The aminobisphosphonate zoledronic acid (ZA) is the most important antiresorptive agent for the treatment of multiple myeloma (MM)-related bone disease (BD). Osteonecrosis of the jaw (ONJ) is an important complication of ZA-treated MM patients (Vannucchi et al, 2005; Filleul et al, 2010). So far, the mechanism of ONJ pathogenesis has not been clearly elucidated. Recently, a genetic susceptibility to ONJ has been suggested and a polymorphism of the cytochrome P450 CYP2C8 has been associated with ZA-related ONJ in MM (Sarasquete et al, 2008).
To further investigate the genetic bases of ONJ, we genotyped in a case-control study a cohort of 19 MM patients treated with ZA who developed [nine cases, median age 66 years (range: 63–79)] or not [10 matched controls, median age 69 years (range: 63–84)] ONJ. We used the novel Affymetrix DMET™ plus platform (Affymetrix, Santa Clara, CA, USA), which interrogates 1936 genetic variations in 225 genes associated with phase I–II drug metabolism, disposition and transport (Deeken, 2009). The study protocol was approved by our University Hospital Bioethical Committee and informed consent was obtained from each patient. All patients received ZA according to the conventional dose and administration schedule; the ONJ group received 20 ± standard deviation (SD) 5·1 treatment courses and the control group underwent 15·1 ± 4·2 courses. MM patients were homogeneous on clinical and pathological characteristics at diagnosis and on their response to treatment. ONJ was diagnosed by clinical examination and imaging, including radiographs and/or computed tomography or magnetic resonance imaging. Peripheral blood was collected and used for DNA extraction. Genotypes were determined for each single nucleotide polymorphism (SNP) site of the 1931 of all interrogated SNPs and for the five Copy Number Variations (CNVs) included in DMET™ Plus GeneChip. Pharmacogenomic profiles were generated by Affymetrix DMET™ Console software®. Statistical analysis was performed by two-tailed Fisher’s exact test. No correction for multiple comparisons was performed. Results are therefore to be interpreted as hypothesis generating.
Eight SNPs were significantly (P ≤ 0·05) associated with ONJ occurrence. Table I shows these SNPs, the reference and variant allele and the genotype and allele frequencies. All alleles were in Hardy-Weinberg equilibrium. The four genes correlated to the eight statistically relevant SNPs were PPARG (peroxisome proliferator-activated receptor gamma), ABP1{amiloride binding protein 1 [amine oxidase (copper-containing)]}, CHST11 [carbohydrate (chondroitin 4) sulfotransferase 11] and CROT (carnitine O-octanoyltransferase). The different distribution of SNP alleles and genotypes between ONJ patients and control cases are reported in Table II. The SNP rs1152003, mapping in PPARG, showed the strongest association with ONJ. We detected a highly significant (P = 0·0055) differential occurrence of the C/C homozygous (HOM) genotype in 77·7% of ONJ cases (7/9) versus only 10% of controls (1/10) (Fig 1A). Moreover, homozygous and heterozygous genotypes for the C variant were differently distributed between ONJ patients and the control group (Table II). The frequency of the C variant allele in the PPARG SNP underlines a highly significant association of the C allele with the ONJ group (P = 0·0064, Table II). No clinical association has been previously reported for these SNPs.
Table I.
SNP polymorphisms associated with ONJ in MM patients.
| Polymorphism Ref>Var | Effect | Ref. SNP alleles (A//B) | Genotype frequencies | Allele frequencies | |||
|---|---|---|---|---|---|---|---|
| Ref | HT | Var | A | B | |||
| PPARG rs 1152003 | NA | C//G | 8 | 9 | 2 | 0·658 | 0·342 |
| ABP1_2357A>G | P545P | G//A | 9 | 9 | 1 | 0·289 | 0·711 |
| ABP1_4064G>A | S630S | G//A | 9 | 9 | 1 | 0·711 | 0·289 |
| ABP1_4107C>G | H645D | C//G | 9 | 9 | 1 | 0·711 | 0·289 |
| CHST11_> (rs2463437) | NA | G//A | 7 | 11 | 1 | 0·342 | 0·658 |
| CHST11_> (rs903247) | NA | C//T | 7 | 10 | 2 | 0·368 | 0·632 |
| CHST11 rs2468110 | NA | G//A | 7 | 10 | 2 | 0·632 | 0·368 |
| CROT_73879> (rs2097937) | NA | G//A | 12 | 7 | 0 | 0·184 | 0·816 |
Distribution data for eight SNPs associated with ONJ in MM patients. Polymorphisms are reported as Reference (Ref), Heterozygosis (HT) and Variant (Var). Allele frequencies were calculated in the population included in the study using the Hardy-Weinberg equation.
