ABSTRACT
Elongation factor P (EF-P) is a universally conserved bacterial translation factor homologous to eukaryotic/archaeal initiation factor 5A. In Salmonella, deletion of the efp gene results in pleiotropic phenotypes, including increased susceptibility to numerous cellular stressors. Only a limited number of proteins are affected by the loss of EF-P, and it has recently been determined that EF-P plays a critical role in rescuing ribosomes stalled at PPP and PPG peptide sequences. Here we present an unbiased in vivo investigation of the specific targets of EF-P by employing stable isotope labeling of amino acids in cell culture (SILAC) to compare the proteomes of wild-type and efp mutant Salmonella. We found that metabolic and motility genes are prominent among the subset of proteins with decreased production in the Δefp mutant. Furthermore, particular tripeptide motifs are statistically overrepresented among the proteins downregulated in efp mutant strains. These include both PPP and PPG but also additional motifs, such as APP and YIRYIR, which were confirmed to induce EF-P dependence by a translational fusion assay. Notably, we found that many proteins containing polyproline motifs are not misregulated in an EF-P-deficient background, suggesting that the factors that govern EF-P-mediated regulation are complex. Finally, we analyzed the specific region of the PoxB protein that is modulated by EF-P and found that mutation of any residue within a specific GSCGPG sequence eliminates the requirement for EF-P. This work expands the known repertoire of EF-P target motifs and implicates factors beyond polyproline motifs that are required for EF-P-mediated regulation.
IMPORTANCE
Bacterial cells regulate gene expression at several points during and after transcription. During protein synthesis, for example, factors can interact with the ribosome to influence the production of specific proteins. Bacterial elongation factor P (EF-P) is a protein that facilitates the synthesis of proteins that contain polyproline motifs by preventing the ribosome from stalling. Bacterial cells that lack EF-P are viable but are sensitive to a large number of stress conditions. In this study, a global analysis of protein synthesis revealed that EF-P regulates many more proteins in the cell than predicted based solely on the prevalence of polyproline motifs. Several new EF-P-regulated motifs were uncovered, thereby providing a more complete picture of how this critical factor influences the cell’s response to stress at the level of protein synthesis.
Introduction
Regulation of gene expression can occur at many steps en route from a DNA-encoded gene to a mature protein. Protein synthesis can be controlled by self-mediated stalling of a peptide chain that exerts a regulatory effect on the translation of the protein or that of downstream genes. For example, the SecM transcript stalls its own translation in a nascent polypeptide-dependent manner via a series of complex interactions with the ribosome (1, 2). This stall leads to remodeling of an mRNA hairpin, thereby revealing the ribosome binding site of the downstream secA gene, which results in the increased production of SecA (3). SecA in turn forms a feedback loop by targeting the SecM-stalled ribosome to the protein export machinery, which allows translation of SecM to resume (2, 4). Though a number of proteins that mediate their own translational stalls have been characterized, each is unique, and common sequence motifs have not been well defined, although certain themes have emerged (5). Stalling can occur, for example, if the transcript contains an internal sequence resembling a Shine-Dalgarno sequence (6). Interaction of the nascent polypeptide with the ribosomal exit tunnel and peptidyl transferase center can also alter the ribosome such that if the A-site aminoacyl-tRNA is a poor acceptor for the particular P-site peptide, peptide bond formation will be stalled (5, 7). Finally, studies have identified C-terminal sequences capable of blocking termination at stop codons by inhibiting the action of ribosomal release factors (5, 8, 9).
Elongation factor P (EF-P) is a universally conserved bacterial translation factor that was originally characterized for its ability to stimulate peptide bond synthesis in vitro. In Escherichia coli and Salmonella, EF-P undergoes a posttranslational modification homologous to hypusination of its eukaryotic homolog, eukaryotic translation initiation factor 5A (eIF5A), wherein a unique β-lysine residue is added to a conserved lysyl residue by the combined activities of PoxA and YjeK (the PYE pathway). The EF-P protein has a shape similar to that of a tRNA and interacts with the ribosome in a unique position contacting the E site of the 30S subunit and the P site on the ribosome. The β-lysylated residue of EF-P is oriented in the ribosome such that it projects into the peptidyl transferase center (PTC), presumably to modulate peptide bond formation. We and other laboratories have noted that deletion of any of the PYE genes in Salmonella results in pleiotropic phenotypes, including increased susceptibility to a range of cellular stressors, as well as motility defects and impaired nutrient utilization relative to findings for the wild-type strain (10–13). It has further been shown that β-lysylated EF-P also undergoes hydroxylation by YfcM, but this additional modification does not appear to be critical for EF-P function based on phenotypic and in vivo analyses (14, 15).
Proteomic analysis of Salmonella poxA and efp mutants and Agrobacterium efp (chvH) mutants suggested that EF-P acts in a targeted manner rather than as a global translation factor (10, 11, 13, 16). Recent work has demonstrated that EF-P rescues the translation of elongating polypeptides that are stalled at particular tripeptide motifs, in particular PPP or PPG (9, 17, 18). EF-P was recently shown to not have an effect at other types of stalling motifs, such as the C-terminal motifs that inhibit release factors (9). The discovery of EF-P target motifs lends insight into why specific proteins are subject to EF-P-mediated regulation. Furthermore, the pleiotropic phenotypes exhibited by bacteria lacking EF-P could likely be explained by the presence of polyproline stretches in factors essential for the phenotype. For instance, a number of flagellar genes contain such motifs, and their misregulation likely contributes to the motility defect observed in PYE mutants (10, 13). However, previous work has focused on in vitro study of particular EF-P targets and has not thoroughly evaluated the role of EF-P in vivo.
Here we present an unbiased in vivo comprehensive analysis of the Salmonella proteins affected by EF-P, using stable-isotope labeling (SILAC) (19, 20). While we confirmed that several proteins containing polyproline motifs are subject to EF-P-mediated regulation, we have found that the presence of polyproline tracts is not sufficient to predict EF-P dependence, since many proteins containing such motifs are unaffected by the absence of EF-P. We also identified alternate target sequences in addition to PPP and PPG that render peptide synthesis dependent on EF-P. Finally, we investigated the determinants of EF-P dependence for pyruvate oxidase (PoxB), which was previously shown to be downregulated approximately 8-fold in E. coli strains lacking PoxA despite the absence of a polyproline sequence (21).
RESULTS
Identification of EF-P-regulated proteins by SILAC.
We previously examined the proteome of the Salmonella enterica poxA mutant using two-dimensional difference gel electrophoresis (2D-DIGE). In that study, total cellular proteins from the wild type and poxA mutants were labeled with fluorescent dyes, mixed in equal amounts, and then separated by 2D-PAGE prior to analysis (11). Using this technique, we determined that a relatively small subset of proteins were affected by perturbations in the PYE pathway—a finding in agreement with earlier work on the efp mutant of Agrobacterium (16). However, we were able to unambiguously identify only a small number of these proteins by mass spectrometry due to crowding on the 2D gel.
