Abstract
BIM is a proapoptotic member of the Bcl-2 family. Here, we investigated the epigenetic status of the BIM locus in NPM/ALK+ anaplastic large cell lymphoma (ALCL) cell lines and in lymph node biopsies from NPM/ALK+ ALCL patients. We show that BIM is epigenetically silenced in cell lines and lymph node specimens and that treatment with the deacetylase inhibitor trichostatin A restores the histone acetylation, strongly upregulates BIM expression, and induces cell death. BIM silencing occurs through recruitment of MeCP2 and the SIN3a/histone deacetylase 1/2 (HDAC1/2) corepressor complex. This event requires BIM CpG methylation/demethylation with 5-azacytidine that leads to detachment of the MeCP2 corepressor complex and reacetylation of the histone tails. Treatment with the ALK inhibitor PF2341066 or with an inducible shRNA targeting NPM/ALK does not restore BIM locus reacetylation; however, enforced expression of NPM/ALK in an NPM/ALK-negative cell line significantly increases the methylation at the BIM locus. This study demonstrates that BIM is epigenetically silenced in NPM/ALK-positive cells through recruitment of the SIN3a/HDAC1/2 corepressor complex and that NPM/ALK is dispensable to maintain BIM epigenetic silencing but is able to act as an inducer of BIM methylation.
Introduction
BIM is a proapoptotic Bcl-2 homology-only member of the Bcl-2 family. Its apoptotic activities are exerted through interactions with other proapoptotic and antiapoptotic Bcl-2 family proteins that result in activation of the proapoptotic proteins Bax and Bak [1]. In a mouse model, loss of both BIM alleles led to a marked protection from several proapoptotic stimuli in pre-T cells [2]. Notably, the protective effect in BIM+/- mice was intermediate between BIM-/- and BIM+/+, thus showing a “gene dosing” effect. In Eµ-Myc-transgenic mice, the inactivation of even asingle BIM allele accelerated Myc-induced development of tumors, particularly acute B cell leukemia, suggesting a key role for BIM as a tumor suppressor in B lymphocytes and showing that BIM is haploinsufficient [3]. A central role for BIM silencing in lymphoid malignancies was demonstrated in mantle cell lymphoma, in which BIM homozygous deletion was identified with high frequency using a genome-wide array-based comparative genomic hybridization analysis [4].
DNA methylation abnormalities have been identified as one of the most frequent epigenetic modifications in cancer. The DNA methylation profile of tumors is frequently characterized by global hypomethylation and simultaneous hypermethylation of CpG islands [5,6]. It is known that, when methylation is localized in the promoter [7], in the 5′ untranslated region (5′UTR), or in the first exons/introns [8–10] of a gene, methylation and transcription of that gene are usually inversely correlated. Methylation of tumor suppressor genes often leads to their down-regulation, potentially playing a pathogenic role in the development of cancer and in resistance to apoptosis-inducing drugs [11]. Methylation may inhibit transcription through two main mechanisms: either by directly blocking the binding of transcription factors to DNA or through recruitment of methyl-CpG-binding proteins (MBPs), which in turn recruit multiprotein corepressor complexes carrying histone deacetylase (HDAC) activity.
We and others recently demonstrated that BIM can be the target of epigenetic silencing in specific types of cancer as leukemia, lymphomas, and solid tumors, but the molecular mechanisms driving this phenomenon are poorly understood [12–18].
It is known that cells expressing the NPM/ALK fusion oncogene can drive BIM down-modulation through a phosphatidylinositide 3-kinase (PI3K)-mediated phosphorylation of the transcription factor FOXO3a [19], which is a known activator of BIM expression [20]. This phosphorylation causes FOXO3a translocation to the cytoplasm, thus causing a decrease of BIM expression. NPM/ALK transfection in Ba/F3 cells showed that FOXO3a phosphorylation can reduce BIM protein levels but is not able to completely abrogate its expression [19]. Since in cell lines derived from patients affected by NPM/ALK-positive anaplastic large cell lymphomas (ALCLs) BIM expression was dramatically reduced (R.G.P. and C.G.-P., unpublished observations), here we investigated the existence of additional FOXO3a-independent mechanisms responsible for strong BIM down-modulation in NPM/ALK-positive cells, demonstrating that BIM is epigenetically silenced through a SIN3a/HDAC1/2 corepressor complex in NPM/ALK-driven tumors and that this phenomenon can be reverted by deacetylase inhibitors or demethylating drugs, thus leading to apoptosis of NPM/ALK-positive cancer cells.
Materials and Methods
Cell Lines
All the cell lines were cultured in RPMI 1640 (BioWhittaker, Cambrex Biosciences, East Rutherford, NJ) supplemented with 10% FBS (BioWhittaker, Cambrex Biosciences), 100 Units/ml penicillin, 100 µg/ml gentamicin, and 2 mM L-glutamine (BioWhittaker, Cambrex Biosciences). The cells were cultured in a humidified incubator at 37°C in an atmosphere of 5% CO2. Trypan blue dye (Sigma, St Louis, MO) exclusion test was used to count viable cells.
Treatment with Methylase Inhibitors
Cells (106) for each cell line were seeded and treated with and without 1 µM 5-azacytidine (AZA) or 5-aza-deoxycytidine (dAZA; Sigma); fresh inhibitor was added every 48 hours. After 5 days of treatment, the cells were harvested for the analysis of BIM expression. Genomic DNA, RNA, and proteins were extracted using TRIzol Reagent (Life Technologies, Paisley, United Kingdom) according to the manufacturer's protocol.
Patients
Informed consent was obtained from each subject involved in the study. All the human investigations were performed in accordance with the principles embodied in the declaration of Helsinki.
Treatment with ALK Inhibitor
Cells (106 SU-DHL-1) were treated with and without 1.2 µM PF2341066 (Pfizer, New York, NY). After 5 hours, the acetylation status of histone H3 tails at the BIM locus was analyzed by chromatin immunoprecipitation (ChIP).
RNA Interference
Inducible silencing of NPM/ALK in SU-DHL-1 cells was generated as previously described [21]. Cells were analyzed after a 3-day incubation with 1 µg/ml doxycycline. For BIM silencing experiments, SU-DHL-1 cells were infected with lentivirus obtained from MISSION-shRNA pLKO-based vectors (Sigma; TRCN0000001054 NM_138621.X-522s1c1) and packaged using 293FT cell line. As a control, a pLKO. 1MISSION non-target control vector (Sigma; SHC002) was used.
Generation of an Inducible BIM Expression System
BimS cDNA was amplified from LAMA-84-S RNA using Titan OneTubeRT-PCRSystem(RocheApplied Science, Mannheim, Germany). The primers used were BimCI-EL-For 5′ATGGCAAAGCAACCTTCTGATG3′ and BimCI-EL-Rev 5′CTCCGCAAAGAACCTGTCAATG3′. BimS polymerase chain reaction (PCR) product was then cloned into PCR 4-TOPO (Life Technologies) to obtain PCR 4-TOPO-BimS. To set up an inducible system for BimS, the luciferase ORF was excised from pcDNA4/TO-luc and a multiple cloning site, excised from pcDNA3 with a HindIII/XbaI double digestion, was subcloned into the vector backbone, giving rise to pcDNA4/TO-MCS. BimS cDNA was excised from PCR 4-TOPO-BimS with an EcoRI digestion and inserted into pcDNA4/TO-MCS, linearized using the same restriction enzyme (pcDNA4/TO-MCS-BimS). KARPAS-299 cell lines stably expressing a Tet repressor (KARPAS299 TR1) encoded by pcDNA6/TR (Life Technologies) were transfected with pcDNA4/TO-MCS-BimS using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. KARPAS-299 double stable cell line (KARPAS-299 BimS) was selected with 10 µg/ml blasticidin and 100 µg/ml Zeocin (Life Technologies). BimS expression was confirmed by Western blot analysis and real-time PCR after induction with 1 µg/ml doxycycline (Clontech, Palo Alto, CA).
Generation of an NPM/ALK-Expressing LAMA-84 Cell Line
Cells (107) were electroporated with 30 µg of pcDNA3 plasmid containing wild-type NPM/ALK (pcDNA3-NA) obtained as previously described [22]. After electroporation, cells were allowed to recover for 48 hours and then selected with 800 µg/ml G418-additioned RPMI for 4 weeks.
Bisulfite Conversion
Two micrograms of genomic DNA was bisulfite-converted using a standard protocol. The bisulfite-modified DNA was amplified with a nested PCR protocol. The reactions were performed using FastStart Polymerase (Roche Applied Science). The primers used for the analysis were BIM-CG-Ext-For (5′GTGTGTATTTTAGAGAAGTT3′) and BIM-CG-Ext-Rev (5′CCTTCAAAATTACCTTATAAC3′) for the first PCR (0.4 µM each) and BIM-CG-For (5′GTTAGATTTTTTTTAGATTTGTTG3′) and BIM-CG-Rev (5′CAAACTACAATTATCTACCTTC3′) for the nested PCR (0.2 µM each). PCR amplifications were carried out on a Mastercycler Personal (Eppendorf, Hamburg, Germany).