Table II.
Allele and genotype frequencies of polymorphisms in MM patients.
| SNP and variants | Gene | Allele distribution | P | Genotype | Genotype distribution | P | Clinical association | ||
|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases (%) | Controls (%) | ||||||
| rs 1152003 | |||||||||
| C | PPARG | 16/18 | 9/20 | 0·0064 | CC | 7/9 (77·7) | 1/10 (10·0) | 0·0055 | Unknown |
| G | 2/18 | 11/20 | CG | 2/9 (22·2) | 7/10 (70·0) | ||||
| GG | 0/9 (00·0) | 2/10 (20·0) | |||||||
| rs10893 | |||||||||
| G | ABP1 | 2/18 | 9/20 | 0·0327 | AA | 7/9 (77·7) | 2/10 (20·0) | 0·023 | Unknown |
| A | 16/18 | 11/20 | AG | 2/9 (22·2) | 7/10 (70·0) | ||||
| GG | 0/9 (00·0) | 1/10 (10·0) | |||||||
| rs4725373 | |||||||||
| G | ABP1 | 16/18 | 11/20 | 0·0327 | GG | 7/9 (77·7) | 2/10 (20·0) | 0·023 | Unknown |
| A | 2/18 | 9/20 | AG | 2/9 (22·2) | 7/10 (70·0) | ||||
| AA | 0/9 (00·0) | 1/10 (10·0) | |||||||
| rs1049793 | |||||||||
| C | ABP1 | 16/18 | 11/20 | 0·0327 | CC | 7/9 (77·7) | 2/10 (20·0) | 0·023 | Unknown |
| G | 2/18 | 9/20 | CG | 2/9 (22·2) | 7/10 (70·0) | ||||
| GG | 0/9 (00·0) | 1/10 (10·0) | |||||||
| rs2463437 | |||||||||
| G | CHST11 | 15/18 | 10/20 | 0·0434 | AA | 6/9 (66·6) | 1/10 (10·0) | 0·0198 | Unknown |
| A | 3/18 | 10/20 | AG | 3/9 (33·3) | 8/10 (80·0) | ||||
| GG | 0/9 (00·0) | 1/10 (10·0) | |||||||
| rs903247 | |||||||||
| C | CHST11 | 3/18 | 11/20 | 0·0205 | TT | 6/9 (66·6) | 1/10 (10·0) | 0·0198 | Unknown |
| T | 15/18 | 9/20 | CT | 3/9 (33·3) | 7/10 (70·0) | ||||
| CC | 0/9 (00·0) | 2/10 (20·0) | |||||||
| rs2468110 | |||||||||
| G | CHST11 | 14/18 | 10/20 | 0·1008 | GG | 6/9 (66·6) | 1/10 (10·0) | 0·0198 | Unknown |
| A | 4/18 | 10/20 | AG | 2/9 (22·2) | 8/10 (80·0) | ||||
| AA | 1/9 (11·1) | 1/10 (10·0) | |||||||
| rs2097937 | |||||||||
| G | CROT | 6/18 | 1/20 | 0·0381 | AG | 6/9 (66·6) | 1/10 (10·0) | 0·0198 | Unknown |
| A | 12/18 | 19/20 | AA | 3/9 (33·3) | 9/10 (90·0) | ||||
| GG | 0/9 (00·0) | 0/10 (00·0) | |||||||
Distribution data for eight SNPs associated with ONJ in MM patients. Allele and genotype distribution between case and control groups. Polymorphisms are reported as rs number used in the human SNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/). The P value was calculated by two-tailed Fisher’s exact test.
Fig 1.