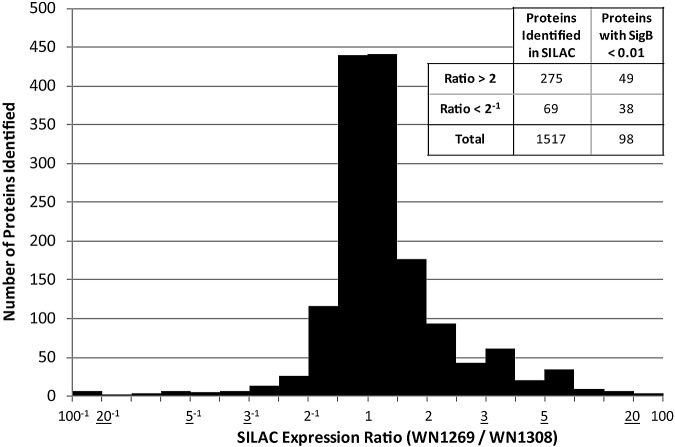
To gain a more comprehensive view of the effect of EF-P on protein levels, we employed stable isotope labeling of amino acids in cell culture (SILAC) in conjunction with quantitative mass spectrometry-based proteomics to examine the proteome of an efp mutant strain of Salmonella enterica serovar Typhimurium strain SL1344 (strain WN1308). We examined the profiles of three biological replicates and in total were able to detect, quantify, and identify a total of 1,517 proteins, or approximately 34% of the 4,514 proteins predicted to be encoded in the S. Typhimurium strain SL1344 genome (Fig. 1). To identify proteins showing altered levels, we first selected candidates that showed a significant difference between the wild-type and efp mutant strains in at least one of the three biological replicates with a significance B (an internal significance score as described in Materials and Methods) cutoff of 0.01. By this criterion, 87 proteins showed changes of 2-fold or greater and 28 displayed a change of greater than 10-fold. Of the 87 significantly misregulated proteins, 49 showed decreased steady-state levels in the efp mutant strain and are more likely to be direct targets of EF-P owing to its characterized stimulatory effect on translation (17, 18).
FIG 1 .
A subset of proteins is significantly misregulated in Δefp Salmonella. The histogram outlines the distribution of protein synthesis ratios identified in SILAC. Columns indicate the number of proteins with an average synthesis ratio between two neighboring x axis values. Underlined values in the x axis indicate a change in scale. The inset table shows the number of SILAC hits demonstrating a greater than 2-fold difference in protein level between the efp+ (WN1269) and ∆efp (WN1308) strains. The second column further indicates the proteins with a significance B value of less than 0.01 in at least one trial. “Total” indicates the number of proteins identified in at least one replicate regardless of expression ratio, and the second column of this row includes proteins with any expression ratio that had a significance B value of less than 0.01 in at least one trial. Synthesis ratios shown are the average normalized heavy/light ratios of three biological replicates.
Identification of amino acid motifs enriched in EF-P-regulated proteins.
To determine if misregulation of specific proteins in efp mutants can be attributed to certain motifs, we analyzed the SILAC data for the prevalence of specific peptide sequences in proteins showing a strong decrease or increase in the Salmonella efp mutant. Since tripeptide motifs (PPP and PPG) had previously demonstrated EF-P-dependent translation, we searched for the frequency of each possible tripeptide motif in the 10% of proteins most strongly affected (either up or down) by EF-P. We compared the actual frequency of each motif to its expected frequency and the frequency observed in the remaining 90% of proteins identified in SILAC. An enrichment score was calculated based on the ratio between observed occurrences of a tripeptide within this group, normalized against the actual-over-expected occurrences in all retrieved SILAC hits (see Materials and Methods). Using this method, we identified a number of tripeptide motifs that were enriched in the 10% of proteins that were most downregulated in the efp mutant (Table 1). The two top-scoring tripeptide sequences identified were PPP and PPG, supporting the recent findings regarding EF-P regulation and also acting as a proof of principle for our analysis. Additionally, a number of previously uncharacterized sequence motifs were also identified, including the third-highest scoring sequence, APP, implicating them as potential EF-P-dependent motifs. While this article was in preparation, Woolstenhulme et al. identified the APP-containing motifs MRAPP and WAPP as sequences capable of stalling ribosomes (9).
TABLE 1 .
Bioinformatic analysis predicts EF-P-regulated tripeptide motifsa
| Motifb | Occhigh | Exphigh | Occmid | Expmid | Occlow | Explow | Enrichhigh | Enrichlow | P valuec |
|---|---|---|---|---|---|---|---|---|---|
| PPP | 18 | 4.96 | 15 | 29.1 | 0 | 3.16 | 4.09 | 0.00 | 2.1E−10 |
| PPG | 28 | 8.48 | 26 | 49.7 | 1 | 5.40 | 3.82 | 0.21 | 5.6E−14 |
| APP | 18 | 11.1 | 22 | 65.1 | 0 | 7.07 | 3.38 | 0.00 | 7.6E−10 |
| RME | 12 | 5.30 | 16 | 31.1 | 2 | 3.38 | 3.00 | 0.79 | 1.3E−04 |
| YIR | 10 | 4.64 | 14 | 27.2 | 1 | 2.96 | 3.00 | 0.47 | 4.7E−04 |
| TQM | 11 | 3.64 | 17 | 21.4 | 0 | 2.32 | 2.95 | 0.00 | 1.4E−04 |
| PFF | 10 | 2.84 | 15 | 16.6 | 2 | 1.81 | 2.78 | 0.87 | 7.4E−05 |
| DPP | 5 | 6.73 | 8 | 39.4 | 1 | 4.29 | 2.68 | 0.84 | 2.0E−07 |
| FFL | 5 | 6.15 | 8 | 36.1 | 1 | 3.92 | 2.68 | 0.84 | 1.4E−06 |
| QNA | 22 | 10.0 | 43 | 58.8 | 5 | 6.39 | 2.36 | 0.84 | 3.0E−05 |
Prevalence among the SILAC hits of all possible tripeptide combinations of the 20 common amino acids. Occ, motif occurrences in the indicated subgroup of the SILAC data; Exp, expected motif occurrences in the indicated subgroup based on amino acid prevalence; Enrich, calculated motif enrichment index in the indicated subgroup of the SILAC data; high, low, and mid, subgroups of SILAC data encompassing the 10% of proteins with the highest (high) or lowest (low) WN1269/WN1308 ratios; “mid” incorporates the remaining 80%.
Motifs shown are those that demonstrated the greatest enrichment index within the 10% of proteins most downregulated in the efp mutant (highest WN1269/WN1308 ratio).
P values were calculated using the chi-square test with Yates’ correction as described in Materials and Methods.
Alternative non-proline-containing motifs can render protein synthesis dependent on EF-P.
Results from the motif analysis lend support to the previously characterized EF-P-dependent sequences but also propose novel motifs that may require EF-P for efficient translation. To assess the possibility that EF-P plays a role in the translation of motifs other than polyproline, we constructed a number of translational fusion plasmids wherein putative target sequences were inserted in-frame at the fourth codon of gfp. Green fluorescent protein (GFP) fluorescence was significantly decreased in an efp mutant relative to that in wild-type E. coli when any of the PPP, PPG, or APP motifs were inserted into GFP (Table 2). Interestingly, insertion of a YIRYIR motif also caused production of the GFP reporter to be dependent on EF-P. This is the first EF-P target motif identified that lacks a proline residue. In contrast, none of the other predicted motifs—including an unrepeated YIR sequence—yielded a significant difference in synthesis. We further analyzed the proteins containing non-EF-P-dependent motifs to determine the reasons they were enriched in our bioinformatic analysis. Of the 10% of proteins that were most downregulated in the efp mutant, 16 of the 29 proteins containing an RME, PFF, or YIR motif also have a PPP, PPG, or APP motif, which may explain their EF-P dependence. For the 13 proteins that contain an RME, PFF, or YIR motif but lack a PPP, PPG, or APP motif, we aligned the residues upstream of the putative regulatory motif but did not identify any conserved sequences (data not shown). The underlying cause of the EF-P dependence of these proteins remains to be investigated.
TABLE 2 .
Verification of predicted EF-P-dependent motifsa
| Motif | GFP fluorescence (WT/Δefp)b |
|---|---|
| Nullc | 0.95 ± 0.09 |
| PPPPPP0d | 20.68 ± 0.38 |
| PPPPPP1d | 18.55 ± 0.38 |
| PPPPPP2d | 18.12 ± 0.20 |
| PPPPPP3d | 15.97 ± 0.08 |
| PPP | 4.72 ± 0.23 |
| PPG | 10.39 ± 0.66 |
| APP | 5.88 ± 0.20 |
| RME | 1.34 ± 0.07 |
| YIR | 0.96 ± 0.13 |
| YIRYIR | 7.61 ± 2.54 |
| PFF | 1.58 ± 0.19 |
Motifs were assayed for EF-P dependence by insertion into the 4th codon position of GFP.
Values at 21 h postinduction were normalized to cotranscriptionally expressed mCherry and are shown as a ratio of wild-type and efp mutant strains expressing the same construct. All values are the averages ± standard deviations for three biological replicates
No motif inserted into GFP.
Sequence of six optimal (CCG; 0) or random (1 to 3) proline codons.
The presence of a PPP, PPG, or APP motif is not sufficient to confer dependence on EF-P.
To investigate whether the characterized target motifs are sufficient for EF-P regulation in vivo, we compared the existence of a PPP, PPG, or APP motif in proteins with their SILAC ratios. Of the 422 proteins that contain one of the three target motifs, 100 were conclusively identified in SILAC (Fig. 2A). Only 20 of these were found to be significantly downregulated in the efp mutant strain. A more conservative analysis of proteins with SILAC ratios of less than 2 (without regard for the significance score) found that there were 45 proteins identified in SILAC that were not downregulated in WN1308 yet have a PPP, PPG, or APP motif (12, 22, and 16 proteins, respectively, contain each motif; 5 proteins have two of the three motifs). This demonstrates that a large percentage of proteins that contain a characterized EF-P target motif are not misregulated in the efp deletion strain. Examples include ZipA, SseA, and YtfM, which were identified to have average expression ratios of 1.07, 0.74, and 0.84, respectively, with significance B scores that were far from significant in all three replicates (see Table S2 in the supplemental material). Though they were not misregulated in the efp mutant, Salmonella ZipA has two distinct APP motifs, as well as an APPP motif, SseA has an APPG motif, and YtfM contains a PPP motif. Taken together, these data suggest that the presence of particular tripeptide motifs may not, in and of itself, be sufficient to cause translational stalling in the absence of EF-P.
FIG 2 .
Comparison of proteins identified in SILAC and those with EF-P target motifs. (A) Venn diagram outlining the overlap of: proteins conclusively identified in SILAC, proteins significantly downregulated in WN1308, and proteins containing a PPP, PPG, or APP motif as annotated in the Salmonella serovar Typhimurium strain SL1344 genome (GenBank accession no. FQ312003.1). (B) DAVID analysis of SILAC hits showing a decreased protein level in the ∆efp strain (ratio > 2) and a significance B score of less than 0.01 in at least one trial. Functional annotation clusters showing significant overrepresentation (P value < 0.05) are shown. Cluster breakdown can be found in Table S4 in the supplemental material. (C) DAVID analysis showing the most significantly overrepresented clusters among the 422 proteins that contain an EF-P target motif. For clarity, only groups with P values < 0.001 are shown here. For a full list, see Table S5. (D) DAVID analysis showing the only significantly overrepresented cluster (P value < 0.05) among the 20 proteins that fall into all three categories (contain an EF-P target motif and were significantly downregulated in the ∆efp strain in SILAC). Cluster breakdown can be found in Table S6 in the supplemental material.
To investigate whether the position of a target motif within a protein affected EF-P regulation, we examined the distribution of the proteins where the target motif occurs in the first 50 residues (early), where it occurs between residues 50 and 200 (middle), and where the motif occurs after the 200th residue (late). For the group of 20 target motif-containing proteins that were significantly downregulated in the efp mutant, we found that 30%, 40%, and 30% of proteins fall into the early, middle, and late categories, respectively. This is comparable to the 80 motif-containing proteins that were not significantly downregulated, for which we find that 25%, 39%, and 36% of proteins are grouped into the early, middle, and late categories, respectively. We also examined sequence context by aligning the motif region of a number of EF-P-dependent and EF-P-independent motif-containing proteins. Results from this analysis did not conclusively identify any upstream sequences consistent with EF-P dependence or independence (data not shown).
EF-P disproportionately affects synthesis of signaling and nucleotide-binding/metabolic proteins.
To examine whether EF-P affected the synthesis of all classes of proteins similarly or if there was a particular bias toward a specific subset of proteins, the SILAC data were subjected to a functional annotation analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) software package (22–24). The program compares a list of genes with functional annotation databases, including GO (gene ontology) terms, KEGG pathways, and SP-PIR (Swiss-Prot and Protein Information Resource) keywords, among others. By comparing the prevalences of proteins belonging to these categories in their respective database and in the input gene list, DAVID generates a P value highlighting significantly overrepresented annotation terms.
Upon analysis of the 49 significantly downregulated proteins identified in SILAC, we find that four clusters demonstrate overrepresentation, with a cluster P value of less than 0.05 (Fig. 2B; see also Table S4 in the supplemental material). Most prominent among these terms were two-component regulatory systems, with particular emphasis on proteins involved in chemotaxis and motility. Furthermore, metabolic proteins were also abundantly downregulated as annotated by functions in nucleotide binding, oxidative phosphorylation, or proteolysis.
We subsequently examined the S. Typhimurium SL1344 genome for proteins containing EF-P-dependent sequences and found 422 open reading frames (ORFs) encoding a PPP, PPG, or APP motif. These motifs occurred in 112 (PPP), 195 (PPG), and 185 (APP) proteins, with 70 of those proteins containing more than one of these motifs. In contrast, the sequence YIRYIR is not present in any Salmonella protein. To examine the physiological processes involving proteins with EF-P-dependent motifs, we conducted a DAVID cluster analysis and found that the overrepresented functional groups were similar to those identified among proteins that were significantly downregulated in SILAC (Fig. 2C). Many of the identified proteins with a PPP/PPG/APP motif are predicted membrane proteins, a group we previously investigated by comparative gel electrophoresis. We previously identified the outer membrane porin, KdgM, as significantly upregulated in the efp mutant, which we observed to occur at the level of transcription (13). The SILAC analysis confirms KdgM as a protein that is 38-fold more abundant in the efp mutant (see Table S2 in the supplemental material). However, the SILAC data did not identify any EF-P-dependent regulators (e.g., transcriptional repressors) that would easily explain why certain proteins, such as KdgM, are observed to have higher levels in the efp mutant strain.