PCR products were separated by electrophoresis on agarose gel, purified with QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany), and cloned using the TOPO Cloning Kit for sequencing (Life Technologies). Sequences were analyzed using Vector NTI 7.0 (Life Technologies).
Real-time Methylated DNA Immunoprecipitation Analysis
Twenty micrograms of genomic DNA extracted with TRIzol Reagent (Life Technologies) were sonicated using a Bandelin Sonoplus HD 2070 sonicator. The average size of the sheared chromatin was ∼500 bp. One microgram of sonicated DNA was immunoprecipitated over-night at 4°C with rotation using 30 µg of the sheep polyclonal 5-methyl-cytosine antibody ab1884 (Abcam, Cambridge, United Kingdom) in an immunoprecipitation (IP) buffer containing 0.01% sodium dodecyl sulfate (SDS; wt/vol), 1% Triton X-100 (wt/wt), 1.1 mM EDTA, 15 mM Tris-HCl, 150 mM NaCl, and 7 mM NaOH. The complex was immunoprecipitated using 60 µl of Salmon Sperm DNA/Protein A Agarose (Upstate, Lake Placid, NY) for 1 hour at 4°C with rotation. The immunocomplex was then washed once with 1 ml of 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 10 mM Tris-HCl, 150 mM NaCl (pH 8.1), once with 1 ml of 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 10 mM Tris-HCl, 500 mM NaCl (pH 8.1), once with 1 ml of 0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl (pH 8.1), and twice with 1 ml of 10 mM Tris-HCl, 1 mM EDTA (pH 8.0). The immunoprecipitated DNA was then eluted incubating twice with 250 µl of freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3) at room temperature for 15 minutes. The DNA was subsequently purified using QIAquick PCR Purification Kit (Qiagen) and quantified using SYBR Green quantitative PCR (QPCR). Five microliters of immunoprecipitated DNA and a corresponding amount of INPUT DNA were used. All the analyses were performed in triplicate. To assess the reproducibility of the data, methylated DNA immunoprecipitation (MeDIP) and real-time analyses were performed at least three times for every cell line. The resulting values were standardized using glyceraldehyde-3-phosphate dehydrogenase (GADPH) as a reference gene. The primers used for the analysis were BIM_mDip1Fw (5′ACTCGGTGAAGGATGATGCC3′) and BIM_mDip1Rev (5′CACACCCGTTAGAGCTTGGC3′); BIM_mDIP1.5Fw (5′AGGAACAGACGACAAGAAATAG3′) and BIM_mDIP1.5Rev (5′GGACACGGCTAAGTAGACTC3 ′); BIM_mDIP2bisFw (5′GGGAGGCTAGGGTACACTTCGG3′) and BIM_mDIP2bisRev (5′GCTCCTACGCCCAATCACTGC3′); BIM_mDIP3bisFw (5′AAGTCCTGCTTTGTCTCCAG3′) and BIM_mDIP3bisRev (5′AAGGCGAGGCGATTGTTGAC3′); BIM_mDIP4Fw (5′GCCTGCAATCGCTGCATCTG3′) and BIM_mDIP4Rev (5′GTCAACAGCTTGCGGAACTGG3′); GAPDH_SYBR3_For (5′TGCTTCTCTGCTGTAGGCTC3′) and GAPDH_SYBR3_Rev (5′AGCGTGTCCATAGGGTGCCA3′). All the primers were used at 160 nM final concentration and were tested for the absence of primer dimers and a specific products using a negative first-derivative melting curve analysis as well as direct visualization on agarose gel stained with ethidium bromide.
Methylation-specific PCR
Five microliters of bisulfite-treated DNA was amplified with a nested PCR protocol. The reactions were performed in 50-µl volume containing GeneAmp PCR buffer (Life Technologies), 200 µM each dNTP, 2.5U Amplitaq (Life Technologies). The primers used for the first reaction were BIM-MSPext-For (5′GGATTGGGTTTGGGGATGGTTT3′) and BIM-MSPext-Rev (5′ATCCCACAAACCCTCCCCTCAA3′). The product of the first reaction was diluted 1:10 and 1 µl of the dilution was used as template for the nested PCR, using the same enzyme. The primers used for the detection of the unmethylated and methylated DNA were, respectively, BIM-MSP-Uint-For (5′TTTTTGATGAAGTGGTAGTT3′) and BIM-MSP-Uint-Rev (5′AACAAAACCCAAAACTCAAA3′) and BIM-MSP-Mint-For (5′TTTTCGACGAAGCGGTAGTC3′) and BIM-MSP-Mint-Rev (5′AACAAAACCCGAAACTCGAA3′).
Chromatin Immunoprecipitation
ChIP was performed using a standard protocol. The immunoprecipitated DNA was amplified with FastStart Polymerase (Roche Applied Science) with a standard protocol. The primers used for the reactions were BIM-ChIP-Int-For 5′GCCTGCAATCGCTGCATCTG3′ and BIM-ChIP-Int-Rev 5′GTCAACAGCTTGCGGAACTGG3′ for BIM; GAPDH_ChIP_For 5′CCCAACTTTCCCGCCTCTC3′ and GAPDH_ChIP_Rev 5′CAGCCGCCTGGTTCAACTG3′ for GAPDH. Densitometric analyses were performed using Kodak 1D 3.5 Image System (Eastman Kodak Company, Rochester, NY).
Real-time QPCR
cDNA was synthesized from 200 ng of total RNA, using TaqMan Reverse Transcription Reagents (Life Technologies). The RNA from transfectant cells was pretreated with DnaseI (Life Technologies) to avoid contamination from genomic DNA. QPCR was performed using TaqMan Universal PCR Master Mix (Life Technologies) for TaqMan or with Brilliant SYBR Green QPCR Master Mix (Stratagene, La Jolla, CA) for SYBR Green real-time PCR on a 7900HT Sequence Detection System (Life Technologies) under standard conditions. All the QPCR experiments were performed in triplicate. The housekeeping gene β-glucuronidase (GUS) was used as an internal reference. The sequences of BIM and GUS probes were, respectively, BIMEx4_Rev Probe (5′FAM-CCGCAACTCTTGGGCGATCCATATCTCTCTAMRA3′) and GUS-probe (5′FAM-CCAGCACTCTCGTCGGTGACTGTTCA-TAMRA3′). The forward and reverse primers for BIM and GUS were, respectively, BIMEx4_For (5′TTCCATGAGGCAGGCTGAAC3′) and BIMEx8_Rev (5′GGTGGTCTTCGGCTGCTTGG3′) and GUS-for (5′GAAAATATGTGGTTGGAGAGCTCATT3′) and GUS-rev (5′CCGAGTGAAGATCCCCTTTTTA3′).
The primer pair for NPM/ALK SYBR Green QPCR analysis was NPM1-SYBR-for (5′TGCATATTAGTGGACAGCAC3′) and ALK1-SYBR-rev (5′CAGCTTGTACTCAGGGCTCTG3′).
Antibodies and Western Blot Analysis
Rabbit anti-BIM was purchased from Abcam (ab32158), anti-p-ERK (9101) was from Cell Signaling Technology (Danvers, MA), and anti-actin was from Sigma-Aldrich (St Louis, MO). Protein lysates, prepared as reported above, were separated by 15% SDS-polyacrylamide gel electrophoresis (Bio-Rad Laboratories, Palm Springs, CA) and blotted onto Hybond ECL nitrocellulose membrane (Amersham Biosciences, Uppsala, Sweden) following a standard protocol. Proteins were visualized by chemiluminescence (Super Signal; Pierce, Rockford, IL) with Image Station 440 CF (Eastman Kodak Company).
Detection of Apoptosis
Quantification of apoptotic cells was performed by TUNEL assay using DeadEnd Fluorometric terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) System (Promega, Madison, WI) using a standard protocol. Flow cytometry analysis was performed on a Becton Dickinson FACSort by CellQuest software (Becton Dickinson Immunocytometry Systems, Mountain View, CA).
Immunohistochemistry
Representative formalin-fixed paraffin-embedded tissue blocks were sectioned (2-µm thickness). Sections were dewaxed in xylene before rehydration through graded alcohol to water. Antigen retrieval was performed in autoclave at 95°C for 6 minutes in 0.05 M citrate buffer. The p80 anti-ALK polyclonal serum (Sanbio, Uden, The Netherlands) was used at a dilution of 1:10. A streptavidin-biotin system (Dako, Glostrup, Denmark) with development in aminoethyl-carbazole was applied.
Thymidine Uptake Assay
Exponentially growing SU-DHL-1 transfectants were plated in five replicates in 96-well plates at a density of 5000 cells/well. Cells were treated with 500 nM trichostatin A (TSA) or vehicle alone for 48 hours. [3H]thymidine (1 µCi; Amersham Biosciences) was added to each well 8 hours before harvesting onto glass fiber filters by a Tomtec automated cell harvester (Tomtec, Hamden, CT). Incorporation of [3H]thymidine was measured using a filter scintillation counter (1430 MicroBeta; Amersham Biosciences).
Results
Identification of BIM 5′UTR Methylation in ALCL Cells
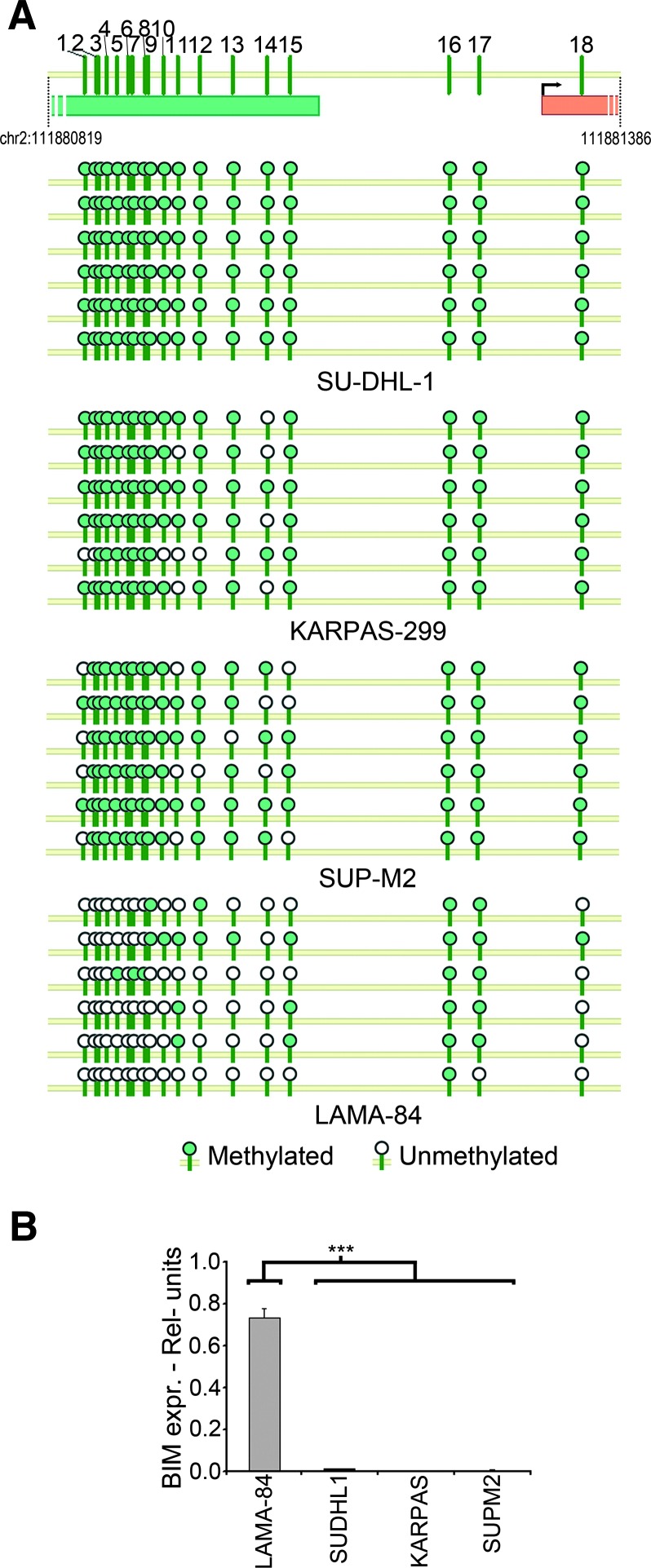
We analyzed the methylation status of 18 CpG sites in the 5′UTR of BIM locus, in the human NPM/ALK+ ALCL cell lines SU-DHL-1, KARPAS-299, and SUP-M2 and in the NPM/ALK-negative chronic myeloid leukemia cell line LAMA-84 as a negative control, using a bisulfite clonal sequencing technique. Globally, we identified a homogeneous, very high level of methylation (92.3%) in all the NPM/ALK+ cell lines (Figure 1A). In the SU-DHL-1 cell line, all the 18 CpG sites were found to be methylated. Only a limited evidence of CpG methylation (25.0%) could be detected in the NPM/ALK-negative LAMA-84 cell line (Figure 1A). This correlated with an almost complete silencing of BIM expression in all the NPM/ALK+ cell lines but not in LAMA-84, as assessed by QPCR (P < .0001; Figure 1B). Similar results were obtained with additional myeloid cell lines (Figure W1).
Figure 1.
(A) Methylation pattern of BIM 5′UTR in SU-DHL-1, KARPAS299, and SUP-M2 and in the NPM/ALK-LAMA-84. The horizontal 5′ bar represents part of the BIM CpG Island; the vertical ticks represent individual CpG sites; the horizontal 3′ bar indicates the first coding exon. Filled circles represent methylated CpG sites; white circles refer to unmethylated ones. Horizontal bullet series represent sequential CpG sites; vertical series show different clones. (B) BIM mRNA levels in the LAMA-84, SU-DHL-1, KARPAS-299, and SUP-M2 cell lines. QPCR quantification of BIM relative to the housekeeping gene GUS is presented. The error bars represent the SD of three replicates.
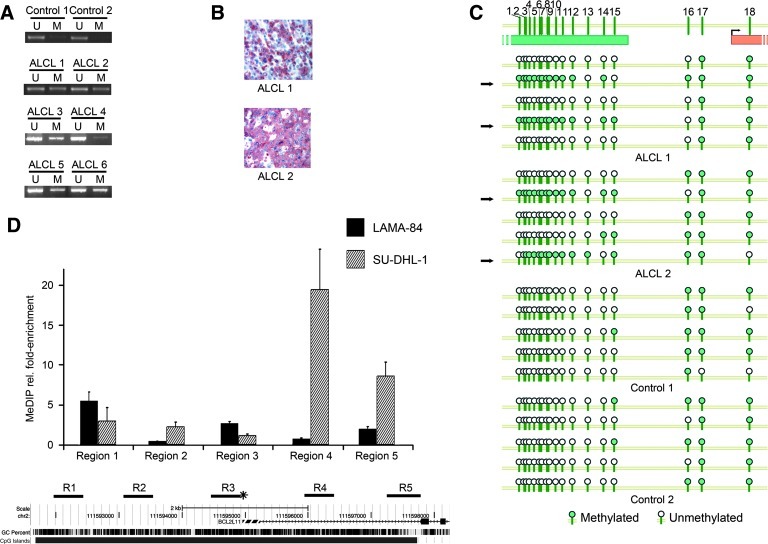
To assess whether BIM methylation was present also in NPM/ALK+ ALCL in vivo, biologic samples taken from lymph nodes of six patients affected by an NPM/ALK+ ALCL were analyzed using methylation-specific PCR (MSP). Despite the likely presence of a mixed population of neoplastic and normal cells in lymph nodes samples, as shown by immunohistochemistry, evidence for BIM methylation was detected in all the cases (Figure 2, A and B). Conversely, no evidence of BIM methylation could be found in lymphocytes from five healthy donors (Figure 2A; controls 1 and 2 are shown). To further characterize BIM methylation, two ALCL samples (samples 1 and 2) and lymphocytes from two healthy donors (controls 1 and 2) were also analyzed by bisulfite clonal sequencing (Figure 2C), confirming the presence of dense methylation in two of five clones in ALCL 1 and 2 (42.2% and 45.6% methylated CpG sites, respectively).
Figure 2.
(A) MSP analysis of BIM 5′UTR in six ALCL samples and in total lymphocytes from two healthy donors (controls 1 and 2). PCR products in lane U indicate the presence of unmethylated alleles, whereas PCR products in lane M indicate the presence of methylated alleles. (B) Immunoreactivity of neoplastic cells for anti-ALK p80 antibody (red) in lymph nodes of two NPM/ALK+ ALCL patients. (C) BIM methylation in lymph nodes from two primary ALCL patients (ALCL 1 and 2) and in total lymphocytes from two healthy donors (controls 1 and 2). Black arrows indicate highly methylated clones. (D) RT-MeDIP analysis of the BIM locus. In the upper panel, the results of the RT-MeDIP for SU-DHL-1 and LAMA-84 are shown. The histogram bars represent the relative enrichment for methylated DNA in five different regions of the BIM locus. In the lower panel, the schematic structure of the BIM genomic locus is shown: the gray boxes represent differently spliced exons; the striped gray box indicates the noncoding exon 1. The lower horizontal box represents the CpG island; the vertical black lines indicate the percentage of G and C bases in a five-base sliding window. The amplified regions are shown as thick black lines and labeled as regions R1 to R5. Asterisk indicates the first (5′) transcription start site.
To further analyze the methylation pattern of the whole BIM CpG island, we carried out real-time MeDIP (RT-MeDIP) profiling on SU-DHL-1 (NPM/ALK+) and LAMA-84 (NPM/ALK-) cell lines as a negative control. In SU-DHL-1, the analysis revealed a high enrichment for methylated DNA in the whole 5′UTR (Figure 2D; regions 4 and 5), while 5′ from the transcription start site (Figure 2D; regions 1, 2, and 3) the enrichment level quickly decreased to levels similar to the control. As expected, no significant enrichment was detected in LAMA-84.
Treatment with Demethylating Agents Leads to BIM Up-Regulation and Induction of Apoptosis
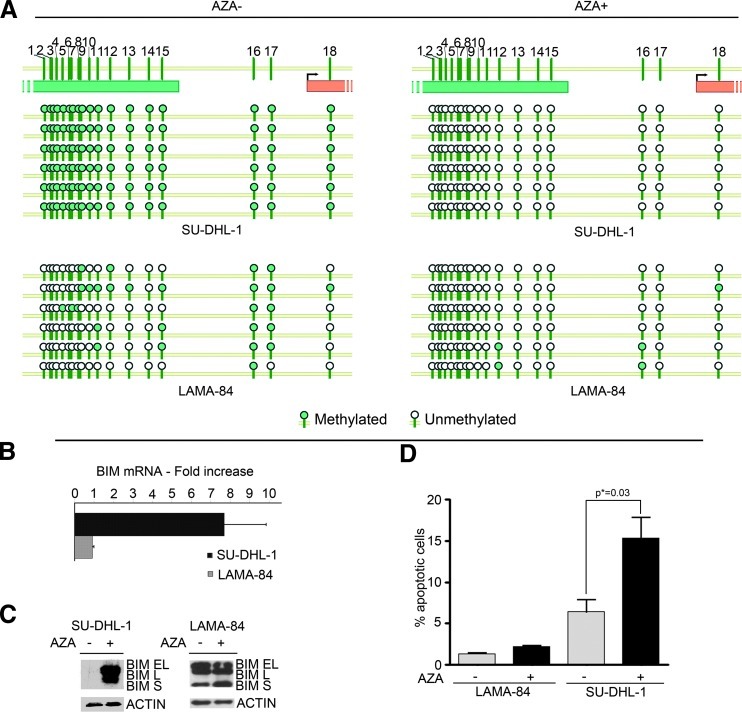
To characterize the contribution of BIM promoter methylation to gene silencing, the SU-DHL-1 and LAMA-84 cell lines were treated with the demethylating agent AZA. Following treatment, methylation decreased from 100% to 0% in SU-DHL-1, while BIM epigenetic status was minimally affected in LAMA-84 (Figure 3A). Change in the methylation pattern of BIM was associated with BIM up-regulation at mRNA (7.7-fold; Figure 3B) and protein levels (Figure 3C) in SU-DHL-1 cell line, whereas, as expected, treatment with AZA was unable to induce BIM up-regulation in LAMA-84 cells (Figure 3, B and C). This correlated with a significant induction of apoptosis in SU-DHL-1 (Figure 3D) but not in LAMA-84 (Figure 3D). Similar results were obtained on the NPM/ALK+ KARPAS-299 cell line, where the treatment with AZA led to a partial demethylation (Figure W2) and to a 2.9-fold increase in BIM expression (not shown).
Figure 3.
(A) BIM methylation pattern in SU-DHL-1 and LAMA-84 untreated (-) or treated (+) with AZA. Horizontal circle series represent sequential CpG sites; vertical series represent different clones. (B) QPCR analysis of BIM expression in SU-DHL-1 and LAMA-84 cell lines after treatment with 1 µM AZA. Results are shown as fold increase relative to the untreated samples. Error bars represent the SD of three replicates. The LAMA-84 cell line, in which BIM locus is largely unmethylated, is used as a control. (C) Western blot analysis of BIM expression in SU-DHL-1 and LAMA-84 cell lines untreated (-) or treated (+) with 1 µM AZA for 5 days. All the three major BIM isoforms are shown. (D) TUNEL assay of SU-DHL-1 and LAMA-84 cells untreated or treated with 1 µM AZA. The graph shows the percentage of apoptotic cells as an average of three independent experiments.
BIM Is Silenced through Histone Deacetylation and Chromatin Condensation
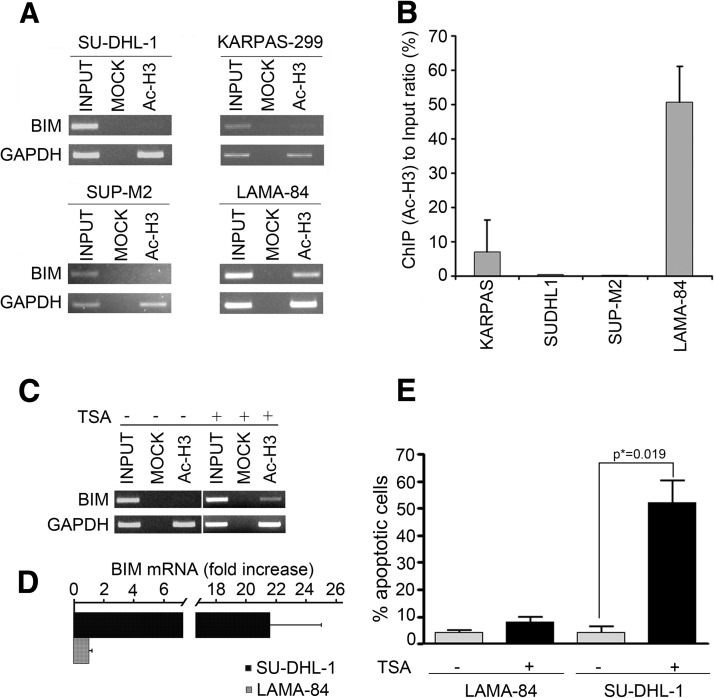
To assess whether deacetylation of histone tails and chromatin condensation are involved in BIM epigenetic silencing, we analyzed the acetylation status of histone H3 tails at the BIM locus in the NPM/ALK+ SU-DHL-1, KARPAS-299, and SUP-M2 and in the NPM/ALK-LAMA-84 cell lines. ChIP analysis showed that histone H3 tails at the BIM locus are strongly deacetylated in all the NPM/ALK+ cell lines (Figure 4, A and B). Conversely, abundant histone H3 tail acetylation was apparent in LAMA-84 cells (Figure 4, A and B).
Figure 4.
(A) α-Acetylated H3 ChIP analysis on SU-DHL-1, KARPAS-299, SUP-M2, and LAMA-84 cell lines. Immunoprecipitates (Ac-H3) were subjected to PCR with primer pairs specific for BIM and for GAPDH, as a positive control. PCRs were performed also using total chromatin input (INPUT) as template. (B) Densitometric analysis of acetylated H3 enrichment of BIM locus in the same cell lines (bands shown in panel). (C) α-Acetylated H3 ChIP analysis before and after treatment with 500 nM TSA. (D) BIM mRNA levels assessed by QPCR before and after treatment with 500 nM TSA. (E) TUNEL assay of SU-DHL-1 and LAMA-84 cells untreated or treated with 500 nM TSA for 3 days. The graph shows the percentage of apoptotic cells as an average of three independent experiments.
To characterize the contribution of histone tail deacetylation to BIM silencing, SU-DHL-1 cells were treated with 500 nM TSA, a potent inhibitor of class I and II HDACs. ChIP analysis following treatment with TSA confirmed the reacetylation of H3 tails (Figures 4C and 7C). We subsequently analyzed the effect of histone reacetylation on BIM expression by QPCR. Treatment with TSA enhanced BIM expression 21.6-fold (Figure 4D). The up-regulation of BIM in the SU-DHL-1 cell line was accompanied by the induction of massive apoptosis, as assessed by TUNEL assay (Figure 4E). In the NPM/ ALK-negative LAMA-84 cell line, no up-regulation of BIM following treatment with TSA could be demonstrated (Figure 4D) and this correlated with a very limited proapoptotic effect (Figure 4E).
Figure 7.
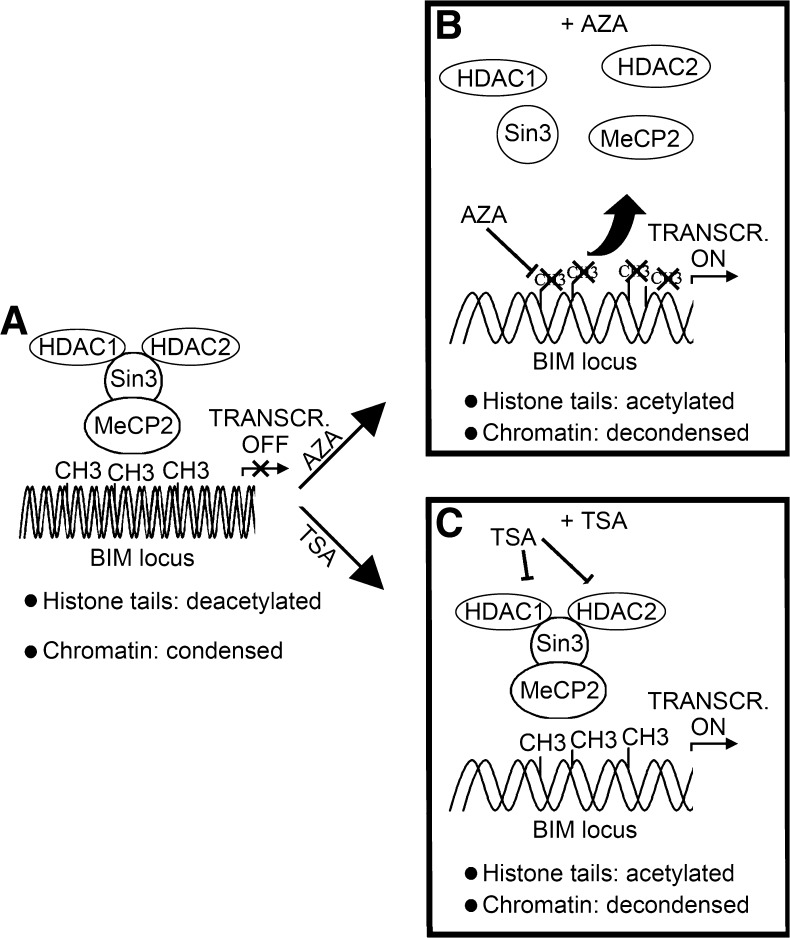
(A) Outline of the proposed model for the epigenetic silencing of BIM: NPM/ALK triggers BIM CpG methylation through a yet unknown pathway. Then, the methyl-CpG-binding protein MeCP2 binds to the 5-methylcytosines and recruits HDAC1, HDAC2, and the corepressor Sin3a. The activity of this complex leads to histone tail deacetylation, chromatin condensation, and BIM transcriptional repression. (B) Treatment with the demethylating agent AZA leads to BIM CpG island demethylation, detachment of MeCP2, disruption of the deacetylating complex, and reactivation of BIM transcription. (C) Treatment with the HDACi TSA causes direct inhibition of HDAC1/2 enzymatic activity, chromatin decondensation, and reactivation of BIM transcription.
BIM Protein Levels Affect Viability of NPM/ALK+ Lymphoid Cell Lines
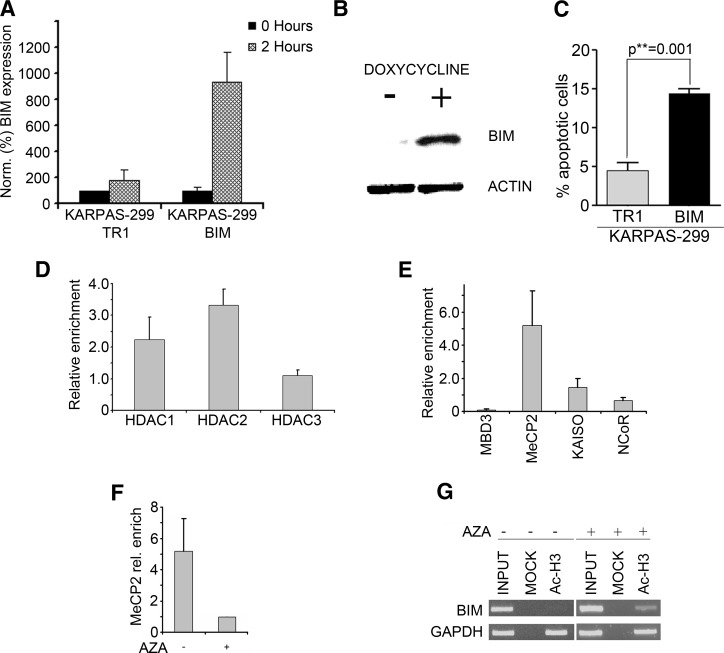
To assess the effect of BIM modulation in NPM/ALK+ cells, we generated an inducible BIM expression system in the highly methylated cell line KARPAS-299 (KARPAS-299 BIM), where BIM is virtually undetectable, as assessed by QPCR (Figure 1B).
Two hours after the addition of 1 µM doxycycline, induction of BIM expression was detected by real-time PCR (Figure 5A) that was followed by an increase in the expression of BIM protein, as assessed by Western blot in the KARPAS-299 BIM cell line (Figure 5B). This correlated with a significant induction of apoptosis in KARPAS-299 BIM compared to control the induction of apoptosis (Figure 5C), suggesting that KARPAS-299 are sensitive to the proapoptotic effect of BIM.
Figure 5.
(A) BIM mRNA level after the induction with doxycycline. (B) Expression level of Bim 12 hours after the induction, as assessed by Western blot. (C) Percentage of apoptosis after 12 hours of induction in KARPAS-299 TR1 and in KARPAS-299 Bim, as assessed by TUNEL assay. (D and E) Identification of the repressor complex associated with the epigenetic silencing of BIM locus. (D) ChIP analyses using α-HDAC 1, 2, and 3 antibodies were performed in SU-DHL-1 to identify the HDACs involved in BIM silencing. The immunoprecipitates were analyzed using QPCR. The error bars represent the SD of three replicates. (E) Characterization of the repressor complex involved in BIM silencing with ChIP experiments using α-MBD3, MeCP2, Kaiso, and NCoR antibodies. The immunoprecipitates were analyzed using QPCR. (F) α-MeCP2 ChIP analysis was performed in untreated cells (-) and in cells treated with 1 µM AZA (+), and immunoprecipitates were then analyzed using QPCR. (G) The effect of a demethylating agent on the acetylation status of histone H3 tails at BIM locus was investigated with α-acetylated H3 ChIP analysis in SU-DHL-1 in the absence (-) and in the presence (+) of 1 µM AZA. Immunoprecipitates (Ac-H3) were subjected to PCR with primer pairs specific for BIM and for GAPDH, as a positive control. PCRs were performed also using total chromatin input (INPUT) as template.
To assess if BIM up-regulation could play a significant role in TSA-mediated apoptosis, we generated a stable BIM silencing model in SU-DHL-1 cells and we compared the effect of TSA in both silenced cells and in a scrambled control. BIM silenced cells showed a higher proliferation rate (P = .03) when treated with TSA in comparison with the control, although no significant difference in the induction of apoptosis, at least up to 72 hours, could be detected (Figure W3, A–C).
BIM Deacetylation Occurs through Recruitment of an MeCP2-SIN3 Methyl-Binding Protein/Corepressor Complex
The finding that treatment with HDAC inhibitors (HDACis) up-regulates BIM suggests that histone tail deacetylation plays an important role in BIM silencing. It is known that class I HDACs, most notably HDAC1, 2, and 3, are frequently associated with gene silencing in human cancer [23–25]. Due to the lack of DNA binding domains in HDACs, deacetylases require the presence of MBPs and corepressor complexes to properly bind methylated DNA and deacetylate histone tails.
HDAC1 and 2 are commonly found as a part of two multiprotein corepressor complexes: SIN3 and NuRD [26,27]. Conversely, HDAC3 typically associates with the NCoR/SMRT complex [28], which is known to play a crucial role in hormone receptor signaling [29]. To study the involvement of specific HDACs in BIM silencing, we performed ChIP analyses with antibodies directed against HDAC1, 2, and 3 on SU-DHL-1 and we analyzed the results by QPCR. As shown in Figure 5D, HDAC1 and 2, but not HDAC3, associate with the methylated CpG island at the BIM genomic locus.
The identification of HDAC1 and 2 as a part of the BIM silencing machinery was suggestive for the involvement of SIN3 or NuRD corepressor complexes. To further investigate these findings, we performed ChIPs against MeCP2, an MBP that recruits SIN3 [30] and, less commonly, NCoR [31], and against MBD3, which is a core subunit of the multiprotein NuRD complex [27]. To confirm the absence of complexes involving HDAC3, we also set up ChIPs against NCoR, which is part of the NCoR/SMRT complex, and against Kaiso, a BTB/poxvirus-zinc finger family member able to bind NCoR [32]. As expected, the only significant enrichment was found for MeCP2 (Figure 5E), thus strengthening the hypothesis that silencing of BIM occurs through recruitment of an MeCP2-SIN3 methyl-binding protein/ corepressor complex (Figure 7A).