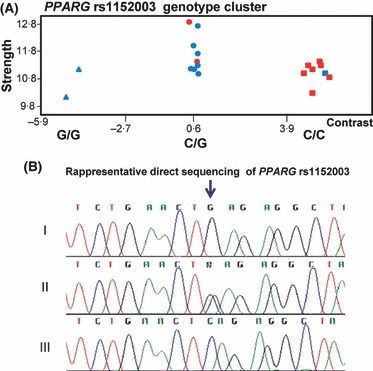
SNP rs1152003 genotype clustering of MM patients. (A) The red colour symbols are MM patients with ONJ. Blue colour symbols are matched MM control patients. Genotypes are identified as homozygote reference allele  , heterozygote (•) and homozygote variant allele (▪). (B) Representative electropherogram of direct sequencing analysis of a homozygote reference allele (I), heterozygote (II) and homozygote variant allele (III). The variant rs1152003 in PPARG gene was analyzed using the following PCR primers: Forward: 5′-TCCTTCTGCAAGGCAGTTTT-3′ and Reverse: 5′-CACGCCTTCAGGGAACTTAG-3′. The black arrow indicates the G, C or G/C sequences.
, heterozygote (•) and homozygote variant allele (▪). (B) Representative electropherogram of direct sequencing analysis of a homozygote reference allele (I), heterozygote (II) and homozygote variant allele (III). The variant rs1152003 in PPARG gene was analyzed using the following PCR primers: Forward: 5′-TCCTTCTGCAAGGCAGTTTT-3′ and Reverse: 5′-CACGCCTTCAGGGAACTTAG-3′. The black arrow indicates the G, C or G/C sequences.
Direct nucleotide sequencing was carried out on patient specimens to further confirm the presence of genetic variations, using an Applied Biosystems ABI 3100 Genetic Analyser. We found a concordance rate of 100% between DMET genotyping and sequence analysis (Fig 1B).
The rs1152003 SNP maps in the 3′UTR region of PPARG, at position 12477055 of chromosome 3 (Genome Build 37.1). Although no clinical correlation has been reported for the rs1152003 variant, polymorphisms in PPARG have been associated with increased risk of a variety of diseases (Dallongeville et al, 2009). PPARG is located in the human chromosome 3, band 3p25. Chromosomal abnormalities, such as 3p deletion, have been identified in several hematologic malignancies. PPARG is involved in adipocyte differentiation and in angiogenesis (Rosen & Spiegelman, 2001). Recently, the PPARG pathway has been recognized as key mechanism for bone remodelling. It acts on mesenchymal stem cell differentiation by increasing adipogenesis but also inhibiting osteoblast and osteoclast formation. Moreover, PPARG polymorphisms correlate with the bone mass density (Ackert-Bicknell et al, 2008). However, a recent study on a wide cohort of Korean individuals, with idiopathic, steroid-induced or alcohol-induced osteonecrosis of the femoral head, failed to demonstrate a significant correlation with three common PPARG polymorphisms (Kim et al, 2007). Interestingly, modulation of PPARG activity within the bone marrow microenvironment has been recently shown to interfere with cytokines such as IL6, which is involved with a central role in the pathogenesis of MM (Wang et al, 2004), suggesting also that PPARG may represent a valuable therapeutic target in MM (Garcia-Bates et al, 2008).
The present study also showed that three SNPs identified in ABP1 were associated with ONJ and were in linkage disequilibrium (data not shown). ABP1 encodes a membrane glycoprotein that is expressed in many epithelial and haematopoietic tissues. Moreover, a further three ONJ-associated SNPs map to CHST11, which was recently described as a factor required for proper chondroitin sulfation and cartilage morphogenesis. Expression of the chondroitin sulfotransferase genes is crucial for the correct mammalian bone morphogenesis. Finally, the ONJ-associated rs2097937 maps to CROT, whose protein is involved in the trans-esterification of acyl-CoA molecules.
Our findings indicate that genetic polymorphisms are involved in the pathogenesis of ONJ in MM patients. The highly significant association of ONJ with the rs1152003 SNP polymorphism in PPARG strongly suggests this genetic variant as candidate biomarker for the identification of MM patients at risk of ONJ if treated with ZA. In fact, the C/C genotype demonstrated an odds ratio of 31·5 (95% confidence interval, 2·35–422·32) for developing ONJ following ZA treatment. Differently from the recent report (Sarasquete et al, 2008), where the study was based on the 500K Affymetrix high density array, we used the DMET platform that interrogates only highly selective SNPs associated with drug toxicity and has the advantage of avoiding an extremely high number of comparisons, which requires statistical corrections and large patient cohorts. We propose the rs1152003 C/C genotype as a candidate genetic biomarker for ONJ, which warrants validation in larger series.
