Abstract
Esophageal cancer is the eighth most common cancer in the world and has an extremely dismal prognosis, with a 5-year survival of less than 20%. Current treatment options are limited, and thus identifying new molecular targets and pathways is critical to derive novel therapies. Worldwide, more than 90% of esophageal cancers are esophageal squamous cell cancer (ESCC). Previously, we identified that Krüppel-like factor 5 (KLF5), a key transcriptional regulator normally expressed in esophageal squamous epithelial cells, is lost in human ESCC. To examine the effects of restoring KLF5 in ESCC, we transduced the human ESCC cell lines TE7 and TE15, both of which lack KLF5 expression, with retrovirus to express KLF5 upon doxycycline induction. When KLF5 was induced, ESCC cells demonstrated increased apoptosis and decreased viability, with up-regulation of the proapoptotic factor BAX. Interestingly, c-Jun N-terminal kinase (JNK) signaling, an important upstream mediator of proapoptotic pathways including BAX, was also activated following KLF5 induction. KLF5 activation of JNK signaling was mediated by KLF5 transactivation of two key upstream regulators of the JNK pathway, ASK1 and MKK4, and inhibition of JNK blocked apoptosis and normalized cell survival following KLF5 induction. Thus, restoring KLF5 in ESCC cells promotes apoptosis and decreases cell survival in a JNK-dependent manner, providing a potential therapeutic target for human ESCC.
Introduction
Esophageal cancer is the eighth most common cancer in the world, with more than 480,000 new cases annually, and is responsible for more than 400,000 deaths, making esophageal cancer the sixth most common cause of cancer death [1]. Worldwide, more than 90% of esophageal cancers are esophageal squamous cell cancer (ESCC) [2]. Despite improvements in surgical therapy, ESCC still has a 5-year survival rate below 20% [2,3]. Neoadjuvant chemotherapy has been proposed to improve survival rates in selected patients [4], but targeted therapies for ESCC are still lacking. Potentially, these treatments could be directed against factors and pathways involved in cell proliferation and/or apoptosis, including targeting proapoptotic and antiapoptotic factors and various cell cycle regulators [5]. However, many of these factors, as well as the key epithelial transcriptional regulators underlying these processes have not yet been delineated.
Krüppel-like factor 5 (KLF5) is a DNA-binding transcriptional regulator highly expressed in epithelial cells, including in the proliferating basal layer of the esophagus [6,7]. Within basal epithelial cells, KLF5 controls normal proliferation and migration, but KLF5 expression is lost in ESCC [8–10]. In ESCC cells, KLF5 expression inhibits proliferation, promotes apoptosis, and decreases invasion [11]. Interestingly, KLF5 loss alone in the context of p53 mutation can transform primary human esophageal keratinocytes, demonstrating an important function for KLF5 in the development of human ESCC [9]. p53 mutation also appears to be critical for the context-dependent role of KLF5 on proliferation seen in esophageal and other epithelia [12,13]. KLF5 effects on cell transformation and invasion appear to be mediated by direct transcriptional regulation of the tumor suppressor NOTCH1 [9,14]. Yet, while the mechanisms of KLF5 function in ESCC proliferation and invasion are beginning to be elucidated, less is understood about the effects on apoptosis. Notably, KLF5 does not trigger apoptosis in normal esophageal epithelial cells [12]. In ESCC cells, KLF5 induces the proapoptotic factor BAX following UV irradiation, but the mechanism of this induction is not known [11]. Since Klf5 overexpression has few consequences in normal esophageal epithelia [7] and KLF5 appears to be silenced epigenetically in at least a subset of ESCC [9], reactivation of KLF5 or otherwise restoring KLF5 is enticing as a therapeutic approach for ESCC. In addition, KLF5 loss has been implicated in several other cancers, including those of the breast and prostate [15,16], and restoring KLF5 expression may therefore be beneficial in these tumors as well.
The c-Jun N-terminal kinase ( JNK) pathway, a subgroup of the mitogen-activated protein kinase (MAPK) superfamily, is an important stress-induced proapoptotic pathway upstream of BAX [17–19]. The MAPK kinases (MAP2Ks) MKK4 and MKK7 phosphorylate and activate JNK [17,20,21] and are a “bottleneck” for JNK signaling [21]. In turn, MKK4 and MKK7 are activated by ASK1, a MAPK kinase kinase (MAP3K) induced by various types of cellular stress [22]. The response to JNK activation, however, is influenced by the duration of activation, with short-term activation leading to increased cell survival, while prolonged activation induces proapoptotic pathways [23]. Thus, prolonged activation of JNK in cancer, as by the up-regulation of key upstream regulators, could be a valuable therapeutic approach [24]. As such, an understanding of the transcriptional regulation of these upstream kinases is essential.
Here, we employ an inducible retroviral system to express KLF5 in human ESCC cells. We demonstrate that restoring KLF5 induces apoptosis and diminishes cell survival in ESCC. Moreover, we define JNK activation as critical for the proapoptotic function of KLF5 in ESCC.
Methods
Cell Culture
The human ESCC cell lines TE7 and TE15 [25,26] were cultured at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium/F12 media (Life Technologies, Grand Island, NY) supplemented with 5% BSA (Life Technologies), 100 units/ml penicillin, and 100 µg/ml streptomycin (Life Technologies). For JNK inhibition, SP600125 (Enzo Life Sciences, Farmingdale, NY) was dissolved in DMSO, and cells were treated at 10 µM for 0, 4, 8, and 24 hours. To block MKK4 phosphorylation, cells were treated for 5 hours with 50 µM PD98059 (Sigma-Aldrich, St Louis, MO), a potent MAP2K inhibitor [27,28], solubilized in DMSO.
Viral Constructs and Infection
KLF5 cDNA was subcloned into the inducible pRevTre retroviral vector (Clontech, Mountain View, CA). pRevTre and pRevTet-on retroviral vectors were packaged by transfecting into AmphoPhoenix cells (National Gene Vector Biorepository, Indianapolis, IN) using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. Virus-containing media were harvested 48 and 72 hours after transfection and filtered with a 0.45-µM MicroFunnel Filter (Paul Life Sciences, Port Washington, NY), aliquoted, and stored at -80°C until needed. TE7 and TE15 cells were infected with culture supernatants from induced AmphoPhoenix cells at a 1:6 dilution. Cells were passaged for 24 hours and selected with 400 µg/ml G418 (Life Technologies) and 3 µg/ml hygromycin (Mediatech Inc, Manassas, VA) for 14 days. KLF5 was induced by treating cells with 4 µg/ml doxycycline.
RNA Analysis
RNA was extracted from ESCC cells using the RNeasy Mini Kit (Qiagen, Valencia, CA), and cDNA was synthesized with Superscript II Reverse Transcriptase (Life Technologies) following the manufacturer's instructions. Quantitative real-time polymerase chain reaction (qPCR) was carried out in triplicate on three samples for each experimental condition using an ABI StepOne Plus (Life Technologies) and SYBR Green PCR Master Mix (Life Technologies). TATA box-binding protein was used as internal control. Primer sequences are listed in Table W1.
Immunoblot Analysis
For each sample, 40 µg of total protein was separated on a NuPage 4% to 12% tris-acrylamide gel (Life Technologies) and transferred onto a polyvinylidene difluoride membrane (EMD Millipore, Billercia, MA), as described previously [8]. The membrane was blocked with 5% nonfat dry milk in tris-buffered saline with tween 20 (TBST) for 2 hours at room temperature and incubated overnight at 4°C with 1:10,000 rabbit anti-KLF5 [8] or 1:1000 dilution of anti-cleaved caspase 3 (Cell Signaling Technology, Danvers, MA), anti-cleaved Poly (ADP-ribose) polymerase (PARP; Cell Signaling Technology), anti-phospho-JNK (Cell Signaling Technology), anti-JNK (Cell Signaling Technology), anti-Ask1 (Cell Signaling Technology), anti-phospho-MKK4 (Cell Signaling Technology), or anti-MKK4 (Cell Signaling Technology). Membranes were then incubated for 1 hour at room temperature with a 1:3000 dilution of anti-rabbit HRP (GE Healthcare Life Sciences, Piscataway, NJ) and developed with Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore). Rabbit anti-β-actin (Sigma-Aldrich) at 1:5000 served an internal control. Western blots were representative of three separate experiments.
MTT Assay
Cell growth rate was evaluated by MTT assay as described previously [11]. In brief, 1 x 104 cells were seeded onto each well of a 48-well plate. After 24 hours, KLF5 was induced with doxycycline. Medium was removed after an additional 24 and 48 hours, and cells were washed in phosphate-buffered saline. MTT reagent (USB, Cleveland, Ohio) was added at 2 mg/ml and incubated for 3 hours. The dark blue crystals formed were dissolved in DMSO and the absorbance measured at 570 nm with background subtracted at 650 nm in a Beckman DU 600 spectrometer. Results represented the mean of three separate experiments, each repeated in eight wells, and were expressed as mean of absorbance relative to time zero.
Annexin V Staining
Cells were plated onto four-well Lab-Tek chamber slides (Nunc, Rochester, NY), and KLF5 was induced with doxycycline. At 24 hours after induction, cells were washed with phosphate-buffered saline, and the Annexin V-FLUOS Staining Kit (Roche Applied Biosciences, Indianapolis, IN) was used for the detection of apoptotic cells as per the manufacturer's instructions. Slides were mounted with Prolong Gold with 4′,6-diamidino-2-phenylindole (DAPI) mounting medium (Life Technologies), and images were captured on a Nikon Eclipse E600 microscope (Nikon Instruments, Melville, NY) with a Photometrics CoolSNAP charge-coupled device camera (Roper Scientific, Tucson, AZ).
Luciferase Assay
Cells were induced with doxycycline and then transfected with pGL3-Bax luciferase reporter (gift of Dr Moshe Oren, Weizman Institute, Rehovot, Israel) [29] and pGL3-Bax-mut using Lipofectamine 2000 (Life Technologies), as per the manufacturer's instructions. pGL3-Bax-mut, containing a mutant KLF5 binding site, was created using the Stratagene QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) by mutating the sequence CCCCTCCCCT in pGL3-Bax to ATTTCTTTTC. Cells were lysed with passive lysis buffer, and luciferase reporter activity was analyzed with Dual-Luciferase Reporter Assay System (Promega, Madison, WI) on a Glomax Multi-Detection Luminometer System (Promega). Luciferase activity was normalized to renilla activity and expressed as relative luciferase activity.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assays were performed with ChIP Assay Kit (EMD Millipore) according to the manufacturer's instructions. Following KLF5 induction, cells were treated with 1% formaldehyde for 10 minutes to cross-link associated protein to DNA. Cells were lysed with sodium dodecyl sulfate buffer and sonicated with an Ultrasonic Processor (Sonics and Materials, Newtown, CT) for four sets of 20-second pulses at 30% power. After a 10-fold dilution, samples were precleared with protein A-agarose/salmon sperm DNA for 30 minutes at 4°C and incubated overnight at 4°C with 1:500 anti-KLF5 or 1:500 anti-rabbit IgG (Sigma-Aldrich), as a negative control. Cells were then precipitated with protein A-agarose for 1 hour, heated at 65°C for 4 hours, and treated with proteinase K. DNA was purified with the QiaQuick PCT Purification Kit (Qiagen), and PCR was performed for BAX, ASK1, and MKK4 using primers listed in Table W2. Putative binding sites were identified using the Transcription Element Search System [30].
Densitometry Analysis
Immunoblots were scanned on a CanoScanLide 50 scanner (Canon U.S.A., Lake Success, NY), and densitometry measurements of the scanned bands were performed using the digitalized scientific software program ImageJ (National Institutes of Health, Bethesda, MD). Data were normalized to β-actin and expressed as means ± SEM.
Statistical Analysis
Data were analyzed for statistical significance with the Student's paired t test using Excel (Microsoft, Seattle, WA) and expressed as means ± SEM. Values of P < .05 were considered statistically significant.
Results
KLF5 Decreases Viability and Induces Apoptosis in ESCC Cells
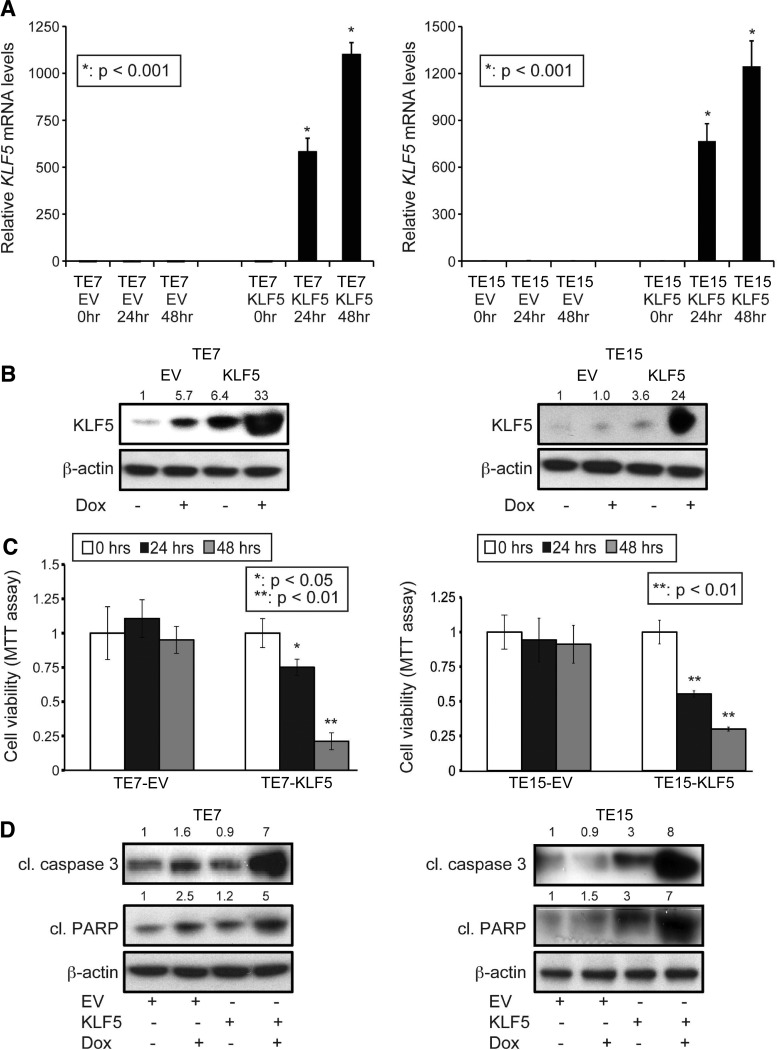
KLF5 expression is markedly decreased or absent in invasive ESCC and in a majority of human ESCC cell lines [9]. We hypothesized that loss of KLF5 was necessary for ESCC and that restoring KLF5 would have a negative effect on ESCC cell survival. To evaluate the role of KLF5 in ESCC cell survival, we stably infected the human ESCC cell lines TE7 and TE15, both of which have no detectable KLF5 expression [9], with doxycycline-inducible retroviral vectors to express KLF5. By quantitative PCR (Figure 1A) and immunoblot analyses (Figure 1B), we confirmed successful KLF5 expression following doxycycline treatment. To examine cell viability following KLF5 induction, we performed MTT assays. KLF5-expressing cancer cells showed a dramatic decrease in viability compared with controls [empty vector (EV); Figure 1C]. Importantly, KLF5 expression triggers considerable apoptosis in ESCC cells, as demonstrated by large increases in annexin V staining (Figure W1) and marked elevation of cleaved PARP [31] and cleaved caspase 3 [32], distinct executioners of the apoptotic machinery (Figure 1D).
Figure 1.
KLF5 decreases ESCC cell viability and induces apoptosis. (A) Stably infected TE7 and TE15 cells were treated with doxycycline for 24 and 48 hours, leading to KLF5 mRNA induction (*P < .001). (B) By Western blot, treatment of TE7 and TE15 cells with doxycycline for 24 hours induced KLF5 protein. (C) By MTT assay, KLF5 induction with doxycycline for 24 or 48 hours decreased ESCC cell viability (*P < .05; **P < .01). No significant changes in survival were seen with EV control. (D) Western blot demonstrated a marked increase in the apoptotic markers cleaved (cl) caspase 3 and cleaved (cl) PARP following 24 hours of KLF5 induction.
KLF5 Upregulates BAX Expression in ESCC Cells
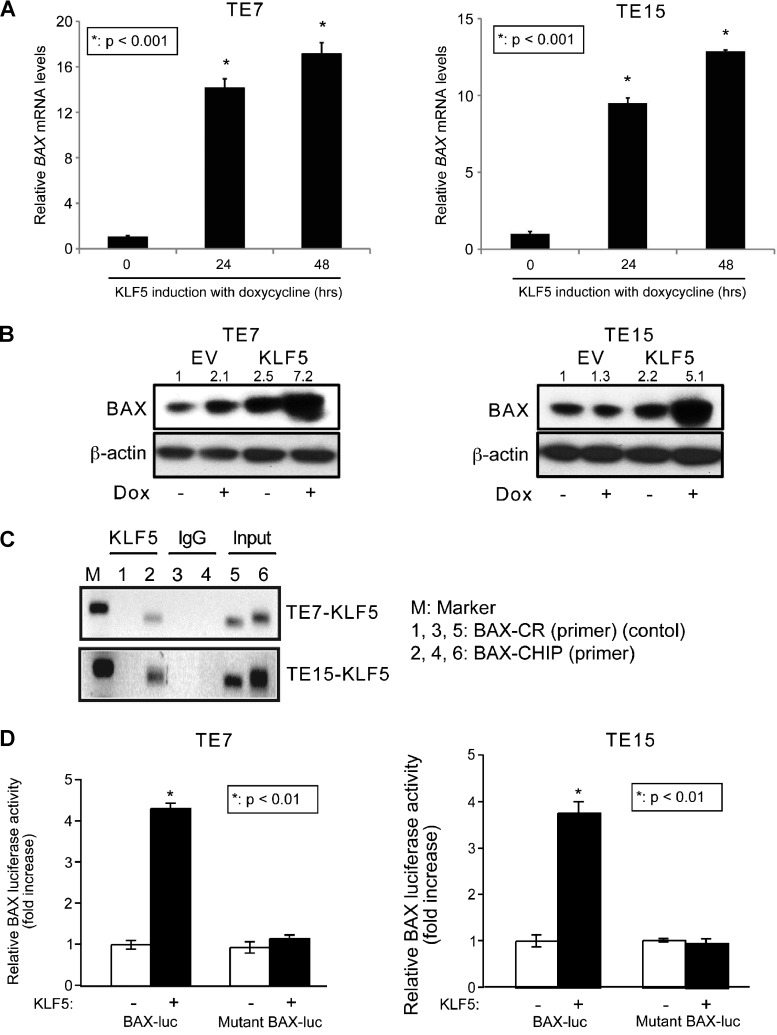
To define the mechanisms of increased apoptosis by KLF5 in ESCC, we focused initially on the proapoptotic Bcl-2 family member BAX, which has been shown to be upregulated by stable expression of KLF5 in ESCC cells [11]. However, the mechanism of BAX regulation by KLF5 is not known. Consistent with this, when KLF5 was induced by doxycycline in TE7 and TE15 ESCC cells, we observed marked induction of BAX, both at the RNA (Figure 2A) and protein (Figure 2B) levels. Using the Transcription Element Search System [30], we identified a putative KLF5 binding site between -980 and -971 upstream of the BAX translational start site. By ChIP assay, KLF5 bound to the 5′ regulatory region of BAX within the region of the putative KLF5 binding site (Figure 2C). Luciferase reporter assays demonstrated BAX trans-activation upon KLF5 induction in TE7 and TE15 cells, and this activation was completely lost following mutation of the KLF5 binding site (Figure 2D).
Figure 2.
KLF5 transactives BAX in human ESCC cells. (A) KLF5 induction with doxycycline for 24 and 48 hours in TE7 and TE15 ESCC cell lines increased BAX mRNA (*P < .001). (B) KLF5 induction also increased BAX protein levels at 24 hours. (C) ChIP assays demonstrated KLF5 binding to the 5′ regulatory region of BAX. IgG served as a negative control, and input DNA was a positive control. BAX ChIP primers spanned the region from -1047 to -931 upstream of the translation start site and control primers spanned the region from -952 to -785. (D) In ESCC cells, BAX promoter activity, assessed with a BAX-luciferase reporter, was increased four-fold by KLF5 following 24 hours of induction; mutation of the putative KLF5 binding site on BAX abolished this increase (*P < .01).
KLF5 Activates JNK Signaling in ESCC Cells
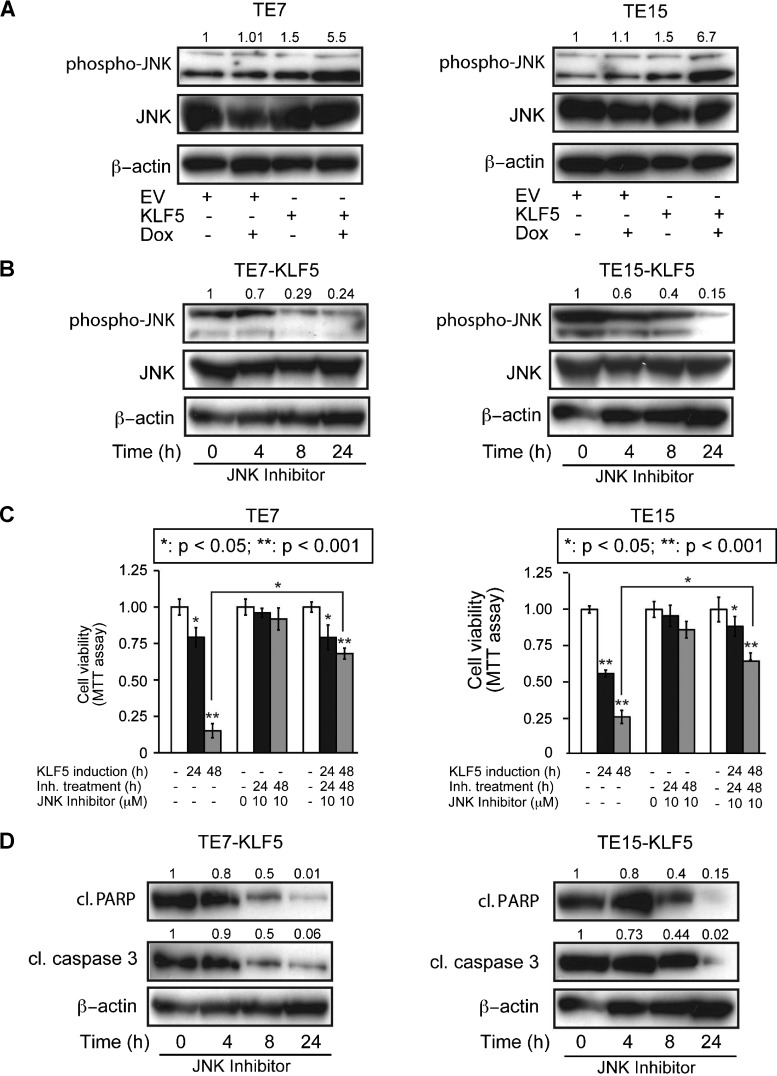
JNK signaling, a subset of the MAPK pathway, triggers apoptosis in response to stress, reactive oxygen species, and other signals [17–19]. We hypothesized that the JNK pathway is activated by KLF5 in ESCC cells, contributing to the increased apoptosis following KLF5 induction in ESCC cells. In support of this, KLF5 induction increased phosphorylated JNK but did not alter levels of total JNK in TE7 and TE15 cells (Figure 3A). Treatment of cells with the small molecule, ATP-competitive JNK inhibitor SP600125 [33,34] successfully blocked JNK phosphorylation upon KLF5 induction (Figure 3B). These data suggested that KLF5 activated JNK signaling upstream of JNK and not by transcriptional regulation of JNK.
Figure 3.
KLF5 activates JNK signaling in ESCC. (A) By Western blot, phosphorylation of JNK increased five-fold to seven-fold in TE7 and TE15 cells after KLF5 induction for 24 hours, while total JNK was unchanged. (B) Treatment of TE7 and TE15 cells with the small molecule JNK inhibitor SP600125 blocked JNK phosphorylation following KLF5 induction, as indicated by Western blot. (C) When TE7 and TE15 were induced with doxycycline for 24 or 48 hours to express KLF5, treatment with JNK inhibitor inhibited the ability of KLF5 to decrease cell viability, as assessed by MTT assay (*P < .05; **P < .001). (D) Treatment with JNK inhibitor also blocked the proapoptotic effects of KLF5 in TE7 and TE15 cells, as demonstrated by levels of cleaved (cl) caspase 3 and cleaved (cl) PARP. KLF5 was induced for the indicated times.
To determine the role of KLF5-mediated JNK activation in ESCC cells, we examined the impact of JNK inhibition on ESCC cell viability and apoptosis following KLF5 induction. Interestingly, treatment of TE7 and TE15 cells with SP600125 following KLF5 induction resulted in markedly elevated cell viability, compared to cells with KLF5 induction alone (Figure 3C); these effects were not seen with JNK inhibition alone, indicating that changes in cell viability were not due to the inhibitor itself. JNK inhibition also decreased apoptosis following KLF5 induction, as indicated by reduced expression of cleaved PARP and cleaved caspase 3 (Figure 3D). Of note, changes in the expression of apoptotic markers appeared to precede changes in cell viability; this may be due to the time required for full activation of apoptotic pathways or to limitations in the ability of the MTT assay to detect changes in cell viability in real time. KLF5 induction also altered the expression of several other apoptotic and survival factors (Figure W2), providing a potential explanation for the failure of JNK inhibition to fully restore ESCC cell viability following KLF5 induction, and KLF5 decreased expression of the KLF family member KLF4, particularly relevant since KLF5 and KLF4 may be yin-yang partners [35]. Nonetheless, JNK activation by KLF5 upstream of BAX played an important role in the apoptotic response.
KLF5 Regulates Upstream Mediators of JNK Signaling
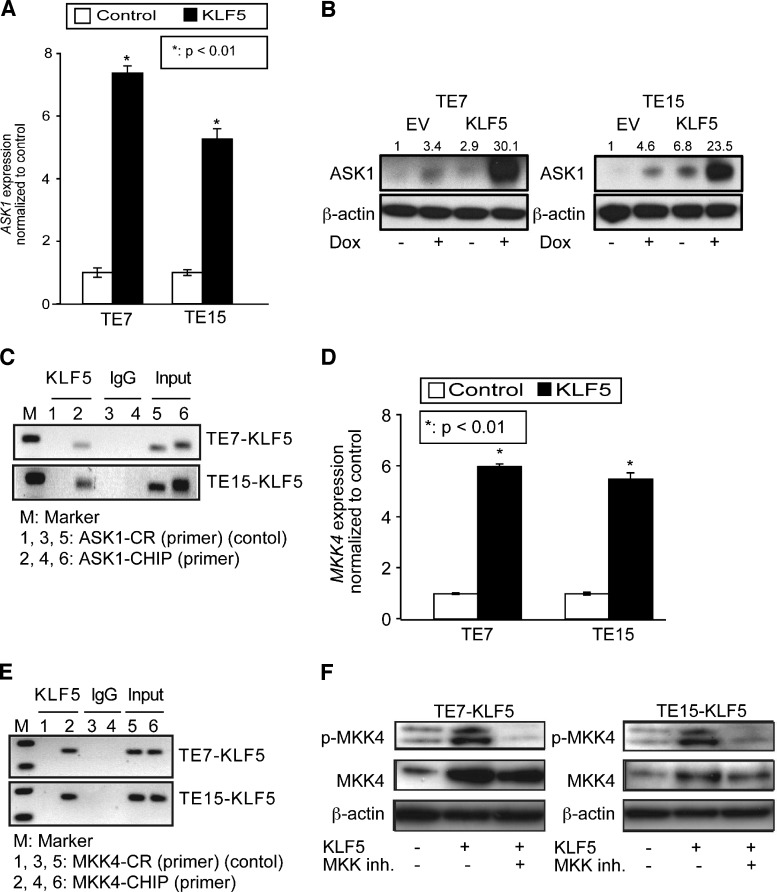
Since JNK signaling is activated at the posttranslational level [19,36], the mechanism of JNK activation by KLF5 is likely indirect. Consistent with this, KLF5 upregulates phospho-JNK but not total JNK. To identify the mechanism of JNK pathway regulation in ESCC cells by KLF5, we examined levels of MKK4 and MKK7, the predominant MAP2Ks upstream of JNK [37], and ASK1, a MAP3K that can directly phosphorylate MKK4 and MKK7 [38]. Of note, different MAP3Ks predominate in the activation of MKKs and JNK in response to various stimuli [18]. Interestingly, KLF5 induction in TE7 and TE15 cells resulted in increased expression of both ASK1 mRNA (Figure 4A) and protein (Figure 4B). To determine whether ASK1 was a direct transcriptional target for KLF5, we examined the 5′ regulatory region of ASK1 for putative KLF5 binding sites. We identified a single putative KLF5 binding site from -449 to -437 upstream of the translation start site and, by ChIP assay, demonstrated KLF5 binding to ASK1 in the vicinity of this putative binding site (Figure 4C).
Figure 4.
KLF5 upregulates upstream mediators of the JNK pathway. (A and B) When KLF5 was induced for 24 hours in TE7 and TE15 ESCC cells, levels of ASK1 mRNA (A) and protein (B) increased. (C) ChIP assays demonstrated KLF5 binding to the 5′ regulatory region of ASK1, in the vicinity of a predicted KLF5 binding site. IgG was a negative control, and input DNA served as a positive control. ASK1 ChIP primers spanned the region from -502 to -280 upstream of the translation start site and control primers spanned the region from -1833 to -1653. (D) By qPCR, KLF5 induction for 24 hours in ESCC cells resulted in a six-fold increase in MKK4 mRNA expression as demonstrated by qPCR. (E) KLF5 bound to a region on MKK4 predicted to contain multiple KLF5 binding sites. IgG and input DNA served as controls. Primers for MKK4 ChIP and control spanned the regions -226 to +4 and -1436 to -1266, respectively, upstream of the translation start site. (F) As seen on Western blot, MKK phosphorylation was increased following KLF5 induction for 24 hours; this increase was blocked by treatment with the MAP2K inhibitor PD98059. Note that total MKK4 is also increased by KLF5 induction, indicative of both transcriptional and posttranslational regulation of MKK4.
The ASK1 target MKK4 was also increased at both the mRNA (Figure 4D) and protein levels (Figure 4E) following KLF5 induction. However, no significant increase in MKK7 was observed upon KLF5 induction (Figure W3), indicating the specificity for MKK4. Surprisingly, by ChIP (Figure 4E), KLF5 bound to the 5′ regulatory region of MKK4 in an area from -126 to -72 predicted to have six KLF5 binding sites. At the protein level, KLF5 induction increased both total MKK4 and MKK4 phosphorylation (Figure 4F), the former likely by direct transactivation of MKK4 and the latter through ASK1 up-regulation. Consistent with this, treatment of cells with PD98059, a small molecule inhibitor of MKK4 phosphorylation, blocked MKK4 phosphorylation but did not affect total MKK4.
Discussion
The development and progression of cancers, including ESCC, require several key steps including alteration in the control of cell proliferation, survival, metastasis, and evasion of apoptosis [39]. Recently, we defined KLF5 loss as a key step in the development of ESCC [9] and identified KLF5, through the cyclin-dependent kinase inhibitor p21Waf1/Cip1, as an important brake on an aberrant cell cycle [12]. The functions of KLF5 in these processes are generally mediated by direct transcriptional regulation of its target genes, and KLF5 may have both transactivating and repressive functions [40]. Here, we define a novel and important function for KLF5 in the activation of JNK signaling to control ESCC cell viability and apoptosis. Of note, we have previously examined the effects of KLF5 on apoptosis in ESCC cells and found similar consequences [11], and subtle differences here may be due to inducible rather than constitutive KLF5 expression.
Transcriptional control of multiple steps in the JNK pathway by KLF5 is characteristic of a coherent feed-forward loop [41] and is indicative of the critical role of KLF5 in the regulation of this signaling network (Figure 5). When KLF5 is induced in ESCC cells, JNK inhibition substantially restores but does not fully rescue cell viability. These data suggest that, while JNK signaling is the major mediator of cell viability and apoptosis induced by KLF5 in ESCC cells, KLF5 transcriptional regulation of BAX and potentially other genes may be functionally relevant. In fact, we find that a number of other apoptotic and survival factors are also altered by KLF5 induction in ESCC cells. In addition, ASK1 and MKK4 can also activate p38 MAPK [37,38], and PD98059 can also inhibit other MAP2Ks [27]. As such, future studies will be directed toward understanding the role of KLF5 in the activation of other MAPK pathways in ESCC and in the transcriptional regulation of other proapoptotic and antiapoptotic factors.
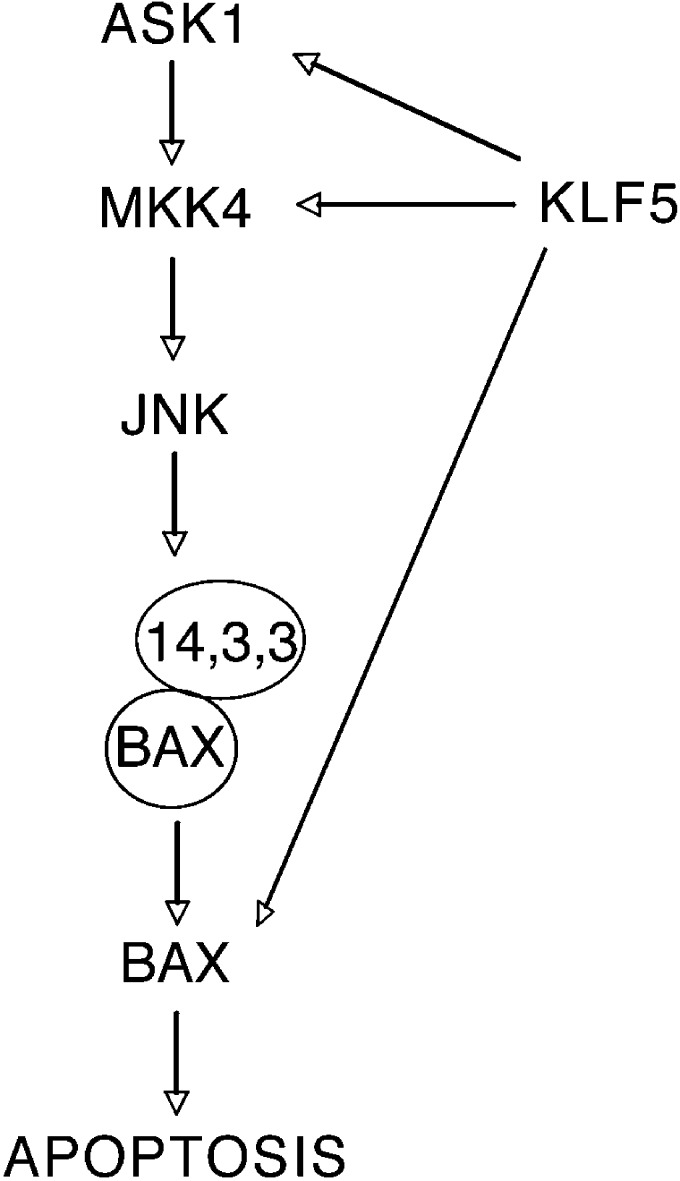
Figure 5.