Twenty proteins that both contained a target motif and were significantly downregulated in SILAC were identified. Of these, 7 were identified as nucleotide binding proteins—the only overrepresented category in this subset of 20 proteins (Fig. 2D). Some of these proteins are involved in central metabolism. AtpD, the catalytic subunit of the F0F1 ATPase, had previously been found to be downregulated in the proteome of poxA mutants (11). PfkB is a phosphofructokinase that functions in glycolysis. Three of the other EF-P-dependent nucleotide binding proteins (HflB/FtsH, HslU, and ClpB) play a role in protein stability and turnover. The function of the polyproline motifs in these proteins is unlikely to be universally conserved. For example, in AtpD (25), ClpB (26), and PfkB (PDB identifier: 3UMP), the putative EF-P-dependent motifs are not proximal to the region of the protein that interacts with ATP, whereas in HflB/FtsH, the putative EF-P-dependent motif (GPPG) makes contact with AMP (27).
Some proteins identified as EF-P dependent by SILAC or by the presence of a polyproline motif can provide a parsimonious explanation for previously described phenotypes of Salmonella strains lacking PoxA or EF-P. We observed, for example, that the enzyme gamma-glutamyl transferase (Ggt) is present at a level approximately 16-fold lower in the efp mutant than in wild-type Salmonella (see Table S2 in the supplemental material). Ggt contains a short polyproline motif (residues 291 to 293), and its strongly reduced synthesis in mutant cells likely explains why Salmonella strains lacking EF-P, PoxA, or YjeK are simultaneously unable to utilize γ-glutamyl-glycine as a nitrogen source and are resistant to the compound S-nitrosoglutathione (GSNO) (13, 28). The misregulation of proteins involved in motility and chemotaxis, such as CheA, which is downregulated approximately 4-fold in the efp mutant, likely contributes to the observed motility defect in poxA, yjeK, and efp mutant strains (13).
Effect of EF-P on synthesis of PoxB and identification of the putative regulatory motif.
Pyruvate oxidase (PoxB) was implicated as a protein dependent on EF-P for its synthesis when it was noted that levels of this enzyme were decreased 5- to 8-fold in E. coli poxA mutants (21, 29). The PoxB protein, however, lacks any of the previously described EF-P-dependent motifs, which leaves open the question as to whether the PoxB protein is indeed a target of EF-P and, if so, what particular sequence renders it dependent on EF-P for its synthesis. We were unable to detect PoxB in our SILAC assay due to the particular growth conditions employed. To circumvent this issue, we constructed a reporter vector wherein the PoxB 5′ untranslated region (UTR) and open reading frame were fused to a “superfolder” variant of green fluorescent protein (sfGFP). Production of the chimeric protein was driven from a constitutive tet promoter, which enabled us to measure translation independently of transcription. In a Salmonella background, the full-length PoxB protein fused to sfGFP showed a marked and reproducible dependence on EF-P for synthesis (Fig. 3) despite the fact that transcription of this chimeric construct was similar between the mutant and wild-type strains as measured by quantitative real-time reverse transcriptase PCR (see Fig. S1 in the supplemental material). Interestingly, though similar poxB transcript levels indicate that transcription is not altered, gfp mRNA levels were decreased in strains expressing EF-P-dependent constructs. However, since previous in vitro data have shown EF-P rescuing ribosome stalls in the absence of nucleases (9, 17, 18), we believe that this degradation is not the cause of decreased GFP fluorescence but rather is an effect of ribosome stalling at EF-P-dependent motifs leading to reduced ribosome-mediated nuclease protection of the 3′ end of the poxB-gfp transcript.
FIG 3 .
The GSCGPG motif of PoxB renders it dependent on EF-P. (A) Outline of translational fusion constructs expressing full-length (FL) PoxB or C-terminal truncations fused to sfGFP. Construct designations are shown in bold at left as a figure key and indicate the length in codons of each truncated poxB gene. The arrow indicates the transcriptional start site under the control of the PLtet0-1 constitutively active promoter, and “fMet” indicates the ATG start codon of PoxB. Numbers indicate poxB lengths in codons. For clarity, only a selection of constructs is illustrated. (B) Sequence of amino acids 69 to 82 of PoxB. The GSCGPG motif is bold and underlined. (C) Relative GFP fluorescence of PoxB truncations. Values were taken at 10 h postinoculation, were normalized to the optical density at 600 nm (OD600), and are shown relative to those of the wild-type strain expressing the same construct. The same plasmid encoding the first 186 codons of lacZ instead of poxB is included as a control. (D) Relative GFP fluorescence (as in panel C) for single-residue mutations to leucine in the full-length PoxB construct. All values are the averages for at least 3 biological replicates. Each error bar shows 1 standard deviation.
Decreased fluorescence was observed not only in an efp mutant but also in poxA and yjeK mutants (see Fig. S2 in the supplemental material). In contrast, a strain lacking the YfcM protein did not show any significant difference in synthesis of the PoxB-sfGFP fusion construct. These results are consistent with previous data showing that YfcM-mediated hydroxylation of EF-P is not critical for its function (14).
To identify the region essential for EF-P dependence, we systematically generated C-terminal truncations of the poxB sequence fused to sfGFP (Fig. 3A). By comparing fluorescence of these constructs in wild-type and efp mutant Salmonella, we narrowed the critical region to a 14-amino-acid stretch between residues 69 and 82 with the sequence VCAGSCGPGNLHLI (Fig. 3B and C). We confirmed that the amino acid (as opposed to the nucleotide) sequence in this region was critical for EF-P dependence by generating a construct with this region shifted to the +1 reading frame and observing an abolition of the requirement for EF-P (see Fig. S3 in the supplemental material).
Residues 69 to 82 of PoxB contain a GPG motif that we hypothesized could, like PPP, mediate the EF-P dependence of the PoxB protein. To address this, we performed site-directed mutagenesis on the GPG motif in the full-length PoxB-sfGFP construct and on additional upstream and downstream residues, converting each residue to a leucine. Leucine was chosen because of its absence from the top-scoring motifs predicted by our analysis of proteins downregulated in the efp mutant (Table 1). We found that mutation of any residue in the GPG motif or in the upstream three residues (sequence GSC) restored GFP fluorescence in the efp mutant (Fig. 3D). In contrast, mutation of residues either downstream or upstream of the GSCGPG motif had negligible effects on the synthesis of the reporter construct. Notably, the GSCGPG sequence is found only in PoxB in Salmonella. These data elucidate a novel EF-P-dependent motif that, like the artificially derived YIRYIR, is considerably larger than the previously described tripeptide motifs and can cause a protein to be dependent on EF-P for proper synthesis in vivo.
DISCUSSION
The recent finding that EF-P alleviates stalls caused by polyproline motifs during translational elongation provides a unifying mechanism for the physiological consequences that are triggered by the loss of this factor. Here we conclusively identified and assessed the steady-state levels of more than 1,500 Salmonella proteins using SILAC and found that less than 6% were significantly misregulated by more than 2-fold in a strain lacking EF-P. A functional annotation analysis of the corresponding downregulated genes revealed that proteins involved in responding to environmental stimuli, motility, metabolism, and proteolysis were overrepresented among the group affected by the loss of EF-P. These data lend insight into the causes underlying the pleiotropic phenotypes exhibited by efp mutants by implicating particular pathway components. For instance, the finding that two-component systems were the most highly overrepresented group among the downregulated proteins suggests that impaired responses to external stimuli contribute to the observed sensitivity of the efp mutant to a range of pharmacologically unrelated antibiotics. Determining the specific defective pathways in future studies may point to potential targets for novel antimicrobials that would act synergistically with existing drugs.