Acknowledgments
This work was partially supported by AIRC (Associazione Italiana per la Ricerca sul cancro) (P.T. project 2007–2009), Milan, and PRIN (P.T. 2007–2009), Italy.
Author contributions
M.T.D., PS.T. and PF.T. designed the study and performed data interpretation; M.T.D. wrote the manuscript; M.T.D and M.A. generated datasets. M.T.D. provided statistical analysis and generated figures; P.H.G., P.V. and M.C. performed data mining; E.P., T.P., I.C., T.C. and M.R. provided blood samples and clinical data; E.L. and F.B. performed sequence analysis; PS.T. and PF.T. reviewed the manuscript.
Disclosures
The authors declare no conflicts of interest to disclose.
References
- Ackert-Bicknell CL, Demissie S, Marín de Evsikova C, Hsu YH, DeMambro VE, Karasik D, Cupples LA, Ordovas JM, Tucker KL, Cho K, Canalis E, Paigen B, Churchill GA, Forejt J, Beamer WG, Ferrari S, Bouxsein ML, Kiel DP, Rosen CJ. PPARG by dietary fat interaction influences bone mass in mice and humans. Journal of Bone and Mineral Research. 2008;23:1398–1408. doi: 10.1359/JBMR.080419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallongeville J, Iribarren C, Ferrières J, Lyon L, Evans A, Go AS, Arveiler D, Fortmann SP, Ducimetière P, Hlatky MA, Amouyel P, Southwick A, Quertermous T, Meirhaeghe A. Peroxisome proliferator-activated receptor gamma polymorphisms and coronary heart disease. PPAR Research. 2009;2009:11. doi: 10.1155/2009/543746. doi: 10.1155/2009/543746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeken J. The Affymetrix DMET platform and pharmacogenetics in drug development. Current Opinion in Molecular Therapeutics. 2009;11:260–268. [PubMed] [Google Scholar]
- Filleul O, Crompot E, Saussez S. Bisphosphonate-induced osteonecrosis of the jaw: a review of 2,400 patient cases. Journal of Cancer Research and Clinical Oncology. 2010;136:1117–1124. doi: 10.1007/s00432-010-0907-7. [DOI] [PubMed] [Google Scholar]
- Garcia-Bates TM, Bernstein SH, Phipps RP. Peroxisome proliferator-activated receptor gamma overexpression suppresses growth and induces apoptosis in human multiple myeloma cells. Clinical Cancer Research. 2008;14:6414–6425. doi: 10.1158/1078-0432.CCR-08-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Hong JM, Park EK, Kim SY. Peroxisome proliferator-activated receptor-gamma gene polymorphisms are not associated with osteonecrosis of the femoral head in the Korean population. Molecules and Cells. 2007;24:388–393. [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM. PPARgamma : a nuclear regulator of metabolism, differentiation, and cell growth. Journal of Biological Chemistry. 2001;276:37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- Sarasquete ME, García-Sanz R, Marín L, Alcoceba M, Chillón MC, Balanzategui A, Santamaria C, Rosiñol L, de la Rubia J, Hernandez MT, Garcia-Navarro I, Lahuerta JJ, González M, San Miguel JF. Bisphosphonate-related osteonecrosis of the jaw is associated with polymorphisms of the cytochrome P450 CYP2C8 in multiple myeloma: a genome-wide single nucleotide polymorphism analysis. Blood. 2008;112:2709–2712. doi: 10.1182/blood-2008-04-147884. [DOI] [PubMed] [Google Scholar]
- Vannucchi AM, Ficarra G, Antonioli E, Bosi A. Osteonecrosis of the jaw associated with zoledronate therapy in a patient with multiple myeloma. British Journal of Haematology. 2005;128:738. doi: 10.1111/j.1365-2141.2005.05382.x. [DOI] [PubMed] [Google Scholar]
- Wang LH, Yang XY, Zhang X, Huang J, Hou J, Li J, