gene); a π-fimbrial operon (AHA_0521); the N-acetylgalactosamine (aga) phosphotransferase system (PTS); the arabinose utilization operon; the pore-forming hemolysin/cytolysin gene hlyA; the polymyxin B resistance gene cluster arn; as well as several other ABC- and RND-type MDR efflux pumps and a T6SS (Table 3; see Table S1 in the supplemental material). Additionally, the cytotonic heat-stable enterotoxin gene ast was found only in the three A. hydrophila genomes.
As described below, a number of genes present in the genome of strain ATCC 7966T were present in the genome of either strain E1 or E2 but not in both. Common to A. hydrophila ATCC 7966T and strain E1 were genes that encode the cytotoxic enterotoxin Act, a MATE family MDR efflux pump (Table 3), and five small genomic regions of unknown function (see Table S1). Strains E2 and ATCC 7966T shared several metabolic operons and transporters, including those for the utilization of N-acetylmuramic acid, for maltose (or a maltose homolog), and for the utilization and transport of the sulfonic acid taurine (Table 3). They also shared genes that encoded a second α-fimbriae (AHA_1021, usher gene), an RTX toxin (AHA_1359) and its transporter (Table 3), and operons involved in the degradation of chloroaromatics (dienelactone) (Table 3), arsenical compounds (see Table S1), and quaternary ammonium compounds, as well as ethanolamine (Table 3).
There were only 18 genes that were shared by A. hydrophila E1 and E2 but not present in the genome of ATCC 7966T (Fig. 1). Of interest among these were distinct serogroup-specific capsular polysaccharide operons that contained some homologous genes (four out of seven; see Table S1 in the supplemental material).
There were 191 CDSs present in the genome of A. hydrophila ATCC 7966T, which were absent in the genomes of strains E1 and E2 (Fig. 1), including two large prophage-like integrated regions (see Table S1), the serogroup O:1 antigen lipopolysaccharide (LPS) cluster, and a putative LPS modification gene cluster, along with several small genomic regions of unknown function (see Table S1), the tight adherence (TAD) Flp pilus (Table 3), a phosphonate transporter, and an acriflavin RND transporter (Table 3). Of the three A. hydrophila genomes analyzed, that of strain E2 had the fewest unique genes, 71 (Fig. 1), most of which encode proteins of unknown functions. Notable exceptions were a gene cluster that encodes a putative arylsulfatase, the O:18 antigen gene cluster, a type I restriction modification system, several transposons, and a putative MFS xylose transporter (see Table S1).
Virulence factor-encoding genes unique to the genome of A. hydrophila E1.
The genome of A. hydrophila E1 contained a number of virulence factor-encoding genes not found in the genome of A. hydrophila ATCC 7966T or strain E2. These included a T3SS, a lateral flagellar system (Table 3), and several large exoproteins, annotated as hemagglutinins or adhesins/hemolysins, including one that was 64% identical to exotoxin A of Pseudomonas aeruginosa (see Table S1). The T3SS found in the genome of E1 was approximately 26,037 bp in length, was contained on contigs 00088 (bp 1710 to the end) and 00128, and was 97% identical (100% coverage) to the T3SS found in A. hydrophila strain AH1 (GenBank accession no. AY394563.2) (see Fig. S2 and S3A in the supplemental material). The genes that encode the lateral flagella of E1 were contained in a single gene cluster of 35 kb located on contig 00003 (~bp 93,233 to 127,913). This cluster was highly homologous to that of A. salmonicida A449 and A. hydrophila AH3, 85% identical, with 93% coverage (see Fig. S3B). In addition, the genome of strain E1 harbored at least two prophage-like elements, a glutamate-aspartate transporter, several transposases and insertion elements, and a number of genomic regions of unknown function (see Table S1).
A. hydrophila strains E1 and E2 exhibit motility.
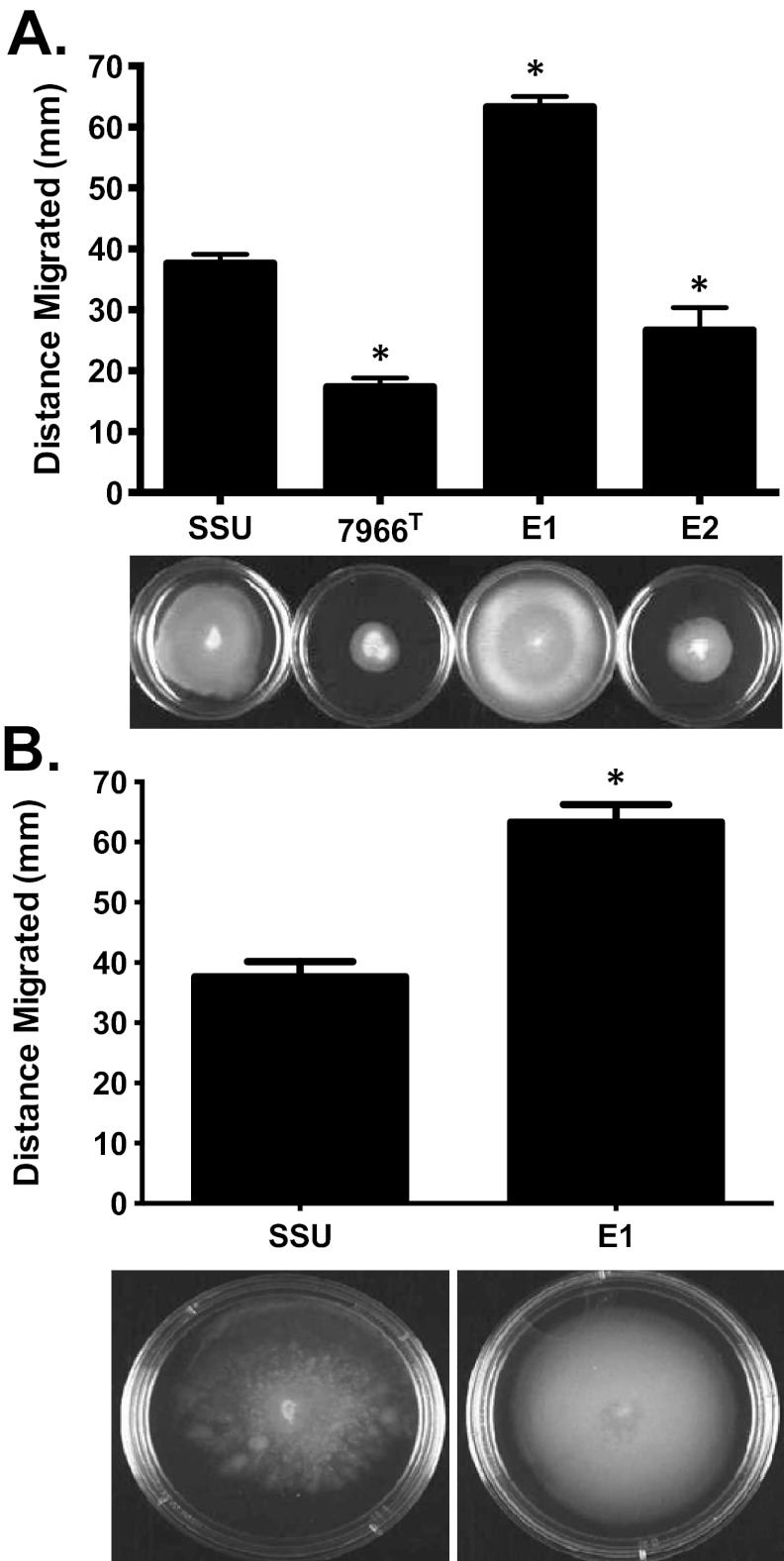
To examine how the genetic differences in motility-associated factors influenced phenotypes, we performed swimming (polar flagellum) and swarming (lateral flagella) motility assays. Strain E1 displayed significantly higher swimming motility than the type strain ATCC 7966T and the virulent diarrhea isolate A. hydrophila SSU, whose motility has been previously demonstrated (10). Strain E2 exhibited less swimming motility than strain SSU but more than ATCC 7966T (Fig. 2A). Since strain E1 was highly motile, we repeated the swimming motility assay with larger petri dishes to better gauge differences in motility between clinical isolates SSU and E1. Indeed, the differences in motility between these two strains increased further because of the increased surface area available for bacteria to swim (Fig. 2B).
FIG 2 .
Swimming motility of A. hydrophila strains SSU, ATCC 7966T, E1, and E2. (A) Strain E1 showed greater swimming motility than SSU (P < 0.001), while strain E2 exhibited less motility than SSU (P < 0.001) (50-mm-diameter petri dishes). (B) To measure exact zones of migration by A. hydrophila strain E1, we used 80-mm-diameter petri dishes. Three independent experiments were performed, and the arithmetic means ± the standard deviations were plotted. An asterisk indicates a P value of <0.001 as determined by one-way ANOVA.