It is known that MeCP2 is able to bind to methylated and also to unmethylated DNA in vitro, albeit with different specificity and affinity [33]. To verify if the recruitment of MeCP2 was dependent on BIM 5′ UTR methylation, we tested its binding to the BIM locus with and without pretreatment with 1 µM AZA. The treatment with the demethylating agent was able to completely disrupt the association of MeCP2 to the BIM locus, thus indicating that MeCP2 acts through a methylation-dependent mechanism (Figures 5F and 7B). Interestingly, the disruption of the MeCP2-SIN3 methyl-binding protein/corepressor complex following treatment with AZA was associated with a prompt reacetylation of the BIM locus (Figures 5G and 7B).
NPM/ALK Kinase Activity Is Not Required to Maintain the Epigenetic Silencing of BIM Locus
The presence of BIM epigenetic silencing in ALK+ cell lines and patient samples suggested that NPM/ALK kinase activity might be the driving force behind this phenomenon.
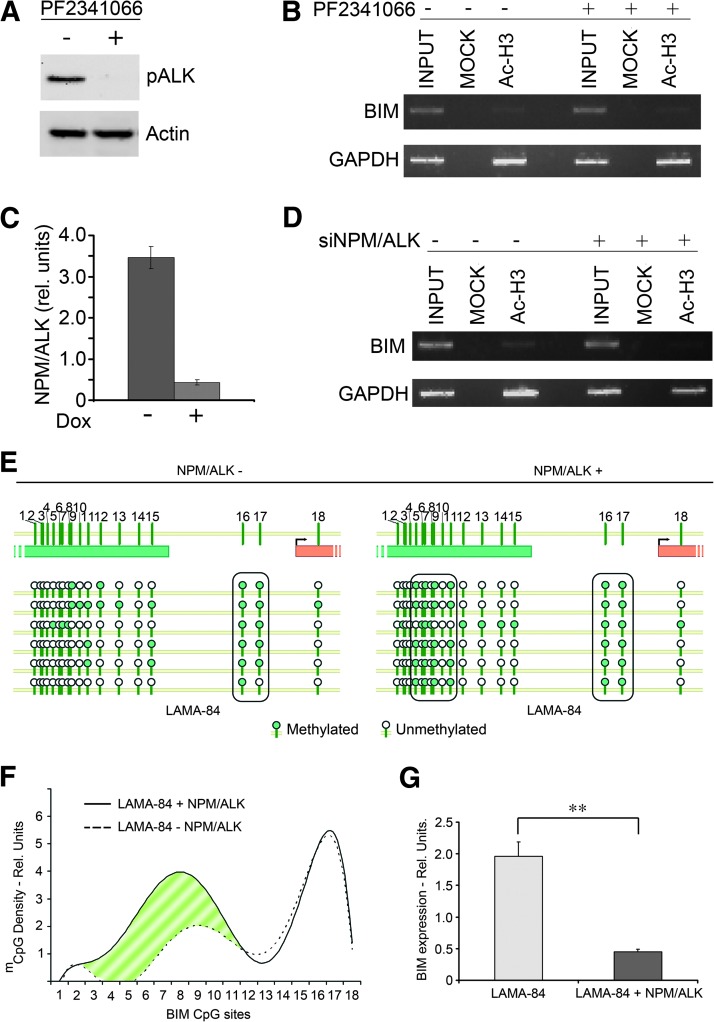
To test whether NPM/ALK could be implicated in the maintenance of BIM epigenetic silencing, we treated the SU-DHL-1 cell line with the ALK inhibitor PF2341066 to assess whether the inhibition of ALK catalytic activity may inactivate the silencing machinery. Although the treatment with PF2341066 led to a prompt inhibition of ALK kinase activity (Figure 6A) and to induction of apoptosis (data not shown), no evidence of reacetylation at BIM locus could be demonstrated (Figure 6B).
Figure 6.
(A) Western blot against phospho-ALK in the presence and absence of the ALK tyrosine kinase inhibitor PF2341066. Results are normalized against β-actin. (B) α-Acetylated H3 ChIP analysis of BIM locus before and after treatment with the ALK tyrosine kinase inhibitor PF2341066. (C) QPCR analysis of NPM/ALK mRNA expression before and after treatment with doxycycline in SU-DHL-1 cells carrying a doxycycline-inducible siRNA against NPM/ALK. (D) α-Acetylated H3 ChIP analysis of BIM locus before and after treatment with doxycycline. (E) Methylation pattern of BIM 5′UTR in LAMA-84 cells expressing (+) or not expressing (-) the NPM/ALK fusion protein. Filled circles represent methylated CpG sites; white circles represent unmethylated ones. Horizontal circle series represent sequential CpG sites; vertical series represent different clones. Black rounded rectangles indicate regions with a regular pattern of highly methylated CpG sites. (F) Polynomial regression of methylated CpG density pattern in LAMA-84 cells expressing (solid line) or not expressing (dashed line) NPM/ALK. The striped area highlights the region with a different methylation density between the two samples. (G) QPCR analysis of BIM expression in LAMA-84 cells transfected with NPM/ALK (+NPM/ALK) or an empty vector (-NPM/ALK). Error bars represent the SD of three replicates.
To further investigate these findings, we generated a doxycycline-inducible silencing system targeting NPM/ALK and we tested the acetylation status of the BIM locus in the presence/absence of doxycycline. As expected, the treatment with doxycycline led to a prompt down-modulation of NPM/ALK expression (Figure 6C); however, in line with the previous findings, no significant evidence of BIM reacetylation could be demonstrated (Figure 6D). Taken together, these data indicate that neither NPM/ALK nor its catalytic activity is required to maintain the epigenetic silencing at BIM locus.
NPM/ALK Induces De Novo BIM 5′UTR Methylation
It is known that the epigenetic silencing of specific genes depends on two biologically distinct processes: 1) initiation, occurring at previously unmethylated loci usually through methylation of a CpG island, and 2) maintenance of DNA methylation throughout cell divisions, typically through recruitment of DNMT1 at hemimethylated sites [34]. When NPM/ALK is silenced or when its catalytic activity is suppressed, no epigenetic effect is detectable at the BIM locus (Figure 6, B and D), which suggests that either NPM/ALK is dispensable for the activation of BIM epigenetic pathway or that it plays a role in initiating the epigenetic process by triggering the methylation of BIM 5′UTR but not in its maintenance. To address this issue, the methylation status of BIM 5′UTR was analyzed (Figure 6E) in the LAMA-84 cell line in the presence and absence of enforced NPM/ALK expression (Figure W4A). In the presence of the fusion protein, clonal bisulfite analysis of BIM 5′UTR revealed a significant increase (Mann-Whitney test, P = .041) in the overall methylation level (Figures 6, E and F, and W4B). Interestingly, the distribution of the methylated CpG sites, which was irregular in the absence of NPM/ALK, acquired a largely reproducible pattern in the presence of the latter (Figure 6, E and F) and this correlated with the down-modulation of BIM expression (Figure 6G; t test, P = .003).
Recent reports showed that the EML4/ALK fusion protein can induce BIM down-regulation through extracellular signal-regulated kinase (ERK) pathway in non-small cell lung cancer [35]. To test the role of the MAP kinase ERK kinase (MEK)/ERK pathway in the regulation of BIM expression in NPM/ALK+ cells, we treated the NPM/ALK+ SU-DHL-1 cell line with the MEK inhibitor CI-1040 [36] and we analyzed BIM expression by means of QPCR: no significant effect on BIM expression could be detected after up to 72 hours of treatment (Figure W5). Although further studies will be required to thoroughly analyze this phenomenon, these data indicate that NPM/ALK is able to modify the status and pattern of BIM 5′UTR methylation, possibly acting as an inducer of this epigenetic process through a MEK/ERK-independent pathway.
Discussion
The BH3-only, Bcl-2 family member BIM is one of the most potent proapoptotic factors involved in the homeostasis of the hematopoietic system [2,37,38]. Several mechanisms of BIM regulation have been described both at the transcriptional and posttranscriptional levels [20,39–44]. Additionally, recent studies showed that BIM expression can be silenced by epigenetic mechanisms in various types of cancers: in a subset of chronic myeloid leukemia patients [12], in renal cell carcinoma samples [14], in a high percentage of Burkitt lymphomas [15], in EBV-infected B cells [13], and in acute lymphoblastic leukemias [16], where it is associated with steroid resistance. Here, we showed for the first time the existence of a BIM epigenetic silencing mechanism in NPM/ALK+ ALCL.
Methylation profiling by RT-MeDIP revealed that methylation is clustered within BIM 5′UTR, at least in SU-DHL-1 (Figure 2D). This finding suggests that the 5′UTR of BIM promoter may have an important role in controlling the expression of the gene. It is well known that silencing of a gene through epigenetic mechanisms is a complex phenomenon [45,46]. Commonly, methylation of the promoter or the 5′UTR of a gene is just the first step of a long series of events leading to gene silencing. Epigenetic silencing can be triggered by CpG methylation through two main mechanisms: DNA methylation can repress transcription by directly impairing the binding of transcriptional activators to cognate DNA sequences [47,48] or MBP can bind to methylated DNA sequences, thus recruiting multi-protein corepressor complexes carrying HDAC activity. These complexes silence gene expression by triggering histone tail deacetylation and chromatin condensation [49]. Using ChIP experiments, we demonstrated the involvement of histone H3 tail deacetylation, a well-known marker of chromatin condensation [50], in BIM silencing (Figure 4, A and B). To further characterize the molecular events leading to chromatin condensation, we performed ChIP experiments using anti-HDAC1, HDAC2, HDAC3, MeCP2, MBD3, Kaiso, and NCoR antibodies, demonstrating HDAC1, HDAC2, and MeCP2, but not HDAC3, MBD3, Kaiso, and NCoR, to be involved in BIM silencing (Figure 5, D and E). HDAC1 and 2 are usually found as part of two corepressor complexes: SIN3 and NuRD. However, the identification of MeCP2, an MBD commonly associated with the SIN3 complex, strongly suggests the involvement of the latter. This hypothesis is further supported by the absence of MBD3, which is part of the core NuRD complex, even if its constitutive presence in NuRD corepressor complex has been questioned [51]. The finding that treatment with demethylating agents is able to completely revert the binding of MeCP2 to the BIM locus (Figure 5F) suggests that BIM silencing could occur through the following steps: CpG methylation→binding of MeCP2 to methylated CpGs→recruitment of SIN3 core-pressor complex→deacetylation of histone tails→chromatin condensation (Figure 7). The evidence that a deacetylating complex plays a functional role in BIM down-regulation is supported by the fact that treatment with HDACi is able to restore BIM expression (Figures 4D and 7C). Interestingly, the detachment of MeCP2 following treatment with deacetylating agents is also associated with histone tail reacetylation (Figure 5G), suggesting the presence of a yet unidentified acetylating complex acting in competition with MeCP2/SIN3.
Although BIM epigenetic signaling can be triggered also in clonal disorders not expressing NPM/ALK, the evidence of BIM silencing in NPM/ALK+ cell lines as well as patient samples strongly suggested a direct role for the deregulated kinase in the maintenance of the BIM epigenetic silencing machinery, at least in NPM/ALK+ lymphomas. This hypothesis was further supported by the evidence that NPM/ALK is able to activate an epigenetic STAT3-dependent pathway [21]. Nevertheless, the treatment with the ALK inhibitor PF2341066 was unable to revert BIM epigenetic silencing (Figure 6, A and B). Although it is possible that BIM demethylation could require a longer time than the one allowed by the use of ALK inhibitors, these findings do not support the hypothesis that the maintenance of BIM epigenetic silencing is linked to the NPM/ALK kinase activity. An alternative hypothesis is that this process could be triggered by the presence of the NPM/ALK protein, even in the absence of any kinase activity. However, when NPM/ALK expression was knocked out using an inducible siRNA silencing system, no significant evidence of BIM locus reacetylation could be found (Figure 6, C and D), suggesting that NPM/ALK expression is dispensable from maintaining BIM epigenetic silencing and that other, yet unknown pathways are required to maintain this process, probably involving DNMT1. Nevertheless, the enforced expression of NPM/ALK on the LAMA-84 cells led to an increase in the methylation of BIM 5′UTR (Figures 6, E and F, and W5) and to the stabilization of a specific methylation pattern (Figure 6E). This phenomenon was accompanied by a significant down-modulation of BIM expression (Figure 6G) that cannot be explained by the NPM/ALK-mediated phosphorylation of FOXO3a, because in BCR/ABL-positive cells, such as LAMA-84, FOXO3a is similarly inactivated by serine/threonine phosphorylation and cytoplasmic translocation [52] through a PI3K-dependent mechanism. Therefore, despite the evidence that NPM/ALK is not critical for the maintenance of BIM epigenetic silencing, these findings, albeit preliminary, suggest that NPM/ALK itself may be the initiator of the BIM epigenetic silencing pathway and that other NPM/ALK-independent mechanisms may be then recruited to maintain its repressed status.
While the epigenetic silencing of BIM may play a crucial role in protecting ALCL cells from apoptosis, other mechanisms leading to the down-regulation of BIM have previously been identified. It is known that the transcription factor FOXO3a is able to directly activate the expression of BIM through its binding site on BIM promoter [52,53]. This mechanism of regulation can be inhibited by the phosphorylation of FOXO3a, which results in the exclusion of the transcription factor from the nucleus and thus in its functional inactivation [52,53]. Notably, this mechanism was demonstrated in NPM/ALK-positive cell lines [19], suggesting that the impairment of the BIM pathway is an important step in lymphomagenesis. Therefore, at least in ALCL cells, two different mechanisms, phosphorylation of FOXO3a through the activation of the PI3K-AKT pathway and BIM epigenetic silencing, may converge to inactivate the potent proapoptotic signal of BIM, thus protecting the lymphoma cells from apoptosis.
These findings reinforce the idea that BIM plays a major role in the surveillance against tumorigenesis in NPM/ALK-positive lymphomas and possibly in several lymphoid malignancies. However, the demonstration that in NPM/ALK+ cells TSA is able to induce apoptosis even when BIM expression is silenced suggests that other, yet unknown oncosuppressors are bona fide epigenetically silenced in these cells, therefore suggesting that further studies are required to fully characterize the epigenetic signature of ALK+ lymphomas. The discovery and description of the mechanisms by which BIM, as well as other oncosuppressors, are inactivated in specific types of cancers may be of great importance to define evidence-based cancer treatment protocols. Specifically, this study shows that epigenetic drugs significantly upregulate BIM expression and induce cell death in ALCL cell lines and suggests that the use of epigenetic modulators may play an important role in the treatment of ALCL.
Supplementary Material
Acknowledgments
We thank Michela Viltadi, Barbara Vergani, Giovanni Roncato, and Manuela Scuro for technical assistance.
Abbreviations
- TSA
trichostatin A
- RT-MeDIP
real-time methylated DNA immunoprecipitation
Footnotes
This work was funded by Associazione Italiana per la Ricerca sul Cancro 2010 (IG-10092), Progetti di Ricerca di Interesse Nazionale program (20084XBENM_004), and Fondazione Cariplo (2009-2667). Written consent for publication was obtained from the patients or their relative. The authors declare that they have no competing financial interests in relation to the work described here.
This article refers to supplementary materials, which are designated by Figures W1 to W5 and are available online at www.neoplasia.com.