A model for the effects of KLF5 on cell survival and apoptosis in ESCC. When KLF5 is restored in ESCC cells, KLF5 trans-activates ASK1 and MKK4 to activate the JNK pathway. Activated JNK signaling then phosphorylates 14-3-3 proteins, leading to BAX release and translocation to the mitochondria, to promote apoptosis. In addition, KLF5 can directly transactivate BAX to increase BAX levels.
BAX is activated in response to multiple proapoptotic stimuli and mediates apoptosis through the intrinsic pathway [42]. Proapoptotic stimuli can also activate the JNK pathway, leading to phosphorylation of the BAX repressor 14-3-3, thereby liberating BAX to initiate the apoptotic machinery [43,44]. While JNK signaling is often proapoptotic, the function of JNK, like KLF5, can depend on context [17,45]. p53 status is critical for determining KLF5 function [9,12], and the antiapoptotic function of JNK may be related to p53 status [46]. For example, JNK inhibition suppresses growth and induces apoptosis of human tumor cells in a p53-dependent manner [47]. KLF5 does not trigger apoptosis in nontransformed esophageal epithelial cells [12], and the differences of KLF5 function in these contexts could depend on p53 status as well. These context-dependent functions of KLF5 and JNK on apoptosis merit further study.
In sum, we have defined a novel role for KLF5 in ESCC, an extremely common cancer worldwide with a particularly poor prognosis. Importantly, KLF5 overexpression does not produce dysplasia or cancer in normal esophageal epithelia [7]. In ESCC, KLF5 expression is typically lost, and we demonstrate here that KLF5 inversely affects ESCC cell survival in a JNK-dependent manner, although the effects of KLF5 on apoptosis may be greater than can be attributed to JNK activation alone. This suggests that loss of KLF5 may be necessary for the development and progression of ESCC, and restoring KLF5 function in ESCC may provide a novel therapeutic approach for this deadly cancer. Future investigations will be directed toward fully defining the factors and pathways downstream of KLF5 to better delineate the molecular mechanisms underlying the pathogenesis of ESCC.
Supplementary Material
Abbreviations
- ChIP
chromatin immunoprecipitation
- ESCC
esophageal squamous cell cancer
- JNK
c-Jun N-terminal kinase
- KLF5
Krüppel-like factor 5
- MAP2K
mitogen-activated protein kinase kinase
- qPCR
quantitative real-time polymerase chain reaction
Footnotes
This work was supported by National Institutes of Health (NIH), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01 DK080031 and DK080031-02S1 to J.P.K., by the University of Pennsylvania Center for Molecular Studies in Digestive and Liver Diseases (NIH, NIDDK P30 DK050306) through the Molecular Pathology and Imaging Core, the Cell Culture Core, and the Molecular Biology/Gene Expression Core, and by NIH, NIDDK P01 CA098101 (“Mechanisms of Esophageal Carcinogenesis”).
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 to W3 and are available online at www.neoplasia.com.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 4.van Hagen P, Hulshof MCCM, van Lanschot JJB, Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BPL, Richel DJ, Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med. 2012;366:2074–2084. doi: 10.1056/NEJMoa1112088. [DOI] [PubMed] [Google Scholar]
- 5.Lin J, Beerm DG. Molecular biology of upper gastrointestinal malignancies. Semin Oncol. 2004;31:476–486. doi: 10.1053/j.seminoncol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 6.McConnell BB, Ghaleb AM, Nandan MO, Yang VW. The diverse functions of Krüppel-like factors 4 and 5 in epithelial biology and pathobiology. Bioessays. 2007;29:549–557. doi: 10.1002/bies.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein BG, Chao HH, Yang Y, Yermolina YA, Tobias JW, Katz JP. Overexpression of Kruppel-like factor 5 in esophageal epithelia in vivo leads to increased proliferation in basal but not suprabasal cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1784–G1792. doi: 10.1152/ajpgi.00541.2006. [DOI] [PubMed] [Google Scholar]
- 8.Yang Y, Goldstein BG, Nakagawa H, Katz JP. Krüppel-like factor 5 activates MEK/ERK signaling via EGFR in primary squamous epithelial cells. FASEB J. 2007;21:543–550. doi: 10.1096/fj.06-6694com. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, Nakagawa H, Tetreault MP, Billig J, Victor N, Goyal A, Sepulveda AR, Katz JP. Loss of transcription factor KLF5 in the context of p53 ablation drives invasive progression of human squamous cell cancer. Cancer Res. 2011;71:6475–6484. doi: 10.1158/0008-5472.CAN-11-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Tetreault MP, Yermolina YA, Goldstein BG, Katz JP. Krüppel-like factor 5 controls keratinocyte migration via the integrin-linked kinase. J Biol Chem. 2008;283:18812–18820. doi: 10.1074/jbc.M801384200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y, Tarapore RS, Jarmel MH, Tetreault MP, Katz JP. p53 mutation alters the effect of the esophageal tumor suppressor KLF5 on keratinocyte proliferation. Cell Cycle. 2012;11:4033–4039. doi: 10.4161/cc.22265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bateman NW, Tan D, Pestell RG, Black JD, Black AR. Intestinal tumor progression is associated with altered function of KLF5. J Biol Chem. 2004;279:12093–12101. doi: 10.1074/jbc.M311532200. [DOI] [PubMed] [Google Scholar]
- 14.Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27:5115–5123. doi: 10.1038/onc.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21:6567–6572. doi: 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- 16.Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate. 2003;55:81–88. doi: 10.1002/pros.10205. [DOI] [PubMed] [Google Scholar]
- 17.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 18.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 19.Dhanasekaran DN, Reddy EP. JNK signaling in apoptosis. Oncogene. 2008;27:6245–6251. doi: 10.1038/onc.2008.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tournier C, Dong C, Turner TK, Jones SN, Flavell RA, Davis RJ. MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes Dev. 2001;15:1419–1426. doi: 10.1101/gad.888501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haeusgen W, Herdegen T, Waetzig V. The bottleneck of JNK signaling: molecular and functional characteristics of MKK4 and MKK7. Eur J Cell Biol. 2011;90:536–544. doi: 10.1016/j.ejcb.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Soga M, Matsuzawa A, Ichijo H. Oxidative stress-induced diseases via the ASK1 signaling pathway. Int J Cell Biol. 2012;2012:439587. doi: 10.1155/2012/439587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ventura JJ, Hubner A, Zhang C, Flavell RA, Shokat KM, Davis RJ. Chemical genetic analysis of the time course of signal transduction by JNK. Mol Cell. 2006;21:701–710. doi: 10.1016/j.molcel.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 25.Nishihira T, Hashimoto Y, Katayama M, Mori S, Kuroki T. Molecular and cellular features of esophageal cancer cells. J Cancer Res Clin Oncol. 1993;119:441–449. doi: 10.1007/BF01215923. [DOI] [PubMed] [Google Scholar]
- 26.Boonstra JJ, van der Velden AW, Beerens EC, van Marion R, Morita-Fujimura Y, Matsui Y, Nishihira T, Tselepis C, Hainaut P, Lowe AW, et al. Mistaken identity of widely used esophageal adenocarcinoma cell line TE-7. Cancer Res. 2007;67:7996–8001. doi: 10.1158/0008-5472.CAN-07-2064. [DOI] [PubMed] [Google Scholar]
- 27.Dudley DT, Pang L, Decker SJ, Bridges AJ, Saltiel AR. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee HY, Oh SH, Suh YA, Baek JH, Papadimitrakopoulou V, Huang S, Hong WK. Response of non-small cell lung cancer cells to the inhibitors of phosphatidylinositol 3-kinase/Akt- and MAPK kinase 4/c-Jun NH2-terminal kinase pathways: an effective therapeutic strategy for lung cancer. Clin Cancer Res. 2005;11:6065–6074. doi: 10.1158/1078-0432.CCR-05-0009. [DOI] [PubMed] [Google Scholar]
- 29.Friedlander P, Haupt Y, Prives C, Oren M. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Mol Cell Biol. 1996;16:4961–4971. doi: 10.1128/mcb.16.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schug J. Curr Protoc Bioinformatics. 2008. Using TESS to predict transcription factor binding sites in DNA sequence. Chapter 2, Unit 2.6. [DOI] [PubMed] [Google Scholar]
- 31.Soldani C, Scovassi AI. Poly(ADP-ribose) polymerase-1 cleavage during apoptosis: an update. Apoptosis. 2002;7:321–328. doi: 10.1023/a:1016119328968. [DOI] [PubMed] [Google Scholar]
- 32.Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15:725–731. doi: 10.1016/j.ceb.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 33.Bogoyevitch MA, Arthur PG. Inhibitors of c-Jun N-terminal kinases: JuNK no more? Biochim Biophys Acta. 2008;1784:76–93. doi: 10.1016/j.bbapap.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett BL, Sasaki DT, Murray BW, O'Leary EC, Sakata ST, Xu W, Leisten JC, Motiwala A, Pierce S, Satoh Y, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA. 2001;98:13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Krüppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sabapathy K. Role of the JNK pathway in human diseases. Prog Mol Biol Transl Sci. 2012;106:145–169. doi: 10.1016/B978-0-12-396456-4.00013-4. [DOI] [PubMed] [Google Scholar]
- 37.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 38.Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev. 2012;92:689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66:2691–2706. doi: 10.1007/s00018-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 42.Lindsay J, Esposti MD, Gilmore AP. Bcl-2 proteins and mitochondria—specificity in membrane targeting for death. Biochim Biophys Acta. 2011;1813:532–539. doi: 10.1016/j.bbamcr.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 43.Tsuruta F, Sunayama J, Mori Y, Hattori S, Shimizu S, Tsujimoto Y, Yoshioka K, Masuyama N, Gotoh Y. JNK promotes Bax translocation to mitochondria through phosphorylation of 14-3-3 proteins. EMBO J. 2004;23:1889–1899. doi: 10.1038/sj.emboj.7600194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nomura M, Shimizu S, Sugiyama T, Narita M, Ito T, Matsuda H, Tsujimoto Y. 14-3-3 Interacts directly with and negatively regulates pro-apoptotic Bax. J Biol Chem. 2003;278:2058–2065. doi: 10.1074/jbc.M207880200. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Lin A. Role of JNK activation in apoptosis: a double-edged sword. Cell Res. 2005;15:36–42. doi: 10.1038/sj.cr.7290262. [DOI] [PubMed] [Google Scholar]
- 46.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 47.Potapova O, Gorospe M, Dougherty RH, Dean NM, Gaarde WA, Holbrook NJ. Inhibition of c-Jun N-terminal kinase 2 expression suppresses growth and induces apoptosis of human tumor cells in a p53-dependent manner. Mol Cell Biol. 2000;20:1713–1722. doi: 10.1128/mcb.20.5.1713-1722.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.