Following the discovery that EF-P rescues ribosomes stalled at particular tripeptide motifs, we probed the proteomic data for an abundance of such sequences in downregulated proteins. We found that in addition to the characterized PPP and PPG motifs, APP was also prominent among downregulated proteins, suggesting it as a novel EF-P target, which was confirmed using a translational fusion assay. In addition to the discovery of APP as an EF-P dependent motif, our SILAC data have also led to the identification of the first characterized EF-P target motif that lacks a proline: YIRYIR. Furthermore, our analysis of PoxB identified the motif GSCGPG as responsible for its regulation by EF-P. Cumulatively these data expand the known repertoire of EF-P target motifs and demonstrate, in certain instances, that they may require sequences longer than three amino acids. This is further reinforced by the fact that the tripeptide motifs can be relatively common in the proteome and yet the overall number of proteins affected strongly by the loss of EF-P is considerably smaller. Indeed, of the 100 tripeptide target motif-containing proteins identified in SILAC, only 20 were significantly downregulated. The data suggest that tripeptide motifs, including polyproline tracts, may not always be solely sufficient to mediate a translational stall and may depend on upstream interactions within the exit tunnel as well. Conversely, some weaker tripeptide motifs, such as the GPG of PoxB, may instigate ribosomal stalls if sufficiently strengthened by upstream interactions.
It is clear that not all stalls are created equal, and we note that previously reported peptides that stall during elongation function in wild-type circumstances where EF-P is present and (presumably) active. Multiple groups have reported on proteins that instigate a translational stall via not only a particular sequence in the vicinity of the PTC but also upstream residues in the nascent polypeptide chain that interact with the ribosomal exit tunnel. These stalls may be dependent on external factors, such as the antibiotic erythromycin in the case of ermAL1 and tryptophan for TnaC, or may be self-mediated, as for SecM and MifM (1, 3, 7, 30–32). The laboratory of Allen Buskirk identified a number of stall sequences containing polyproline tracts near the PTC that also require upstream residues for efficient stalling (5, 9). Future work will delineate the specific structural features that enable certain nascent chain-induced stalls to be alleviated by EF-P, while others, as in the case of SecM or TnaC, require alternate factors to rescue the stall. Also unresolved is whether EF-P preferentially targets particular subsets of proteins (e.g., nucleotide binding proteins) for the purpose of regulation or if this EF-P-dependent subset of proteins merely requires difficult-to-translate structural features, such as polyprolines, for functional purposes.
MATERIALS AND METHODS
Bacterial strains and plasmids.
For SILAC, we employed an arginine and lysine auxotroph (ΔargH ΔlysA) derivative of Salmonella enterica serovar Typhimurium strain SL1344 utilized in an earlier study (33). Auxotrophy was confirmed by growth in minimal medium in the absence of lysine or arginine. The Δefp mutation from strain WN934 was transferred into the auxotrophic strain by transduction using the HT105/1 int-201 derivative of phage P22 (34) to generate strain WN1308.
The E. coli strains used for motif verification were derivatives from the Keio knockout collection (35), including Δyfcm, Δyjek, ΔpoxA, and Δefp strains and BW25113 (wild type). Deletion of genes and removal of kanamycin cassettes using FLP recombinase (36) was confirmed by PCR. Plasmids used for motif verification were derivatives of pBAD30 (37) containing a tandem fluorescent fusion cassette composed of green fluorescent protein (GFP) followed immediately by the mCherry Shine-Dalgarno sequence and mCherry, both of which are optimized for synthesis in E. coli (40). This construct is designated pBAD30mw700 and served as the PCR template for construction of other reporters (see Table S1 in the supplemental material). Unless otherwise stated, the codons used for motif inserts were optimized according to tRNA abundance in E. coli (41).
Using the red-gam recombinase protocol described above, we generated strain WN1405 by deleting the majority of the efp gene (base pairs 145 to 424) from S. Typhimurium strain 14028s such that the new mutation would not influence the promoter of the upstream antiparallel yjeK gene. The mutant allele was transduced to a fresh strain background prior to experimentation. Deletions of efp were confirmed by PCR amplification using the primers WNp582 and WNp583 (see Table S1 in the supplemental material). WN1405 and the previously used Δefp mutant, WN934 (42), behave identically under all conditions tested (data not shown).
Plasmids used for the PoxB translational fusion assay were generated by PCR amplification of the 5′ UTR (28 bp upstream of the start codon) and full-length (excluding the stop codon) or C-terminally truncated poxB gene using S. Typhimurium strain 14028s genomic DNA as a template. These amplicons were inserted into the NsiI and NheI sites of pXG10sf (38) to generate translational fusions to “superfolder” GFP (sfGFP). The sfGFP is a variant with improved folding and fluorescence in E. coli (38, 39) and we found that it substantially improved the sensitivity of our assay compared to results with standard GFP variants. The plasmid employs the constitutively active PLtet0-1 promoter and the tightly controlled pSC101 low-copy-number origin of replication to ensure minimal variation in transcript levels. Point mutations were generated by site-directed mutagenesis of the full-length PoxB construct. Plasmids were named according to the truncation site in poxB (i.e., the PoxB 68 construct is truncated after codon 68 of poxB) or the location of a specific point mutation in full-length poxB (e.g., PoxB P76L contains a proline-to-leucine mutation at codon 76). LacZ186, included as a control, consists of the first 186 codons of the E. coli lacZ gene inserted into the NsiI and NheI sites of the pXG10sf plasmid, similar to the PoxB fusions.
SILAC.
For stable isotope labeling of amino acids in cell culture (SILAC), we supplemented morpholinepropanesulfonic acid (MOPS) minimal medium with amino acids at the concentrations previously described (33) and used 0.2% (wt/vol) glycerol as a carbon source. For heavy-isotope-labeled samples, arginine and lysine were replaced with 13C6-Arg and 2H4-Lys isotopes at the equivalent molar concentration. WN1269 (wild type) was grown in heavy arginine and lysine isotopes, and WN1308 (Δefp) was grown in light isotopes in this medium for 16 h to ensure complete labeling of all proteins. Strains were subsequently subcultured 1:200 in the same media and grown to an optical density (600 nm) of 0.5 (mid-log phase), at which point the cells were harvested by centrifugation. The pellets were subjected to lysis by heating to 99°C for 5 min in fresh lysis buffer (1% deoxycholate [DOC] in 50 mM ammonium bicarbonate [NH4HCO3] at pH 8). Cell debris was removed by centrifugation at 13,000 × g for 15 min, and the supernatant was frozen at −80°C until used.
Mass spectrometry and proteomic data analysis.
For analysis of the isotope-labeled lysates, 30 µg (each) of protein from WN1269 (heavy) and WN1308 (light) were combined, fractionated into 12 pieces by gel electrophoresis, and in-gel trypsin digested, following a previously outlined procedure (43). The resulting peptides were subjected to liquid chromatography coupled to tandem mass spectrometry using an Orbitrap XL mass spectrometer (Thermo Scientific) as described previously (44). Data analysis was conducted using the MaxQuant software program (45) to generate an average normalized heavy/light ratio over three biological replicates, and significance B values were calculated using Perseus software (46). To determine significance, we used the cutoff of a significance B score of less than 0.01 in at least one trial. This statistic measures significance within a single trial even for proteins that were identified in only one the three biological replicates. When we assessed significance using the average of significance B scores across all three trials (excluding scores where the protein of interest was not identified in a given trial), we obtained similar percentages of significantly misregulated proteins containing EF-P target motifs. For example, using an average significance B score of less than 0.05, we identified 107 significantly misregulated proteins. Sixty-one of these are greater than 2-fold downregulated in the efp mutant, and 25 of these 61 contain a PPP, PPG, or APP motif. We report all expression ratios and significance B scores in Table S2 in the supplemental material.
DAVID analysis.
Gene lists were generated based on the SILAC data and on the presence of EF-P target motifs in Salmonella proteins, as described in the text and the figure legends. Gene lists were uploaded to the Database for Annotation, Visualization and Integrated Discovery (DAVID) online analysis software program, and we employed the functional annotation tools to determine overrepresented annotation groupings among our input genes (22). DAVID employs a one-tailed Fisher exact probability test to calculate P values for individual annotation groups, and cluster P values are generated as the geometric means of the P values of all constituent groups. Functional annotation clusters with P values less than 0.05 are shown in figures. A full breakdown of clusters can be found in tables in the supplemental material.
Bioinformatic identification of prominent tripeptide motifs.
Out of the 1,517 Salmonella proteins identified by SILAC, 1,294 were retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/), parsed, and processed for further analysis of prominent tripeptide motifs using a customized Perl script. All possible combinations of 20 common amino acids in 3-digit length were enumerated and counted for their respective occurrences in three designated groups, namely, high 10%, middle 80%, and low 10%, ranked by the gene’s WN1269/WN1308 SILAC ratio.
Expected occurrences in each group were calculated by first determining the rate at which each amino acid occurred in the retrieved database (e.g., Ala accounts for 10% of all retrieved amino acids) and then calculating the accumulated probability of each specific tripeptide motif, assuming all combinations are completely unbiased (e.g., Ala-Ala-Ala is expected to occur in [10%]3 = 0.1% of all tripeptides inspected), finally multiplying this probability with the three-digit sliding windows inspected in each group (e.g., if the high-10% group contains 55,650 sliding windows of three consecutive amino acids, the expected occurrence of Ala-Ala-Ala is then 55,650 × 0.1% = 55.65).
The enrichment index of each tripeptide in a group was calculated as the ratio of actual-over-expected occurrences of that tripeptide in that group, normalized by the actual-over-expected occurrence of that tripeptide in total (total, in this case, refers to the 1,294 sequences retrieved from the SILAC data): Enrichmentgroup = (occurrence in one group/expectation in that group)/(occurrence in all groups/expectation in all groups).
The P value of each tripeptide was calculated using a chi-square test with Yates’ correction as follows:
Top candidate motifs most enriched in proteins downregulated in the efp mutant are screened with a P value of <0.0005 and an Enrichmentlowest 10% value of <1 and then ranked by Enrichmenthighest 10%. Similarly, top candidate motifs enriched in upregulated genes are screened by a P value <0.0005 and Enrichementhighest10% value of <1 and then ranked by Enrichmentlowest 10%.
Motif verification.
Luria-Bertani (LB) broth and base M9 minimal salts medium were of standard composition (47). Media were supplemented with 200 µg/ml ampicillin when required. All cultures were incubated at 37°C. Overnight cultures of E. coli strains harboring pBAD30 constructs in LB were diluted to an optical density at 600 nm (OD600) of 0.05 in M9 medium supplemented with 0.2% glycerol. After 2 h or when the OD600 reached 0.1 to 0.15, the culture was supplemented with 0.2% arabinose to induce synthesis of the GFP and mCherry reporter proteins, and fluorescence was assessed using a spectrofluorimeter (Horiba) at designated time points. Cells were analyzed for GFP using excitation at 481 nm and emission at 507 nm and for mCherry with excitation at 587 nm and emission at 610 nm. The background level with blank medium was subtracted, and the ratio of GFP fluorescence over that of mCherry was calculated. Reported values represent averages and standard deviations determined from three independent experimental replicates.
PoxB-sfGFP translational fusion assay.
Wild-type (S. Typhimurium strain 14028s) and isogenic Δefp mutant (WN1405) strains carrying plasmids bearing full-length or truncated poxB translational fusions to sfGFP were grown in MOPS minimal medium supplemented with 0.2% (wt/vol) glucose and 20 µg/ml chloramphenicol. Growth was conducted for 16 h at 37°C in a Tecan Infinite M200 microplate reader with constant aeration. OD600 and GFP fluorescence readings (excitation and emission wavelengths of 475 nm and 511 nm, respectively) were taken every 15 min. For both OD600 and GFP, background values taken from no-cell controls were subtracted from all readings. For clarity, the wild-type/Δefp fluorescence ratios are displayed as GFP fluorescence per OD600 unit at 10 h of growth. Results obtained when cultures were measured at 6, 8, 12, and 16 h of growth were similar to those at 10 h.
SUPPLEMENTAL MATERIAL
PoxB mRNA levels are similar in wild-type and ∆efp strains for both EF-P-dependent and -independent constructs. GFP fluorescence normalized to optical density (black), poxB mRNA transcript levels (light grey), and sfGFP mRNA transcript levels (dark grey) were assayed at mid-log phase (OD600 = 0.5; OD600 ≈ 0.2 as read in the Tecan Infinite M200 instrument). Transcript levels were tested by reverse transcriptase quantitative PCR using gene-specific primers and the technique used previously by Zou et al. (2012) (see reference 13 in the main article). Since the strains used carry poxB in their genomes, the mRNA levels obtained for lacZ186 with the poxB-specific primers were subtracted as background from all other values. Of note, under the conditions tested, the background poxB mRNA level was negligible (greater than 300-fold lower) compared to those for any of the strains expressing PoxB-GFP from a plasmid. All values are shown as the ratios of the result for the efp mutant relative to that for the wild-type strain expressing the same construct. All values are the averages for at least 3 biological replicates with the exception of those for the LacZ186 construct, which are the averages for 2 biological replicates. For the replicate where LacZ186 was not included, background poxB mRNA levels were obtained from another strain expressing a non-poxB construct from the same plasmid. Each error bar shows 1 standard deviation. Download
PoxB-sfGFP translation is impaired in ∆poxA, ∆yjeK, and ∆efp strains but not in the ∆yfcM mutant. Fluorescence of sfGFP fusion constructs in mutant strains lacking an EF-P-modifying protein is shown. Mutant strains are indicated in the top row of the x axis; the sfGFP fusion construct (as outlined in Fig. 3 in the main article) are indicated in the bottom row of the x axis. Values were taken at 10 h postinoculation and are shown in arbitrary fluorescence units (AU) normalized to the optical density at 600 nm (OD600). The “no plasmid” controls do not contain an sfGFP fusion plasmid and were grown in the absence of chloramphenicol. All values are the averages for at least 3 biological replicates. Each error bar shows 1 standard deviation. Download
EF-P regulation of PoxB is determined by the amino acid sequence as opposed to the nucleotides. (A) Nucleotide (upper line) and amino acid (lower line) sequences of the PoxB FL and PoxB Shifted constructs are shown from residues 66 to 85. The PoxB Shifted construct was generated by shifting codons 69 to 82 (bold) of full-length PoxB to the +1 reading frame by introducing an upstream single-base-pair insertion (underlined adenine) and downstream deletion (struck-out cytosine). (B) Relative GFP fluorescence values taken at 10 h postinoculation, normalized to the optical density at 600 nm (OD600) and shown relative to values for the wild-type strain expressing the same construct. P76::CCX constructs are mutated at proline 76 to an alternate proline codon. LacZ186, PoxB FL, PoxB 82, and PoxB 68 are shown for comparison. All values are the averages for at least 3 biological replicates. Each error bar shows 1 standard deviation. Download
PoxB synthesis demonstrates a similar dependence on EF-P when assayed during mid-log phase. Relative GFP fluorescence of PoxB constructs shown at mid-log phase instead of at 10 h postinoculation. Values were taken at the first time point after the culture surpassed an OD600 of 0.2 (mid-log phase as assayed in a Tecan Infinite M200 reader), normalized to the OD600, and are shown relative to those for the wild-type strain expressing the same construct. Data were obtained from the same biological replicates used to generate Fig. 3 in the main text. All values are the averages for at least 3 biological replicates. Each error bar shows 1 standard deviation. Download
Strains, plasmids, and primers
All proteins and expression ratios identified in SILAC
Bioinformatic identification of tripeptide motifs enriched within the 10% of proteins that were most upregulated in the efp mutant (lowest WN1269/WN1308 expression ratio)
DAVID cluster analysis of proteins significantly (significance B value [SigB] < 0.01) downregulated (expression ratio > 2) in WN1308
DAVID cluster analysis of proteins containing a PPP, PPG, or APP motif
DAVID cluster analysis of proteins significantly (SigB < 0.01) downregulated (expression ratio > 2) in WN1308 that also contain a PPP, PPG, or APP motif
ACKNOWLEDGMENTS
W.W.N. received support from the Natural Sciences and Engineering Research Council of Canada (NSERC) (RGPIN 386286-10). S.J.H. and S.B.Z. are each recipients of Vanier Canada Graduate Scholarships from NSERC. M.I. is supported by a grant from the National Institutes of Health (GM065183). L.J.F. is the Canada Research Chair in Quantitative Proteomics and received support from the CIHR for this work (MOP-77688). The mass spectrometry infrastructure used in this work was supported by the Canada Foundation for Innovation, the British Columbia Knowledge Development Fund, and the BC Proteomics Network.
We thank Jörg Vogel for the generous gift of the sfGFP-encoding plasmid pXG10sf. We also thank Brian Coombes and Colin Cooper for the gift of the auxotrophic Salmonella strain used in our SILAC study.
Footnotes
Citation Hersch SJ, Wang M, Zou SB, Moon K, Foster LJ, Ibba M, Navarre WW. 2013. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. mBio 4(2):e00180-13. doi:10.1128/mBio.00180-13.
REFERENCES
- 1. Bhushan S, Hoffmann T, Seidelt B, Frauenfeld J, Mielke T, Berninghausen O, Wilson DN, Beckmann R. 2011. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 9:e1000581 http://dx.doi.org/10.1371/journal.pbio.1000581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nakatogawa H, Ito K. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell 108:629–636 [DOI] [PubMed] [Google Scholar]
- 3. Murakami A, Nakatogawa H, Ito K. 2004. Translation arrest of SecM is essential for the basal and regulated expression of SecA. Proc. Natl. Acad. Sci. U. S. A. 101:12330–12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakatogawa H, Ito K. 2001. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol. Cell 7:185–192 [DOI] [PubMed] [Google Scholar]
- 5. Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR. 2009. Genetic identification of nascent peptides that induce ribosome stalling. J. Biol. Chem. 284:34809–34818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li GW, Oh E, Weissman JS. 2012. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 484:538–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramu H, Vázquez-Laslop N, Klepacki D, Dai Q, Piccirilli J, Micura R, Mankin AS. 2011. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol. Cell 41:321–330 [DOI] [PubMed] [Google Scholar]
- 8. Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H. 2004. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA 10:378–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. 2013. Nascent peptides that block protein synthesis in bacteria. Proc. Natl. Acad. Sci. U.S.A. 110:E878–E887 http://dx.doi.org/10.1073/pnas.1219536110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bearson SM, Bearson BL, Brunelle BW, Sharma VK, Lee IS. 2011. A mutation in the poxA gene of Salmonella enterica serovar typhimurium alters protein production, elevates susceptibility to environmental challenges, and decreases swine colonization. Foodborne Pathog. Dis. 8:725–732 [DOI] [PubMed] [Google Scholar]
- 11. Navarre WW, Zou SB, Roy H, Xie JL, Savchenko A, Singer A, Edvokimova E, Prost LR, Kumar R, Ibba M, Fang FC. 2010. PoxA, YjeK, and elongation factor P coordinately modulate virulence and drug resistance in Salmonella enterica. Mol. Cell 39:209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Dyk TK, Smulski DR, Chang YY. 1987. Pleiotropic effects of poxA regulatory mutations of Escherichia coli and Salmonella typhimurium, mutations conferring sulfometuron methyl and alpha-ketobutyrate hypersensitivity. J. Bacteriol. 169:4540–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zou SB, Hersch SJ, Roy H, Wiggers JB, Leung AS, Buranyi S, Xie JL, Dare K, Ibba M, Navarre WW. 2012. Loss of elongation factor P disrupts bacterial outer membrane integrity. J. Bacteriol. 194:413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bullwinkle TJ, Zou SB, Rajkovic A, Hersch SJ, Elgamal S, Robinson N, Smil D, Bolshan Y, Navarre WW, Ibba M. 2013. (R)-β-lysine-modified elongation factor P functions in translation elongation. J. Biol. Chem. 288:4416–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peil L, Starosta AL, Virumäe K, Atkinson GC, Tenson T, Remme J, Wilson DN. 2012. Lys34 of translation elongation factor EF-P is hydroxylated by YfcM. Nat. Chem. Biol. 8:695–697 [DOI] [PubMed] [Google Scholar]
- 16. Peng WT, Banta LM, Charles TC, Nester EW. 2001. The chvH locus of Agrobacterium encodes a homologue of an elongation factor involved in protein synthesis. J. Bacteriol. 183:36–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339:85–88 [DOI] [PubMed] [Google Scholar]
- 18. Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339:82–85 [DOI] [PubMed] [Google Scholar]
- 19. Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. 2002. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1:376–386 [DOI] [PubMed] [Google Scholar]
- 20. Ong SE, Foster LJ, Mann M. 2003. Mass spectrometric-based approaches in quantitative proteomics. Methods 29:124–130 [DOI] [PubMed] [Google Scholar]
- 21. Chang YY, Cronan JE., Jr. 1982. Mapping nonselectable genes of Escherichia coli by using transposon Tn10: location of a gene affecting pyruvate oxidase. J. Bacteriol. 151:1279–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. 2003. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4:3 http://dx.doi.org/10.1186/gb-2003-4-5-p3 [PubMed] [Google Scholar]