References
- 1.Fletcher JI, Huang DC. Controlling the cell death mediators Bax and Bak: puzzles and conundrums. Cell Cycle. 2008;7:39–44. doi: 10.4161/cc.7.1.5178. E-pub 2007 Oct 2016. [DOI] [PubMed] [Google Scholar]
- 2.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 3.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci USA. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. E-pub 2004 Apr 6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, Morishima Y, Nakamura S, Seto M. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005;24:1348–1358. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 5.Gama-Sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 7.Toyota M, Issa JP. Epigenetic changes in solid and hematopoietic tumors. Semin Oncol. 2005;32:521–530. doi: 10.1053/j.seminoncol.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Strathdee G, Davies BR, Vass JK, Siddiqui N, Brown R. Cell type-specific methylation of an intronic CpG island controls expression of the MCJ gene. Carcinogenesis. 2004;25:693–701. doi: 10.1093/carcin/bgh066. [DOI] [PubMed] [Google Scholar]
- 9.Parolini O, Weinhausel A, Kagerbauer B, Sassmann J, Holter W, Gadner H, Haas OA, Knapp W. Differential methylation pattern of the X-linked lymphoproliferative (XLP) disease gene SH2D1A correlates with the cell lineage-specific transcription. Immunogenetics. 2003;55:116–121. doi: 10.1007/s00251-003-0557-x. [DOI] [PubMed] [Google Scholar]
- 10.Petak I, Danam RP, Tillman DM, Vernes R, Howell SR, Berczi L, Kopper L, Brent TP, Houghton JA. Hypermethylation of the gene promoter and enhancer region can regulate Fas expression and sensitivity in colon carcinoma. Cell Death Differ. 2003;10:211–217. doi: 10.1038/sj.cdd.4401132. [DOI] [PubMed] [Google Scholar]
- 11.Esteller M. Profiling aberrant DNA methylation in hematologic neoplasms: a view from the tip of the iceberg. Clin Immunol. 2003;109:80–88. doi: 10.1016/s1521-6616(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 12.San Jose-Eneriz E, Agirre X, Jimenez-Velasco A, Cordeu L, Martin V, Arqueros V, Garate L, Fresquet V, Cervantes F, Martinez-Climent JA, et al. Epigenetic down-regulation of BIM expression is associated with reduced optimal responses to imatinib treatment in chronic myeloid leukaemia. Eur J Cancer. 2009;45:1877–1889. doi: 10.1016/j.ejca.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Paschos K, Smith P, Anderton E, Middeldorp JM, White RE, Allday MJ. Epstein-Barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathog. 2009;5:e1000492. doi: 10.1371/journal.ppat.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zantl N, Weirich G, Zall H, Seiffert BM, Fischer SF, Kirschnek S, Hartmann C, Fritsch RM, Gillissen B, Daniel PT, et al. Frequent loss of expression of the pro-apoptotic protein Bim in renal cell carcinoma: evidence for contribution to apoptosis resistance. Oncogene. 2007;26:7038–7048. doi: 10.1038/sj.onc.1210510. [DOI] [PubMed] [Google Scholar]
- 15.Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, Siebert R, Climent J, Fresquet V, Beltran E, Agirre X, Marugan I, Marin M, et al. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood. 2007;109:271–280. doi: 10.1182/blood-2006-06-026500. E-pub 2006 Sep 2007. [DOI] [PubMed] [Google Scholar]
- 16.Bachmann PS, Piazza RG, Janes ME, Wong NC, Davies C, Mogavero A, Bhadri VA, Szymanska B, Geninson G, Magistroni V, et al. Epigenetic silencing of BIM in glucocorticoid poor-responsive pediatric acute lymphoblastic leukemia, and its reversal by histone deacetylase inhibition. Blood. 2010;116:3013–3022. doi: 10.1182/blood-2010-05-284968. E-pub 2010 Jul 3020. [DOI] [PubMed] [Google Scholar]
- 17.Leo E, Mancini M, Aluigi M, Castagnetti F, Martinelli G, Barbieri E, Santucci MA. DNA hypermethylation promotes the low expression of pro-apoptotic BCL2L11 associated with BCR-ABL1 fusion gene of chronic myeloid leukaemia. Br J Haematol. 2012;159:373–376. doi: 10.1111/bjh.12023. [DOI] [PubMed] [Google Scholar]
- 18.Richter-Larrea JA, Robles EF, Fresquet V, Beltran E, Rullan AJ, Agirre X, Calasanz MJ, Panizo C, Richter JA, Hernandez JM, et al. Reversion of epigenetically mediated BIM silencing overcomes chemoresistance in Burkitt lymphoma. Blood. 2010;116:2531–2542. doi: 10.1182/blood-2010-02-268003. [DOI] [PubMed] [Google Scholar]
- 19.Gu TL, Tothova Z, Scheijen B, Griffin JD, Gilliland DG, Sternberg DW. NPM-ALK fusion kinase of anaplastic large-cell lymphoma regulates survival and proliferative signaling through modulation of FOXO3a. Blood. 2004;103:4622–4629. doi: 10.1182/blood-2003-03-0820. E-pub 2004 Feb 4612. [DOI] [PubMed] [Google Scholar]
- 20.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 21.Ambrogio C, Martinengo C, Voena C, Tondat F, Riera L, di Celle PF, Inghirami G, Chiarle R. NPM-ALK oncogenictyrosinekinase controls T-cell identity by transcriptional regulation and epigenetic silencing in lymphoma cells. Cancer Res. 2009;69:8611–8619. doi: 10.1158/0008-5472.CAN-09-2655. E-pub 2009 Nov 8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galietta A, Gunby RH, Redaelli S, Stano P, Carniti C, Bachi A, Tucker PW, Tartari CJ, Huang CJ, Colombo E, et al. NPM/ALK binds and phosphorylates the RNA/DNA-binding protein PSF in anaplastic large-cell lymphoma. Blood. 2007;110:2600–2609. doi: 10.1182/blood-2006-01-028647. E-pub 2007 May 2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senese S, Zaragoza K, Minardi S, Muradore I, Ronzoni S, Passafaro A, Bernard L, Draetta GF, Alcalay M, Seiser C, et al. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007;27:4784–4795. doi: 10.1128/MCB.00494-07. E-pub 2007 Apr 4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 25.Brown R, Strathdee G. Epigenomics and epigenetic therapy of cancer. Trends Mol Med. 2002;8:S43–S48. doi: 10.1016/s1471-4914(02)02314-6. [DOI] [PubMed] [Google Scholar]
- 26.Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 30.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 31.Kokura K, Kaul SC, Wadhwa R, Nomura T, Khan MM, Shinagawa T, Yasukawa T, Colmenares C, Ishii S. The Ski protein family is required for MeCP2-mediated transcriptional repression. J Biol Chem. 2001;276:34115–34121. doi: 10.1074/jbc.M105747200. E-pub 32001 Jul 34115. [DOI] [PubMed] [Google Scholar]
- 32.Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell. 2003;12:723–734. doi: 10.1016/j.molcel.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–66. doi: 10.4161/epi.1.2.2767. E-pub 2006 Apr 2005. [DOI] [PubMed] [Google Scholar]
- 35.Takezawa K, Okamoto I, Nishio K, Janne PA, Nakagawa K. Role of ERK-BIM and STAT3-survivin signaling pathways in ALK inhibitor-induced apoptosis in EML4-ALK-positive lung cancer. Clin Cancer Res. 2011;17:2140–2148. doi: 10.1158/1078-0432.CCR-10-2798. [DOI] [PubMed] [Google Scholar]
- 36.Allen LF, Sebolt-Leopold J, Meyer MB. CI-1040 (PD184352), a targeted signal transduction inhibitor of MEK (MAPKK) Semin Oncol. 2003;30:105–116. doi: 10.1053/j.seminoncol.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Bouillet P, Cory S, Zhang LC, Strasser A, Adams JM. Degenerative disorders caused by Bcl-2 deficiency prevented by loss of its BH3-only antagonist Bim. Dev Cell. 2001;1:645–653. doi: 10.1016/s1534-5807(01)00083-1. [DOI] [PubMed] [Google Scholar]
- 38.Hildeman DA, Zhu Y, Mitchell TC, Bouillet P, Strasser A, Kappler J, Marrack P. Activated T cell death in vivo mediated by proapoptotic Bcl-2 family member Bim. Immunity. 2002;16:759–767. doi: 10.1016/s1074-7613(02)00322-9. [DOI] [PubMed] [Google Scholar]
- 39.Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem. 2003;278:18811–18816. doi: 10.1074/jbc.M301010200. [DOI] [PubMed] [Google Scholar]
- 40.Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Chung UI, Fukuda A, Hikita A, Seto H, Okada T, et al. Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J. 2003;22:6653–6664. doi: 10.1093/emboj/cdg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G, Auberger P. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene. 2003;22:6785–6793. doi: 10.1038/sj.onc.1206792. [DOI] [PubMed] [Google Scholar]
- 42.Putcha GV, Le S, Frank S, Besirli CG, Clark K, Chu B, Alix S, Youle RJ, LaMarche A, Maroney AC, et al. JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron. 2003;38:899–914. doi: 10.1016/s0896-6273(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 43.Biswas SC, Greene LA. Nerve growth factor (NGF) down-regulates the Bcl-2 homology 3 (BH3) domain-only protein Bim and suppresses its proapoptotic activity by phosphorylation. J Biol Chem. 2002;277:49511–49516. doi: 10.1074/jbc.M208086200. [DOI] [PubMed] [Google Scholar]
- 44.Shinjyo T, Kuribara R, Inukai T, Hosoi H, Kinoshita T, Miyajima A, Houghton PJ, Look AT, Ozawa K, Inaba T. Downregulation of Bim, a proapoptotic relative of Bcl-2, is a pivotal step in cytokine-initiated survival signaling in murine hematopoietic progenitors. Mol Cell Biol. 2001;21:854–864. doi: 10.1128/MCB.21.3.854-864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome—components and functional correlates. Genes Dev. 2006;20:3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 47.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 48.Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 49.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. E-pub 2006 Jan 2005. [DOI] [PubMed] [Google Scholar]
- 50.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 51.Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, Lasonder E, Stunnenberg HG. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Essafi A, Fernandez de Mattos S, Hassen YA, Soeiro I, Mufti GJ, Thomas NS, Medema RH, Lam EW. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene. 2005;24:2317–2329. doi: 10.1038/sj.onc.1208421. [DOI] [PubMed] [Google Scholar]
- 53.Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, Medema RH. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.