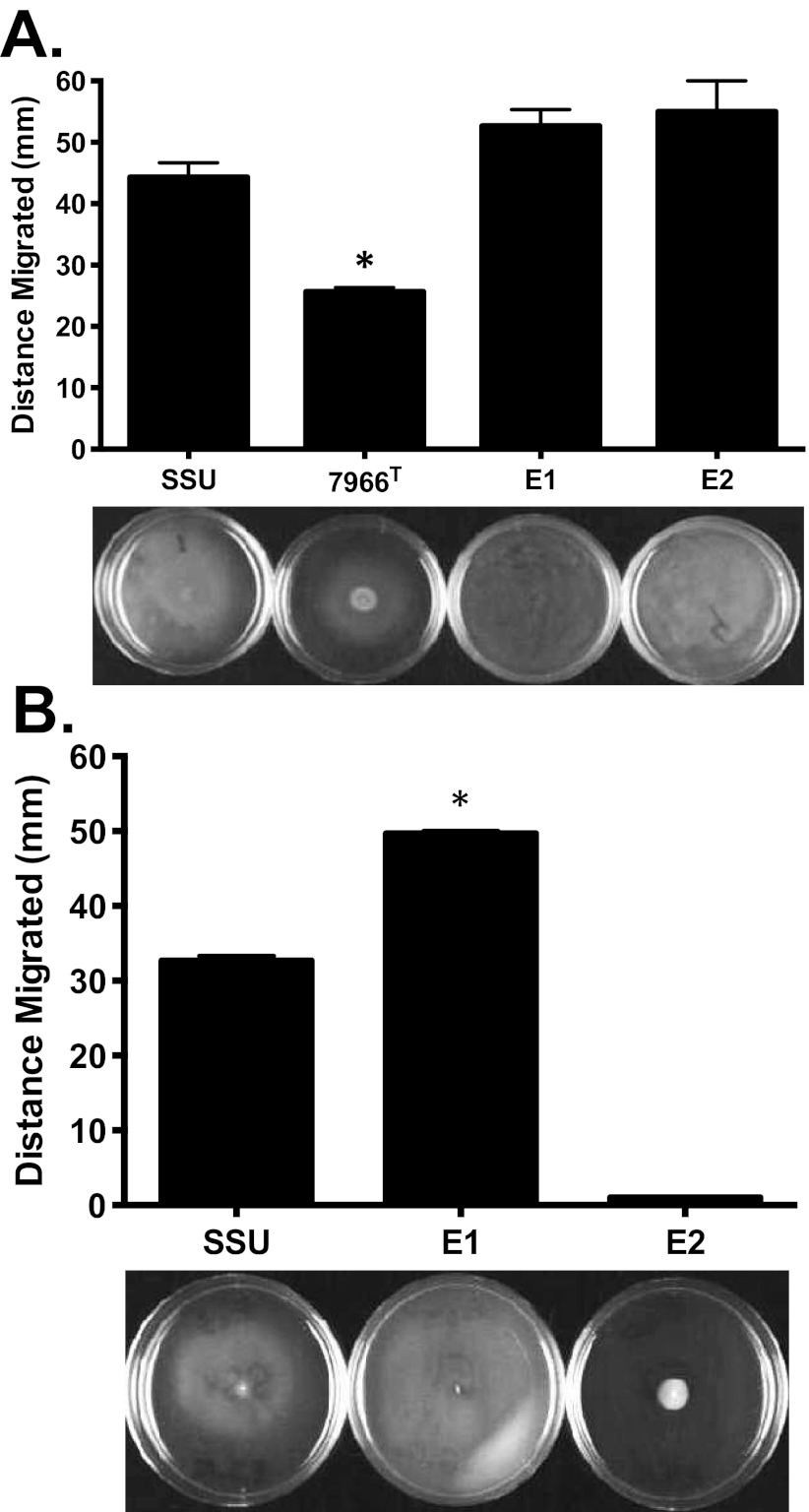
The swarming motility assays were performed as described by Kirov et al. (19), with freshly poured and dried swarming agar plates inoculated from the swimming agar plate culture. Strains E1, E2, and SSU exhibited similar swarming behavior on 0.5% Eiken agar plates at 30°C (Fig. 3A). This was surprising, given the absence of lateral flagella genes in the genome of strain E2. An alternate protocol, detailed in Materials and Methods, was used that gave phenotypic results for strain E2 in agreement with the genomic findings (Fig. 3B) while not affecting the observed swarming phenotypes of strains SSU and E1. Other perturbations of the swarming motility assay protocol had pronounced effects on the outcome, including the temperature of incubation of the plates, the concentration of agar added to the medium, and the brand of agar used, as well as the overall medium formulation (data not shown). For example, when the swarming plates were incubated at 37°C irrespective of the agar used (0.5% Eiken versus 0.8% Bacto agar), none of the bacterial cultures tested (E1, E2, or SSU) exhibited any swarming (data not shown). When a different medium composition was used at 30°C, the swarming pattern changed, with strains E1 and SSU exhibiting swarming, while strain E2 showed a moderate level of motility (data not shown).
FIG 3 .
Swarming motility of A. hydrophila strains SSU, ATCC 7966T, E1, and E2. (A) Strains E1 and E2 had swarming motility comparable to that of strain SSU on Difco nutrient agar plates with 0.5% Eiken agar at 30°C when the bacteria were propagated as described previously (19). (B) By an alternative protocol, strains were subcultured from the freezer onto blood agar plates and then inoculated onto swarming plates. In this assay, strain E1 had swarming motility superior to that of strain SSU, and strain E2 exhibited no swarming motility. Three independent experiments were performed, and the arithmetic means ± the standard deviations were plotted. An asterisk indicates a P value of <0.001 as determined by one-way ANOVA.
Electron microscopy demonstrates lateral flagella of strain E1 and rafting behavior of strain E2.
To further investigate the confounding swarming behavior of strain E2, electron microscopy of cultures taken directly from the swarming plates was performed (see Fig. S4 in the supplemental material). In agreement with the alternate swarming assay and genome analysis, A. hydrophila strain E1 produced lateral flagella (see Fig. S4A) similar to those of A. hydrophila strain SSU (see Fig. S4C). In contrast, A. hydrophila strain E2 did not produce lateral flagella but instead exhibited a “rafting”-like behavior that could mimic swarming because of unidentified cell-cell interactions possibly involving extracellular capsule and one or more types of small fimbriae (see Fig. S4B).
A. hydrophila strains E1 and E2 produce AHLs.
Quorum sensing (QS)- or cell density-dependent regulation is controlled by the concentration of small signal molecules, N-acyl-homoserine lactones (AHLs), termed autoinducers. A common approach used to detect AHLs is via bacterial reporter strains, which do not produce intrinsic AHLs. In the presence of exogenously produced AHLs, these reporter strains display specific, QS-induced phenotypes, such as purple pigment production by Chromobacterium violaceum CV026 (20). A. hydrophila strains E1, E2, and SSU produced similar levels of AHLs (Table 4). Two negative-control A. hydrophila strains, namely, ATCC 7966T (21) and the ∆ahyRI isogenic mutant of strain SSU (10), demonstrated no lactone production in this bioassay.
TABLE 4 .
Lactone production and hemolytic activities of four A. hydrophila isolates
| A. hydrophila strain | Lactone productiona | Mean hemolytic activity ± SD (U/ml/108 CFU) |
|---|---|---|
| SSU | +++ | 163 ± 2.8b |
| SSU∆ahyRI | − | NAc |
| ATCC 7966T | − | 52 ± 4.0 |
| E1 | +++ | 115 ± 3.3 |
| E2 | +++ | 24 ± 4.5 |
Lactone production scored semiquantitatively: − (none), + (weak), ++ (moderate), or +++ (high).
The differences between hemolytic activity titers were statistically significant (P < 0.001), as determined pairwise by t test.
NA, hemolytic activity of strain SSU∆ahyRI was not measured in this study.
Strains E1, E2, and ATCC 7966T form less biofilm biomass than strain SSU does.
Biofilm formation represents a characteristic feature of persistent infections, and 30% of Aeromonas infections are associated with this virulence trait (22–24). To measure solid surface-associated biofilm formation by A. hydrophila strains E1 and E2, we performed a crystal violet (CV) staining assay of biofilms from cultures grown in polystyrene tubes at 37°C after overnight incubation. A. hydrophila E1 and E2 formed significantly less solid surface-associated biofilm in polystyrene tubes, as demonstrated by a more-than-3-fold decrease in CV staining, compared to that of the clinical isolate, SSU (see Fig. S5 in the supplemental material). The biofilms formed by strains E1 and E2 were comparable to that of the type strain, ATCC 7966T.
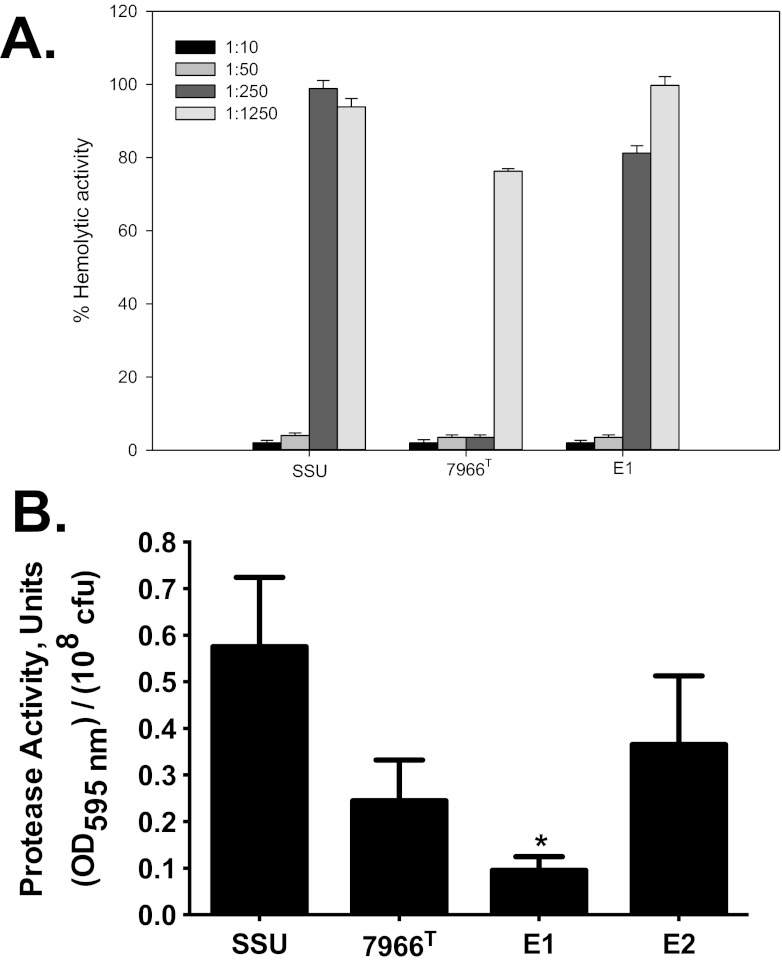
Hemolytic and proteolytic activities of A. hydrophila E1 and E2.
We observed that the hemoglobin release from rabbit red blood cells (RBCs) by the hemolysin(s) produced by strain E1 was comparable to that produced by strain SSU (163 ± 2.8 and 115 ± 3.3, respectively), while that produced by strain E2 (24 ± 4.5 U/ml/108 CFU) was much lower (Table 4), which was considered baseline activity. The hemolytic activity associated with strain ATCC 7966T was 52 ± 4.0 U/ml/108 CFU. To demonstrate that most of the hemolytic activity of strain E1 was associated with Act, we neutralized the toxin with specific antibodies that were serially diluted (5-fold). These antibodies abrogated the hemolytic activity associated with Act in the culture filtrates of strains E1, SSU, and ATCC 7966T in a dose-dependent fashion (Fig. 4A). Strain E2 does possess the hlyA gene, GCAT, and a number of other putative hemolysins (Table 3; see Table S1 in the supplemental material), which may be responsible for the low baseline hemolytic activity observed in Table 4. As noted in Table 3, strain E2 did not possess the Act-encoding gene. Previous studies have indicated that the pathogenic nature of A. hydrophila is, in part, associated with the production of exoenzymes, such as proteases and lipases (25, 26). Consequently, we measured protease production and noted that E1 produced a significantly lower level of protease activity than isolate SSU, while strain E2 had a production level of this enzyme comparable to that of SSU (Fig. 4B).
FIG 4 .
The Act-associated hemolytic activity neutralization assay and the protease activity of A. hydrophila strains E1 and E2 compared to that of A. hydrophila SSU and/or ATCC 7966T. (A) The neutralization of hemolytic activity associated with Act in the culture filtrates of A. hydrophila E1 and ATCC 7966T compared to A. hydrophila SSU. The culture filtrates of the strains studied were mixed with either preimmune rabbit serum (control) or 5-fold dilutions of hyperimmune rabbit serum (laboratory stock) containing antibodies to Act before the measurement of hemolytic activity. (B) Protease activity in the culture supernatants of A. hydrophila E1 and E2 compared to that of A. hydrophila SSU and ATCC 7966T. A. hydrophila E1 demonstrated a statistically significant decrease in protease activity compared to that of E2 and the two control strains, SSU and ATCC 7966T. The data were normalized to 1 × 108 CFU to account for any minor differences in the growth rates. All of the experiments were performed in triplicate, and the data presented are arithmetic means ± standard deviations. OD595, optical density at 595 nm.
Expression of T3SS and T6SS.
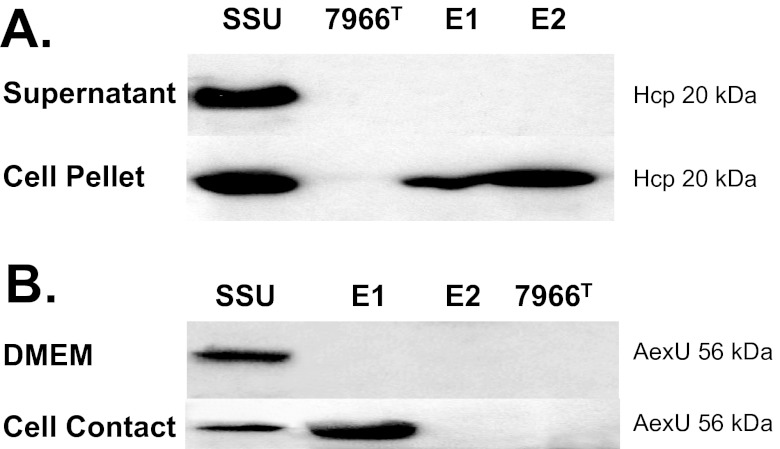
The secretion of hemolysin-coregulated protein (Hcp) has become a reliable indicator of functional T6SS in all bacteria with an intact T6SS structure, even though the gene that encodes Hcp is not always found in T6SS clusters (27). Among the Aeromonas strains analyzed in this study, Hcp production (cell pellet) and secretion (supernatant) were noted only in A. hydrophila strain SSU (Fig. 5A). While strain ATCC 7966T did not produce Hcp, strains E1 and E2 did synthesize Hcp, as it could be detected in the cell pellet, but were unable to secrete it into the medium (Fig. 5A). This raises the question of whether the T6SS is functional in strains E1 and E2.
FIG 5 .
Expression and production of T6SS-associated Hcp and T3SS effector AexU in A. hydrophila strains SSU, ATCC 7966T, E1, and E2. (A) While only SSU secretes Hcp into the supernatant, the Hcp protein is found in the cell pellet of strains E1 and E2. (B) Though SSU is the only strain to express and produce AexU when grown in DMEM, both SSU and E1 expressed and produced AexU when grown in contact with HeLa cells. As expected from the genomic analyses that demonstrated the absence of AexU from strains E2 and ATCC 7966T, neither of these strains produced AexU under either condition.
The AexU toxin is a T3SS effector identified in A. hydrophila SSU whose secretion can be indicative of a functional T3SS (28, 29). While the positive control, A. hydrophila SSU, expressed and produced AexU when grown in Dulbecco’s modified eagle medium (DMEM), strains ATCC 7966T, E1, and E2 were unable to express the gene encoding AexU (Fig. 5B). The results for strain E1 were surprising, given the fact that the genome of strain E1 harbored the T3SS gene cluster, as well as the small accessory aexU gene cluster (Table 3; see Table S1 in the supplemental material). We questioned whether bacterium-host contact might be needed for the expression of the aexU gene in strain E1, and indeed, interaction of E1 with HeLa cells led to the expression and production of AexU (Fig. 5B).
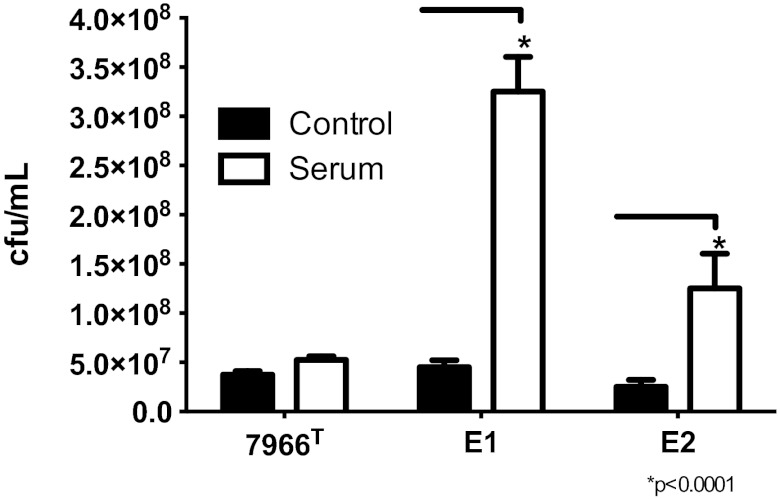
Serum resistance of strains E1 and E2.
The survival of strains E1 and E2 in the presence of naïve-mouse serum was compared with that of A. hydrophila ATCC 7966T (negative control). As expected, no difference in colony counts was noted when strain ATCC 7966T was incubated with either phosphate-buffered saline (PBS) or naive-mouse serum for 1 h (Fig. 6). In contrast, the colony counts obtained with both strains E1 and E2 after incubation with serum were significantly higher than the counts of samples from PBS, and this difference was greater for the E1 strain than for the E2 strain (Fig. 6). These data indicated that strains E1 and E2 were serum resistant, as demonstrated by continued growth during 1 h of incubation with serum.
FIG 6 .
Serum resistance of A. hydrophila E1 and E2 isolates compared to that of A. hydrophila ATCC 7966T. A. hydrophila 7966T, E1, and E2 were grown overnight, harvested, and resuspended in an equivalent amount of PBS. An aliquot (50 µl) of the bacterial cells was mixed with either PBS or serum and incubated at 37°C for 1 h. The CFU counts in the samples were then determined as described in Materials and Methods. A. hydrophila strains E1 and E2 demonstrated statistically significant increases in serum resistance compared to that of strain ATCC 7966T.
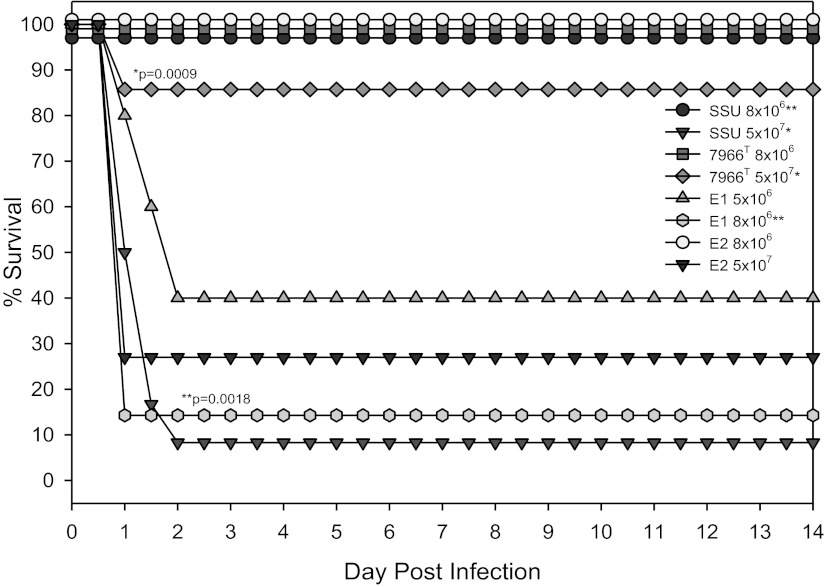
A. hydrophila E1 is highly virulent in an animal model.
To assess the overall virulence potential of A. hydrophila strains E1 and E2, compared to that of strains SSU and ATCC 7966T, we injected various doses of these bacterial isolates into mice via the intraperitoneal (i.p.) route. While a dose of 5 × 107 CFU of strain SSU killed more than 90% of the mice, ATCC 7966T killed <20% of the animals at this dose (Fig. 7). Strain E2 had moderate virulence, as it killed more than 70% of the mice at a dose of 5 × 107 CFU. All of the animals injected with strain E1 at a dose of 1 × 107 CFU died within 2 days, while minimal mortality was noted in mice injected with either strain SSU or E2 at this dose (data not shown). When animals were injected with smaller doses of strain E1, for example, 8 × 106, 5 × 106, or 3 × 106 CFU, 85, 60, and 20% (not shown) mortality, respectively, was observed (Fig. 7). These data indicated that strain E1 was much more lethal than the other A. hydrophila strains tested in this mouse model. For example, strain E1 was approximately 10 times more virulent than strain SSU.
FIG 7 .
Kaplan-Meier survival curves of mice injected with A. hydrophila strains SSU, ATCC 7966T, E1, and E2. Groups of nine Swiss-Webster mice were injected with doses of 5 × 107, 1 × 107, 8 × 106, 5 × 106, and 3 × 106 CFU by the i.p. route. The animals were observed for death over a period of 14 days. The data were statistically analyzed by using a Kaplan-Meier survival estimate. Single and double asterisks indicate the groups compared for statistically significant differences.
DISCUSSION
This is the first genomic analysis of a virulent clinical A. hydrophila isolate, namely, E1. By analyzing the genome and phenotypic characteristics of E1 alongside those of a less virulent clinical isolate and several additional strains of Aeromonas, we have identified key differences that distinguish this disease-causing strain from the ubiquitous environmental isolates common to freshwaters. Among the features that distinguished E1 from the other aeromonads in this study were capsular polysaccharide, enhanced bacterial motility, a functional T3SS, and the presence of Act. Our genomic and phenotypic findings regarding each of these features have important implications for virulence in Aeromonas.
Antigen presentation can be a critical determinant of bacterial pathogenicity. While strain ATCC 7966T is defined as an O:1 serotype, we could deduce from the genomic sequences of the O-antigen and group II capsular polysaccharide genes that strain E2 was an isolate of either the O:18 serogroup (30) or a very closely related one. The serotype of strain E1 was unclear, but it was not O:1, O:18, or O:34, for which we have sequence information for comparison. From genome annotation, we could also deduce that strains E1 and E2 were encapsulated, while the capsular polysaccharide gene cluster was absent from the genome of environmental isolate ATCC 7966T, diarrhea isolate Ae398, and fish isolate A449; however, A. salmonicida isolates typically harbor analogous A(S)-layer protein-encoding genes. From these genomic data and other experimental findings, it is clear that capsule production imparts serum resistance to the bacteria (30). Indeed, the higher level of A. hydrophila strain E1 and E2 resistance to naive-mouse serum than that of ATCC 7966T could also be related to the presence of capsular polysaccharide in the wound isolates. Therefore, we conclude that extracellular layers such as the capsule are important for the survival of bacteria in wounds.
Bacterial motility, enabled by lateral or polar flagella, may facilitate the invasion of human and nonhuman host cellular barriers (31, 32). Our study found that the lateral flagellum in the genome of A. hydrophila E1 was homologous to that found in A. salmonicida A449 (33) and enabled swarming motility (34). Surprisingly, strain E2 mimicked swarming motility under certain assay conditions although it lacks genes for lateral flagella. While electron microscopy revealed lateral flagella on strains E1 and SSU, E2 lacked flagella and exhibited a rafting or “bridging” multicellular behavior that may be driven by LPS, capsule, or cell surface-associated factors such as type IV pili (35). E2’s phenotype may also be explained by “sliding” motility, in which bacteria synthesize and secrete surfactants that allow them to spread over surfaces (34). In all, we found flagellum-enabled “true” motility in Aeromonas strains with enhanced virulence but only rafting or sliding motility in less virulent strains such as E2 and ATCC 7966T.
The ability to form biofilms, multicellular sessile communities (22), was not a distinguishing feature of A. hydrophila E1. Biofilms can facilitate wound chronicity and persistence by creating a barrier against neutrophils, macrophages, and antimicrobials (36). We recently demonstrated that the effect of cyclic di-GMP (c-di-GMP) on motility and biofilm formation in A. hydrophila SSU was dependent on the coexpression of three QS systems, AI-1, AI-2, and AI-3 (37–39). Intriguingly, the genomes of E1 and E2 harbored numerous genes that encode proteins with GGDEF and EAL domains (involved in the synthesis and degradation of c-di-GMP, respectively), as well as N-acyl-homoserine AI-1, S-ribosylhomocysteinase (LuxS)-based AI-2, and QseBC-based AI-3 QS systems. In addition, we examined AHL production, since AHL-mediated QS has been shown to regulate exoprotease production and biofilm formation in A. hydrophila (10, 37–41). While AHL-deficient knockout mutants demonstrated attenuated virulence in a mouse model (10), the role of AHL-mediated QS in wound infection isolates of A. hydrophila such as E1 and E2 has never been tested before. This study demonstrated AHL production by strains E1 and E2, corroborating the presence of an AI-1 QS system. Taken together, these findings indicate that strains E1 and E2 produce less biofilm biomass or that biofilms from these isolates take longer to mature. Aside from regulatory defects unrelated to QS, one possible explanation is that the presence and expression of capsular polysaccharide genes may have a negative effect on biofilm formation rates, as has been demonstrated in various Vibrio species (42, 43). Future studies should include extension of the assay time and the use of static culture for biofilm formation, as well as the examination of other regulatory factors to better understand the role of Aeromonas biofilms in virulence.
In contrast, the ability to form and secrete extracellular toxins appears to be a distinguishing feature of A. hydrophila E1. For example, Act is a potent virulence factor secreted via the T2SS that functions as a hemolysin, a cytotoxin, or an enterotoxin, depending upon the target cells (12, 25, 44–46). In this study, we demonstrated that the significant hemolytic activity of A. hydrophila E1 was due primarily to Act. In contrast, the genome of A. hydrophila strain E2 did not harbor the act gene and it exhibited baseline hemolytic activity. While both strains E1 and E2 exhibited a functional T6SS, as evidenced by the presence of Hcp in the bacterial pellet, no Hcp was found in the supernatant of strain E1 or E2. Therefore, future studies may desire to determine whether E1 and E2 can translocate Hcp into host cells, since the evasion of host innate immunity by strain SSU has been attributed to the secretion of this effector (47). The presence of T3SSs is more strongly correlated with pathogenicity because of the presence of this factor in the genomes of known pathogens, such as Yersinia pestis, Salmonella enterica, Vibrio parahaemolyticus, and Shigella species (48–50). We found T3SS genes only in the genomes of A. hydrophila strain E1 and A. salmonicida A449 and demonstrated the functionality of strain E1’s T3SS by detecting AexU in the presence of host cells. As AexU inhibits macrophage phagocytosis of A. hydrophila SSU and leads to host cell apoptosis (29, 51), it is likely that the T3SS played a role in the virulence of strain E1
Extracellular proteases have also been suggested as a virulence factor of aeromonads (52), and both temperature-labile serine proteases and temperature-stable metalloproteases have been characterized in A. hydrophila (53). In experimental animal models, protease-null mutants of A. hydrophila and A. salmonicida exhibit less virulence than wild-type bacteria (54, 55), and a role for protease in Aeromonas-associated tissue damage has been reported (55). Recently, we showed that the AI-1 QS system positively modulates metalloprotease activity in A. hydrophila SSU (10). Interestingly, we observed a lower level of protease activity in the culture filtrate of A. hydrophila strain E1 than in that of strains SSU and E2. These results raise the possibility that, in contrast to previous studies (25, 26), protease activity and virulence may be inversely correlated in the Aeromonas strains studied—a topic that warrants further study with specific gene knockouts. There were approximately 20 genes annotated as proteases or proteinases in the genome of A. hydrophila strains ATCC 7966T and E2. Only one of these genes was missing from strain E1, specifically, that for a PfpI-like protease. However, it is quite likely that this defect may not be explained by differences in gene content.
When initially isolated, strain E2 exhibited resistance to several aminoglycoside antibiotics, namely, amikacin, tobramycin, and gentamicin, as well as several cephalosporins (8). During the annotation of the genome sequences, we did not find any unique aminoglycoside-modifying enzymes in the genome of strain E2, compared to that of strain E1. Furthermore, upon resuscitation from cold storage, in preparation for whole-genome sequencing, strain E2 was sensitive to the above antimicrobials. Therefore, the loss of a plasmid harboring antibiotic resistance genes is a possible explanation. Previous studies have demonstrated plasmid-borne antimicrobial resistance in Aeromonas; for example, plasmid pAsa4 of A. salmonicida A449, contains a streptomycin/spectinomycin O-adenyltransferase-encoding gene (33). Furthermore, Aeromonas strains have been shown to lose plasmids under stressful conditions (56, 57). Alternatively, initial resistance to several aminoglycosides could be explained by adaptive resistance via increased cell impermeability in this strain, which was induced in the clinical setting (58).
Through comparative genomic and functional analyses of wound isolates along with other clinical and environmental strains of Aeromonas, we not only demonstrated that strain E1 was more virulent than strains E2 and ATCC 7966T in a mouse model of infection but also showed the presence of several functional virulence factors that may be related to its enhanced pathogenicity. Previously, Daily et al. (59) and Joseph and Carnahan (60) hypothesized the possibility of subsets of virulent aeromonads within and between the eight species most frequently associated with disease. The findings of the present study provide the foundation for the establishment of distinct pathotypes within the genus Aeromonas. Although strain E1 produced less protease and biofilm than diarrhea isolate SSU did, it was much more virulent in a mouse model of infection, possibly because it harbored a capsule, flagella, a functional T3SS, and highly hemolytic Act. Furthermore, we demonstrated the ability to detect these virulence-associated factors through genome sequencing and annotation. More extensive genomic and epidemiological investigations will allow us to ascertain the frequency of core and accessory genes in environmental and clinical Aeromonas isolates. In the future, we can aim to detect novel virulence factors through genomic surveillance and estimate the virulence potential of clinical isolates through sequence analysis alone.
MATERIALS AND METHODS
Bacterial strains.
Freezer stocks of A. hydrophila strains E1 and E2, corresponding to Aero 1 and Aero 2 of Shak et al. (8), were streaked and subcultured on Trypticase soy agar plates with 5% sheep blood agar (SBA; BDMS, Sparks, MD). While E1 colonies were flat and grayish in color with a smooth surface, colonies of E2 were raised and whitish yellow in color with a visible mucoidal exterior, suggestive of the presence of an extracellular capsule. Strains E1 and E2 were tested for seven phenotypic traits by using the Aerokey II dichotomous key as previously described (61) and were identified as A. hydrophila subsp. hydrophila. Unless otherwise specified, A. hydrophila here refers to A. hydrophila subsp. hydrophila. For reference, additional strains of A. hydrophila, ATCC 7966T, SSU, and SSU∆ahyRI (10, 62), were included in phenotypic experiments.
Genome sequencing and annotation.
Genome sequencing of A. hydrophila strains E1 and E2 was performed at the Emory Genome Center with a GS Junior pyrosequencer (454 Life Sequencing, Branford, CT). The numbers of reads were 130,920 for strain E1 and 130,449 for strain E2, and the average read lengths were 405 and 338 bp, respectively. The estimated average coverages of the E1 and E2 genomes were 11-fold and 9-fold, respectively. Contigs were assembled by using Newbler (63) and uploaded as multiple-sequence FASTA files to RAST for annotation (64). Further analyses to identify shared and dispensable genetic traits were performed by using the closed genomes of A. hydrophila ATCC 7966T (GenBank accession no. CP000462.1) (17) and A. salmonicida A449 (GenBank BioProject PRJNA58631) (33), as well the draft genomes of A. aquariorum AAK1 (GenBank accession no. BAFL00000000.1) (65), A. veronii B565 (GenBank accession no. CP002607.1) (66), and A. caviae Ae398 (GenBank accession no. WGS CACP00000000) (67). To facilitate comparisons, the genomes of the other five Aeromonas species were also annotated by using the RAST server with small hypothetical protein-encoding genes accepted from the initial RAST annotation only if they were consistently annotated among the majority of the genomes analyzed. Genomic regions and mobile genetic elements were mapped to the syntenic core on the basis of the homology of conserved flanking genes or sequences.
Comparative genomic analyses.
The syntenic core genome of A. hydrophila was determined by using the SEED viewer comparative genomic feature (68). To ensure the most accurate syntenic core gene set, the closed genome of A. hydrophila ATCC 7966T (GenBank accession no. CP000462.1) was used to “map” contigs from draft genomes. For the draft genomes of strains E1 and E2, genes at the end of a contig or interrupted by contig gaps were analyzed by using bidirectional BLASTN analysis against all other genomes. ANI by BLAST was computed with Jspecies (69). Evolutionary analyses were conducted in MEGA5 (70). Comparison of the T3SS gene cluster was performed with the Artemis comparison tool (71).
Motility assays.
Luria-Bertani (LB) medium with 0.35% Bacto agar (Difco Laboratories, Detroit, MI) was used to characterize swimming motility, while Difco nutrient broth with 0.5% Eiken agar (Eiken Chemical Co., Ltd., Tokyo, Japan) was used to measure the swarming motility of A. hydrophila strains. To evaluate swimming motility, the overnight cultures were adjusted to the same optical density and equal numbers (108) of CFU were stabbed onto 0.35% LB agar plates. The plates were incubated at 37°C overnight, and motility was measured by examining the migration of bacteria through the agar from the center toward the periphery of the plate (62). Growth from the edge of the swimming zone within the agar of these plates provided the inocula for the swarming assay. The swarming plates were inoculated by streaking bacteria onto the surface of the agar (19, 72) and then incubated at 30°C overnight. For swarming motility, an alternate protocol was also used (J. G. Shaw, personal communication). Briefly, inocula from cryogenically frozen cultures were subcultured onto SBA plates, which were incubated overnight at 30°C. Growth from SBA was used to inoculate the surfaces of swarming plates as described above or to inoculate an alternative formulation containing 0.8% (wt/vol) Bacto agar; 0.5% (wt/vol) glucose; 1.0% (wt/vol) tryptone; 0.5% (wt/vol) NaCl; and 0.002% (vol/vol) Tween 80 (J. G. Shaw, personal communication). Inoculated swarming plates were then incubated at 30°C and examined after 8, 16, and 24 h of growth for the swarming phenotype.
Electron microscopy.
Cell suspensions of cultures grown overnight at 30°C on swarming plates were added to 200 µl of 0.5% sodium phosphotungstate (pH 6.8). Subsequently, 15 µl of the sample was applied to the surface of a 300-mesh, carbon-coated, Formvar-coated copper grid. Excess stain was removed, and the grids were air dried. A JEOL 1011 transmission electron microscope (JEOL United States, Inc., Peabody, MA) operating at an accelerating voltage of 80 kV was used to examine the bacterial cells for the presence of lateral flagella.
Production of AHLs.
AHL production was detected by cross-streaking A. hydrophila strains against the biosensor strain C. violaceum CV026 on LB agar plates. Positive assays were judged by the degree of induction of the purple violacein pigment in the biosensor strain (73). Pigment production by C. violaceum CV026 was scored on the basis of the intensity of the color after overnight incubation of the plates at 30°C. Lactone production was scored semiquantitatively as follows: −, no lactone production; +, weak; ++, moderate; +++, high. An isogenic ∆ahyRI mutant of A. hydrophila SSU was used as a negative control, as this mutant does not produce lactones and, hence, no violacein is induced in the biosensor strain (62).
Biofilm formation assays.
In a modification of the biofilm ring assay (74), strains of A. hydrophila were transferred from fresh LB agar plates into 3 ml of LB medium contained in polystyrene tubes at 37°C overnight with shaking. Biofilm formation was quantified according to procedures previously described (75). Biofilm formation results were normalized to 1 × 109 CFU to account for any minor differences in the growth rates of the various bacterial strains used.
Measurement of hemolytic activity.
To examine the lysis of RBCs, culture filtrates from A. hydrophila strains grown for 18 h in LB medium at 37°C with shaking (180 rpm) were treated with trypsin (final concentration, 0.05%; to activate Act or other hemolysins) (76, 77) at 37°C for 1 h and then subjected to a hemolytic assay as described previously (12). The number of hemolytic units per milliliter of cell filtrate per 1 × 108 CFU was reported (62). For the neutralization assay, culture filtrates of the strains studied were mixed and incubated for 1 h at 37°C with either preimmune (control) or serially diluted (5-fold) hyperimmune rabbit serum (laboratory stock) containing antibodies to Act before the measurement of hemolytic activity (10).
Measurement of protease activity.
Protease activity was measured in filtrates of A. hydrophila cultures grown overnight as described previously (78). LB medium was inoculated with fresh growth from LB agar plates and incubated overnight at 37°C. Protease activity was calculated per milliliter of culture filtrate per 108 CFU. Hide Powder Azure (Calbiochem, La Jolla, CA) was used as the substrate to measure protease activity because of the sensitivity and rapidity of the assay. The substrate incubated with PBS alone served as a negative control.
Expression and production of AexU.
To examine the expression and production of AexU, cultures of A. hydrophila isolates grown overnight were either reinoculated (1 × 108 CFU) into 2 ml of DMEM with 4 mM glutamine and incubated at 37°C for 4 h in the CO2 incubator or used directly to infect HeLa cells at a multiplicity of infection of 10 for 4 h in a six-well plate. The bacterial cells were then harvested from either the DMEM or HeLa cell culture supernatants by centrifugation and subsequently lysed in SDS-PAGE loading buffer for Western blot analysis with anti-AexU antibodies as previously described (51).
Expression and secretion of Hcp.
Western blot analysis was used to detect T6SS effector protein Hcp expression in and secretion from A. hydrophila isolates. Briefly, the supernatants and cell pellets from overnight broth cultures of Aeromonas strains were separated by centrifugation. The cell pellets were directly lysed in SDS-PAGE loading buffer, while the proteins in the supernatants were first precipitated with 10% trichloroacetic acid and then dissolved in the loading buffer. The samples of supernatants and pellets were subjected to SDS-PAGE, followed by Western blot analysis with specific antibodies to Hcp, according to the procedure described previously (47).
Serum resistance assay.
Pooled sera from naive mice were used in the serum resistance assay. Briefly, overnight A. hydrophila ATCC 7966T, E1, and E2 cultures were harvested and diluted in PBS to an optical density at 600 nm of 0.2 (~1 × 108 CFU/ml). Next, 50 µl of the diluted bacteria (~5 × 106 CFU) was mixed with 200 µl of normal mouse serum or PBS. The samples were incubated at 37°C for 1 h. The number of CFU of surviving bacteria in each sample was determined by serial dilutions and plating on LB plates.
Animal virulence model.
Groups of nine female Swiss Webster mice (Taconic Farms) were injected via the i.p. route with A. hydrophila strains SSU, ATCC 7966T, E1, and E2 in accordance with an approved Institutional Animal Care and Use Committee protocol. The animals were injected with doses of 5 × 107, 1 × 107, and 8 × 106 CFU of all of the strains. Additional animals were also injected at lower doses of 5 × 106 and 3 × 106 CFU of strain E1. Deaths were recorded daily for 14 days postinfection.
Statistical analyses.
All in vitro experiments were performed in triplicate, and differences were analyzed for significance by one-way analysis of variance (ANOVA). The animal data were analyzed by using a Kaplan-Meier survival estimate, and P values of ≤0.05 were considered significant.
Nucleotide sequence accession numbers.
The genome sequences determined in this study were deposited at NCBI under accession numbers SRA063950 (A. hydrophila strain E1) and SRA063951 (A. hydrophila strain E2).
SUPPLEMENTAL MATERIAL
Bidirectional BLASTP heat map of Aeromonas species genomes with that of A. hydrophila ATCC 7966T as the reference for comparison. The comparison genomes included, from the outermost to the innermost, those of A. hydrophila E1, A. hydrophila E2, A. aquariorum AAK1, A. caviae Ae398 A. salmonicida A449, and A. veronii B565. The percent translated protein sequence identity for each gene is color coded according to the scale shown. Regions in white represent genes that are present in the reference genome (that of strain ATCC 7966T) and absent from the others. Download
Comparative genomics of genes that encode T3SSs in six Aeromonas strains. Entire T3SS gene clusters of strains E1 and E2 and GenBank sequences of the T3SSs of A. hydrophila SSU (GenBank accession no. AY763611), A. hydrophila AH3 (GenBank accession no. AY528667), A. veronii biovar Sobria HM21 (GenBank accession no. EF215451), and A. salmonicida A449 (GenBank accession no. CP000646) were compared by using NCBI BLASTn, and the results were visualized by using the Artemis comparison tool (Wellcome Trust Sanger Institute). Regions in red are homologous and located on the same DNA strand. Regions in white indicate regions of divergence. Open reading frames of A. salmonicida A449 are shown at the bottom. Download
Phylogenetic relationship among Aeromonas species genes that encode T3SSs (A) and genes that encode lateral flagella (B). T3SS sequences were retrieved from GenBank for A. hydrophila AH1 (GenBank accession no. AY394563), A. hydrophila SSU (GenBank accession no. AY763611), A. hydrophila AH3 (GenBank accession no. AY528667), A. veronii biovar Sobria HM21 (GenBank accession no. EF215451), A. salmonicida JF2267 (GenBank accession no. AJ616218), A. salmonicida A449 (GenBank accession no. CP000646), and P. aeruginosa PAO1 (GenBank accession no. AE004091). Lateral flagellum sequences were retrieved from GenBank for A. salmonicida A449 (GenBank accession no. CP000644) and A. hydrophila AH3 (GenBank accession no. DQ124694). The evolutionary history was inferred by the neighbor-joining method. The bootstrap consensus tree was inferred from 1,000 replicates. The evolutionary distances were computed by the maximum composite likelihood method, and each value is the number of base substitutions per site. Download
Transmission electron photomicrographs of three A. hydrophila strains. A, A. hydrophila E1; B, A. hydrophila E2; C, A. hydrophila SSU. Staining was done with 0.5% phosphotungstic acid. A. hydrophila E1 and SSU display lateral flagella, while strain E2 exhibits rafting-like behavior. Download
A. hydrophila E1, E2, and ATCC 7966T form significantly less biofilm biomass on polystyrene than A. hydrophila SSU does. Biofilms were quantified by CV staining after 24 h of incubation at 37°C. The results presented represent the mean and standard deviation of three independent experiments. An asterisk indicates a P value of less than 0.001 as determined by one-way ANOVA. Download
Distribution of A. hydrophila genes from the genomes of strains E1, E2, and ATCC 7966T. A comparative genomic analysis of the three A. hydrophila genomes was performed, and the syntenic pangenome is presented as mapped to the closed genome of ATCC 7966T, with start and stop positions, lengths of genes, and putative functions. For core genome homologues in strains E1 and E2, BLASTP scores are shown, in relation to the ATCC 7966T allele. For genes at ends of contigs and interrupted by contig gaps, bidirectional BLASTN was performed by using the gene sequences from the other strains to confirm the presence and inclusion in or exclusion from the core gene set, and these values are underlined. For dispensable genes not found in ATCC 7966T, strain E1 was used as the reference for BLAST analysis. A small subset of unique genes from strains E1 and E2 could not be mapped to the syntenic core genome and are labeled as orphan genes. Certain traits are highlighted for importance.
ACKNOWLEDGMENTS
J.R.S. acknowledges the financial support of the Molecules to Mankind Program and the Medical Scientist Training Program at Emory University. A.K.C. acknowledges funds from his Dr. Leon Bromberg Professorship, University of Texas Medical Branch, to conduct portions of these studies. C.J.G. acknowledges financial support from Oak Ridge Associated Universities.
454 Life Sequencing supplied the GS Junior sequencer and reagents free of charge for testing purposes and had no role in study design, data analysis, manuscript preparation, or the decision to publish.
We thank Bruce Ribner and Jennifer Whitaker for their clinical insights into the original case.
Footnotes
Citation Grim CJ, Kozlova EV, Sha J, Fitts EC, van Lier CJ, Kirtley ML, Joseph SJ, Read TD, Burd EM, Tall BD, Joseph SW, Horneman AJ, Chopra AK, Shak JR. 2013. Characterization of Aeromonas hydrophila wound pathotypes by comparative genomic and functional analyses of virulence genes. mBio 4(2):e00064-13. doi:10.1128/mBio.00064-13.
REFERENCES
- 1. Horneman AJ, Ali A. 2011. Aeromonas, p 658-665 In Versalovic J, Manual of clinical microbiology, 10th ed. ASM Press, Washington, DC [Google Scholar]
- 2. Janda JM, Abbott SL. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23:35–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parker JL, Shaw JG. 2011. Aeromonas spp. Clinical microbiology and disease. J. Infect. 62:109–118 [DOI] [PubMed] [Google Scholar]
- 4. Huang KF, Hung MH, Lin YS, Lu CL, Liu C, Chen CC, Lee YH. 2011. Independent predictors of mortality for necrotizing fasciitis: a retrospective analysis in a single institution. J. Trauma 71:467–473 [DOI] [PubMed] [Google Scholar]
- 5. Roberts MT, Enoch DA, Harris KA, Karas JA. 2006. Aeromonas veronii biovar sobria bacteraemia with septic arthritis confirmed by 16S rDNA PCR in an immunocompetent adult. J. Med. Microbiol. 55:241–243 [DOI] [PubMed] [Google Scholar]
- 6. Abuhammour W, Hasan RA, Rogers D. 2006. Necrotizing fasciitis caused by Aeromonas hydrophilia [sic] in an immunocompetent child. Pediatr. Emerg. Care 22:48–51 [DOI] [PubMed] [Google Scholar]
- 7. Dwivedi M, Mishra A, Prasad A, Azim A, Singh RK, Baronia AK, Prasad KN, Dwivedi UN. 2008. Aeromonas caviae septicemia in immunocompetent gastrointestinal carriers. Braz. J. Infect. Dis. 12:547–548 [DOI] [PubMed] [Google Scholar]
- 8. Shak JR, Whitaker JA, Ribner BS, Burd EM. 2011. Aminoglycoside-resistant Aeromonas hydrophila as part of a polymicrobial infection following a traumatic fall into freshwater. J. Clin. Microbiol. 49:1169–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chopra AK, Houston CW, Peterson JW, Jin GF. 1993. Cloning, expression, and sequence analysis of a cytolytic enterotoxin gene from Aeromonas hydrophila. Can. J. Microbiol. 39:513–523 [DOI] [PubMed] [Google Scholar]
- 10. Khajanchi BK, Sha J, Kozlova EV, Erova TE, Suarez G, Sierra JC, Popov VL, Horneman AJ, Chopra AK. 2009. N-acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology 155:3518–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zacaria J, Delamare AP, Costa SO, Echeverrigaray S. 2010. Diversity of extracellular proteases among Aeromonas determined by zymogram analysis. J. Appl. Microbiol. 109:212–219 [DOI] [PubMed] [Google Scholar]
- 12. Sha J, Kozlova EV, Chopra AK. 2002. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70:1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu HB, Rao PS, Lee HC, Vilches S, Merino S, Tomas JM, Leung KY. 2004. A type III secretion system is required for Aeromonas hydrophila AH-1 pathogenesis. Infect. Immun. 72:1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sen K, Lye D. 2007. Importance of flagella and enterotoxins for Aeromonas virulence in a mouse model. Can. J. Microbiol. 53:261–269 [DOI] [PubMed] [Google Scholar]
- 15. Joseph SW, Daily OP, Hunt WS, Seidler RJ, Allen DA, Colwell RR. 1979. Aeromonas primary wound infection of a diver in polluted waters. J. Clin. Microbiol. 10:46–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Joseph SW, Carnahan AM, Brayton PR, Fanning GR, Almazan R, Drabick C, Trudo EW, Jr, Colwell RR. 1991. Aeromonas jandaei and Aeromonas veronii dual infection of a human wound following aquatic exposure. J. Clin. Microbiol. 29:565–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz MJ, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine OC, Ali A, Horneman AJ, Heidelberg JF. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: Jack of all trades. J. Bacteriol. 188:8272–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morgan DR, Johnson PC, DuPont HL, Satterwhite TK, Wood LV. 1985. Lack of correlation between known virulence properties of Aeromonas hydrophila and enteropathogenicity for humans. Infect. Immun. 50:62–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kirov SM, Tassell BC, Semmler AB, O’Donovan LA, Rabaan AA, Shaw JG. 2002. Lateral flagella and swarming motility in Aeromonas species. J. Bacteriol. 184:547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart GS, Williams P. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703–3711 [DOI] [PubMed] [Google Scholar]
- 21. Jangid K, Kong R, Patole MS, Shouche YS. 2007. luxRI homologs are universally present in the genus Aeromonas. BMC Microbiol. 7:93 http://dx.doi.10.1186/1471-2180-7-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 23. Gracey M, Burke V, Robinson J. 1982. Aeromonas-associated gastroenteritis. Lancet ii:1304–1306 [DOI] [PubMed] [Google Scholar]
- 24. Rautelin H, Hänninen ML, Sivonen A, Turunen U, Valtonen V. 1995. Chronic diarrhea due to a single strain of Aeromonas caviae. Eur. J. Clin. Microbiol. Infect. Dis. 14:51–53 [DOI] [PubMed] [Google Scholar]
- 25. Chopra AK, Houston CW. 1999. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1:1129–1137 [DOI] [PubMed] [Google Scholar]
- 26. Janda JM, Abbott SL. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332–344 [DOI] [PubMed] [Google Scholar]
- 27. Pukatzki S, McAuley SB, Miyata ST. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12:11–17 [DOI] [PubMed] [Google Scholar]
- 28. Sierra JC, Suarez G, Sha J, Baze WB, Foltz SM, Chopra AK. 2010. Unraveling the mechanism of action of a new type III secretion system effector AexU from Aeromonas hydrophila. Microb. Pathog. 49:122–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sierra JC, Suarez G, Sha J, Foltz SM, Popov VL, Galindo CL, Garner HR, Chopra AK. 2007. Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila—part II. Microb. Pathog. 43:147–160 [DOI] [PubMed] [Google Scholar]
- 30. Zhang YL, Arakawa E, Leung KY. 2002. Novel Aeromonas hydrophila PPD134/91 genes involved in O-antigen and capsule biosynthesis. Infect. Immun. 70:2326–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tomich M, Herfst CA, Golden JW, Mohr CD. 2002. Role of flagella in host cell invasion by Burkholderia cepacia. Infect. Immun. 70:1799–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Merino S, Rubires X, Aguilar A, Tomás JM. 1997. The role of flagella and motility in the adherence and invasion to fish cell lines by Aeromonas hydrophila serogroup O:34 strains. FEMS Microbiol. Lett. 151:213–217 [DOI] [PubMed] [Google Scholar]
- 33. Reith ME, Singh RK, Curtis B, Boyd JM, Bouevitch A, Kimball J, Munholland J, Murphy C, Sarty D, Williams J, Nash JH, Johnson SC, Brown LL. 2008. The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9:427 http://dx.doi.10.1186/1471-2164-9-427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kearns DB. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8:634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mattick JS. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289–314 [DOI] [PubMed] [Google Scholar]
- 36. Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. 2011. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 186:6585–6596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kozlova EV, Khajanchi BK, Popov VL, Wen J, Chopra AK. 2012. Impact of QseBC system in c-di-GMP-dependent quorum sensing regulatory network in a clinical isolate SSU of Aeromonas hydrophila. Microb. Pathog. 53:115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kozlova EV, Khajanchi BK, Sha J, Chopra AK. 2011. Quorum sensing and c-di-GMP-dependent alterations in gene transcripts and virulence-associated phenotypes in a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 50:213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozlova EV, Popov VL, Sha J, Foltz SM, Erova TE, Agar SL, Horneman AJ, Chopra AK. 2008. Mutation in the S-ribosylhomocysteinase (luxS) gene involved in quorum sensing affects biofilm formation and virulence in a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 45:343–354 [DOI] [PubMed] [Google Scholar]
- 40. Swift S, Lynch MJ, Fish L, Kirke DF, Tomás JM, Stewart GS, Williams P. 1999. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect. Immun. 67:5192–5199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lynch MJ, Swift S, Kirke DF, Keevil CW, Dodd CE, Williams P. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18–28 [DOI] [PubMed] [Google Scholar]
- 42. Joseph LA, Wright AC. 2004. Expression of Vibrio vulnificus capsular polysaccharide inhibits biofilm formation. J. Bacteriol. 186:889–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Enos-Berlage JL, Guvener ZT, Keenan CE, McCarter LL. 2005. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 55:1160–1182 [DOI] [PubMed] [Google Scholar]
- 44. Chopra AK, Xu X, Ribardo D, Gonzalez M, Kuhl K, Peterson JW, Houston CW. 2000. The cytotoxic enterotoxin of Aeromonas hydrophila induces proinflammatory cytokine production and activates arachidonic acid metabolism in macrophages. Infect. Immun. 68:2808–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Galindo CL, Fadl AA, Sha J, Gutierrez C, Jr., Popov VL, Boldogh I, Aggarwal BB, Chopra AK. 2004. Aeromonas hydrophila cytotoxic enterotoxin activates mitogen-activated protein kinases and induces apoptosis in murine macrophages and human intestinal epithelial cells. J. Biol. Chem. 279:37597–37612 [DOI] [PubMed] [Google Scholar]
- 46. Ferguson MR, Xu XJ, Houston CW, Peterson JW, Coppenhaver DH, Popov VL, Chopra AK. 1997. Hyperproduction, purification, and mechanism of action of the cytotoxic enterotoxin produced by Aeromonas hydrophila. Infect. Immun. 65:4299–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. 2010. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192:155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ly KT, Casanova JE. 2007. Mechanisms of Salmonella entry into host cells. Cell. Microbiol. 9:2103–2111 [DOI] [PubMed] [Google Scholar]