- 23. Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 25. Biter AB, Lee S, Sung N, Tsai FT. 2012. Structural basis for intersubunit signaling in a protein disaggregating machine. Proc. Natl. Acad. Sci. U. S. A. 109:12515–12520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rees DM, Montgomery MG, Leslie AG, Walker JE. 2012. Structural evidence of a new catalytic intermediate in the pathway of ATP hydrolysis by F1-ATPase from bovine heart mitochondria. Proc. Natl. Acad. Sci. U. S. A. 109:11139–11143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Niwa H, Tsuchiya D, Makyio H, Yoshida M, Morikawa K. 2002. Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8. Structure 10:1415–1423 [DOI] [PubMed] [Google Scholar]
- 28. Navarre WW, Halsey TA, Walthers D, Frye J, McClelland M, Potter JL, Kenney LJ, Gunn JS, Fang FC, Libby SJ. 2005. Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol. Microbiol. 56:492–508 [DOI] [PubMed] [Google Scholar]
- 29. Chang YY, Cronan JE., Jr. 1983. Genetic and biochemical analyses of Escherichia coli strains having a mutation in the structural gene (poxB) for pyruvate oxidase. J. Bacteriol. 154:756–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chiba S, Ito K. 2012. Multisite ribosomal stalling: a unique mode of regulatory nascent chain action revealed for MifM. Mol. Cell 47:863–872 [DOI] [PubMed] [Google Scholar]
- 31. Gong F, Yanofsky C. 2002. Instruction of translating ribosome by nascent peptide. Science 297:1864–1867 [DOI] [PubMed] [Google Scholar]
- 32. Sandler P, Weisblum B. 1989. Erythromycin-induced ribosome stall in the ermA leader: a barricade to 5′-to-3′ nucleolytic cleavage of the ermA transcript. J. Bacteriol. 171:6680–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cooper CA, Zhang K, Andres SN, Fang Y, Kaniuk NA, Hannemann M, Brumell JH, Foster LJ, Junop MS, Coombes BK. 2010. Structural and biochemical characterization of SrcA, a multi-cargo type III secretion chaperone in Salmonella required for pathogenic association with a host. PLoS Pathog. 6:e1000751 http://dx.doi.org/10.1371/journal.ppat.1000751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schmieger H. 1971. A method for detection of phage mutants with altered transducing ability. Mol. Gen. Genet. 110:378–381 [DOI] [PubMed] [Google Scholar]
- 35. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 http://dx.doi.org/10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Corcoran CP, Podkaminski D, Papenfort K, Urban JH, Hinton JC, Vogel J. 2012. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 84:428–445 [DOI] [PubMed] [Google Scholar]
- 39. Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24:79–88 [DOI] [PubMed] [Google Scholar]
- 40. Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 [DOI] [PubMed] [Google Scholar]
- 41. Sharp PM, Cowe E, Higgins DG, Shields DC, Wolfe KH, Wright F. 1988. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 16:8207–8211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zou SB, Roy H, Ibba M, Navarre WW. 2011. Elongation factor P mediates a novel post-transcriptional regulatory pathway critical for bacterial virulence. Virulence 2:147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850–858 [DOI] [PubMed] [Google Scholar]
- 44. Chan QW, Howes CG, Foster LJ. 2006. Quantitative comparison of caste differences in honeybee hemolymph. Mol. Cell. Proteomics 5:2252–2262 [DOI] [PubMed] [Google Scholar]
- 45. de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M. 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455:1251–1254 [DOI] [PubMed] [Google Scholar]
- 46. Cox J, Mann M. 2011. Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80:273–299 [DOI] [PubMed] [Google Scholar]
- 47. Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PoxB mRNA levels are similar in wild-type and ∆efp strains for both EF-P-dependent and -independent constructs. GFP fluorescence normalized to optical density (black), poxB mRNA transcript levels (light grey), and sfGFP mRNA transcript levels (dark grey) were assayed at mid-log phase (OD600 = 0.5; OD600 ≈ 0.2 as read in the Tecan Infinite M200 instrument). Transcript levels were tested by reverse transcriptase quantitative PCR using gene-specific primers and the technique used previously by Zou et al. (2012) (see reference 13 in the main article). Since the strains used carry poxB in their genomes, the mRNA levels obtained for lacZ186 with the poxB-specific primers were subtracted as background from all other values. Of note, under the conditions tested, the background poxB mRNA level was negligible (greater than 300-fold lower) compared to those for any of the strains expressing PoxB-GFP from a plasmid. All values are shown as the ratios of the result for the efp mutant relative to that for the wild-type strain expressing the same construct. All values are the averages for at least 3 biological replicates with the exception of those for the LacZ186 construct, which are the averages for 2 biological replicates. For the replicate where LacZ186 was not included, background poxB mRNA levels were obtained from another strain expressing a non-poxB construct from the same plasmid. Each error bar shows 1 standard deviation. Download
PoxB-sfGFP translation is impaired in ∆poxA, ∆yjeK, and ∆efp strains but not in the ∆yfcM mutant. Fluorescence of sfGFP fusion constructs in mutant strains lacking an EF-P-modifying protein is shown. Mutant strains are indicated in the top row of the x axis; the sfGFP fusion construct (as outlined in Fig. 3 in the main article) are indicated in the bottom row of the x axis. Values were taken at 10 h postinoculation and are shown in arbitrary fluorescence units (AU) normalized to the optical density at 600 nm (OD600). The “no plasmid” controls do not contain an sfGFP fusion plasmid and were grown in the absence of chloramphenicol. All values are the averages for at least 3 biological replicates. Each error bar shows 1 standard deviation. Download
EF-P regulation of PoxB is determined by the amino acid sequence as opposed to the nucleotides. (A) Nucleotide (upper line) and amino acid (lower line) sequences of the PoxB FL and PoxB Shifted constructs are shown from residues 66 to 85. The PoxB Shifted construct was generated by shifting codons 69 to 82 (bold) of full-length PoxB to the +1 reading frame by introducing an upstream single-base-pair insertion (underlined adenine) and downstream deletion (struck-out cytosine). (B) Relative GFP fluorescence values taken at 10 h postinoculation, normalized to the optical density at 600 nm (OD600) and shown relative to values for the wild-type strain expressing the same construct. P76::CCX constructs are mutated at proline 76 to an alternate proline codon. LacZ186, PoxB FL, PoxB 82, and PoxB 68 are shown for comparison. All values are the averages for at least 3 biological replicates. Each error bar shows 1 standard deviation. Download
PoxB synthesis demonstrates a similar dependence on EF-P when assayed during mid-log phase. Relative GFP fluorescence of PoxB constructs shown at mid-log phase instead of at 10 h postinoculation. Values were taken at the first time point after the culture surpassed an OD600 of 0.2 (mid-log phase as assayed in a Tecan Infinite M200 reader), normalized to the OD600, and are shown relative to those for the wild-type strain expressing the same construct. Data were obtained from the same biological replicates used to generate Fig. 3 in the main text. All values are the averages for at least 3 biological replicates. Each error bar shows 1 standard deviation. Download
Strains, plasmids, and primers
All proteins and expression ratios identified in SILAC
Bioinformatic identification of tripeptide motifs enriched within the 10% of proteins that were most upregulated in the efp mutant (lowest WN1269/WN1308 expression ratio)
DAVID cluster analysis of proteins significantly (significance B value [SigB] < 0.01) downregulated (expression ratio > 2) in WN1308
DAVID cluster analysis of proteins containing a PPP, PPG, or APP motif
DAVID cluster analysis of proteins significantly (SigB < 0.01) downregulated (expression ratio > 2) in WN1308 that also contain a PPP, PPG, or APP motif