- 49. Burdette DL, Yarbrough ML, Orvedahl A, Gilpin CJ, Orth K. 2008. Vibrio parahaemolyticus orchestrates a multifaceted host cell infection by induction of autophagy, cell rounding, and then cell lysis. Proc. Natl. Acad. Sci. U. S. A. 105:12497–12502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Konradt C, Frigimelica E, Nothelfer K, Puhar A, Salgado-Pabon W, di Bartolo V, Scott-Algara D, Rodrigues CD, Sansonetti PJ, Phalipon A. 2011. The Shigella flexneri type three secretion system effector IpgD inhibits T cell migration by manipulating host phosphoinositide metabolism. Cell Host Microbe 9:263–272 [DOI] [PubMed] [Google Scholar]
- 51. Sha J, Wang SF, Suarez G, Sierra JC, Fadl AA, Erova TE, Foltz SM, Khajanchi BK, Silver A, Graf J, Schein CH, Chopra AK. 2007. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila—part I. Microb. Pathog. 43:127–146 [DOI] [PubMed] [Google Scholar]
- 52. Ljungh A, Wadström T. 1981. Aeromonas toxins. Pharmacol. Ther. 15:339–354 [DOI] [PubMed] [Google Scholar]
- 53. Nieto TP, Ellis AE. 1986. Characterization of extracellular metallo- and serine-proteases of Aeromonas hydrophila strain B51. J. Gen. Microbiol. 132:1975–1979 [DOI] [PubMed] [Google Scholar]
- 54. Leung KY, Stevenson RM. 1988. Tn5-induced protease-deficient strains of Aeromonas hydrophila with reduced virulence for fish. Infect. Immun. 56:2639–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sakai DK. 1985. Loss of virulence in a protease-deficient mutant of Aeromonas salmonicida. Infect. Immun. 48:146–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Daher RK, Filion G, Tan SG, Dallaire-Dufresne S, Paquet VE, Charette SJ. 2011. Alteration of virulence factors and rearrangement of pAsa5 plasmid caused by the growth of Aeromonas salmonicida in stressful conditions. Vet. Microbiol. 152:353–360 [DOI] [PubMed] [Google Scholar]
- 57. Tanaka KH, Dallaire-Dufresne S, Daher RK, Frenette M, Charette SJ. 2012. An insertion sequence-dependent plasmid rearrangement in Aeromonas salmonicida causes the loss of the type three secretion system. PLoS One 7:e33725 http://dx.doi.10.1371/journal.pone.0033725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Poole K. 2012. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 67:2069–2089 [DOI] [PubMed] [Google Scholar]
- 59. Daily OP, Joseph SW, Coolbaugh JC, Walker RI, Merrell BR, Rollins DM, Seidler RJ, Colwell RR, Lissner CR. 1981. Association of Aeromonas sobria with human infection. J. Clin. Microbiol. 13:769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Joseph SW, Carnahan AM. 2000. Update on the genus Aeromonas. ASM News 66:218–223 [Google Scholar]
- 61. Carnahan AM, Behram S, Joseph SW. 1991. Aerokey II: a flexible key for identifying clinical Aeromonas species. J. Clin. Microbiol. 29:2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Khajanchi BK, Kozlova EV, Sha J, Popov VL, Chopra AK. 2012. The two-component QseBC signalling system regulates in vitro and in vivo virulence of Aeromonas hydrophila. Microbiology 158:259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75 http://dx.doi.10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu CJ, Wang HC, Chen CS, Shu HY, Kao AW, Chen PL, Ko WC. 2012. Genome sequence of a novel human pathogen, Aeromonas aquariorum. J. Bacteriol. 194:4114–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li Y, Liu Y, Zhou Z, Huang H, Ren Y, Zhang Y, Li G, Zhou Z, Wang L. 2011. Complete genome sequence of Aeromonas veronii strain B565. J. Bacteriol. 193:3389–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Beatson SA, das Graças de Luna M, Bachmann NL, Alikhan NF, Hanks KR, Sullivan MJ, Wee BA, Freitas-Almeida AC, Dos Santos PA, de Melo JT, Squire DJ, Cunningham AF, Fitzgerald JR, Henderson IR. 2011. Genome sequence of the emerging pathogen Aeromonas caviae. J. Bacteriol. 193:1286–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33:5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106:19126–19131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 72. Gavín R, Rabaan AA, Merino S, Tomás JM, Gryllos I, Shaw JG. 2002. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43:383–397 [DOI] [PubMed] [Google Scholar]
- 73. Swift S, Karlyshev AV, Fish L, Durant EL, Winson MK, Chhabra SR, Williams P, Macintyre S, Stewart GS. 1997. Quorum sensing in Aeromonas hydrophila and Aeromonas salmonicida: identification of the LuxRI homologs AhyRI and AsaRI and their cognate N-acylhomoserine lactone signal molecules. J. Bacteriol. 179:5271–5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. O’Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 75. Morohoshi T, Shiono T, Takidouchi K, Kato M, Kato N, Kato J, Ikeda T. 2007. Inhibition of quorum sensing in Serratia marcescens AS-1 by synthetic analogs of N-acylhomoserine lactone. Appl. Environ. Microbiol. 73:6339–6344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sha J, Lu M, Chopra AK. 2001. Regulation of the cytotoxic enterotoxin gene in Aeromonas hydrophila: characterization of an iron uptake regulator. Infect. Immun. 69:6370–6381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Erova TE, Sha J, Horneman AJ, Borchardt MA, Khajanchi BK, Fadl AA, Chopra AK. 2007. Identification of a new hemolysin from diarrheal isolate SSU of Aeromonas hydrophila. FEMS Microbiol. Lett. 275:301–311 [DOI] [PubMed] [Google Scholar]
- 78. Erova TE, Pillai L, Fadl AA, Sha J, Wang S, Galindo CL, Chopra AK. 2006. DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect. Immun. 74:410–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bidirectional BLASTP heat map of Aeromonas species genomes with that of A. hydrophila ATCC 7966T as the reference for comparison. The comparison genomes included, from the outermost to the innermost, those of A. hydrophila E1, A. hydrophila E2, A. aquariorum AAK1, A. caviae Ae398 A. salmonicida A449, and A. veronii B565. The percent translated protein sequence identity for each gene is color coded according to the scale shown. Regions in white represent genes that are present in the reference genome (that of strain ATCC 7966T) and absent from the others. Download
Comparative genomics of genes that encode T3SSs in six Aeromonas strains. Entire T3SS gene clusters of strains E1 and E2 and GenBank sequences of the T3SSs of A. hydrophila SSU (GenBank accession no. AY763611), A. hydrophila AH3 (GenBank accession no. AY528667), A. veronii biovar Sobria HM21 (GenBank accession no. EF215451), and A. salmonicida A449 (GenBank accession no. CP000646) were compared by using NCBI BLASTn, and the results were visualized by using the Artemis comparison tool (Wellcome Trust Sanger Institute). Regions in red are homologous and located on the same DNA strand. Regions in white indicate regions of divergence. Open reading frames of A. salmonicida A449 are shown at the bottom. Download
Phylogenetic relationship among Aeromonas species genes that encode T3SSs (A) and genes that encode lateral flagella (B). T3SS sequences were retrieved from GenBank for A. hydrophila AH1 (GenBank accession no. AY394563), A. hydrophila SSU (GenBank accession no. AY763611), A. hydrophila AH3 (GenBank accession no. AY528667), A. veronii biovar Sobria HM21 (GenBank accession no. EF215451), A. salmonicida JF2267 (GenBank accession no. AJ616218), A. salmonicida A449 (GenBank accession no. CP000646), and P. aeruginosa PAO1 (GenBank accession no. AE004091). Lateral flagellum sequences were retrieved from GenBank for A. salmonicida A449 (GenBank accession no. CP000644) and A. hydrophila AH3 (GenBank accession no. DQ124694). The evolutionary history was inferred by the neighbor-joining method. The bootstrap consensus tree was inferred from 1,000 replicates. The evolutionary distances were computed by the maximum composite likelihood method, and each value is the number of base substitutions per site. Download
Transmission electron photomicrographs of three A. hydrophila strains. A, A. hydrophila E1; B, A. hydrophila E2; C, A. hydrophila SSU. Staining was done with 0.5% phosphotungstic acid. A. hydrophila E1 and SSU display lateral flagella, while strain E2 exhibits rafting-like behavior. Download
A. hydrophila E1, E2, and ATCC 7966T form significantly less biofilm biomass on polystyrene than A. hydrophila SSU does. Biofilms were quantified by CV staining after 24 h of incubation at 37°C. The results presented represent the mean and standard deviation of three independent experiments. An asterisk indicates a P value of less than 0.001 as determined by one-way ANOVA. Download
Distribution of A. hydrophila genes from the genomes of strains E1, E2, and ATCC 7966T. A comparative genomic analysis of the three A. hydrophila genomes was performed, and the syntenic pangenome is presented as mapped to the closed genome of ATCC 7966T, with start and stop positions, lengths of genes, and putative functions. For core genome homologues in strains E1 and E2, BLASTP scores are shown, in relation to the ATCC 7966T allele. For genes at ends of contigs and interrupted by contig gaps, bidirectional BLASTN was performed by using the gene sequences from the other strains to confirm the presence and inclusion in or exclusion from the core gene set, and these values are underlined. For dispensable genes not found in ATCC 7966T, strain E1 was used as the reference for BLAST analysis. A small subset of unique genes from strains E1 and E2 could not be mapped to the syntenic core genome and are labeled as orphan genes. Certain traits are highlighted for importance.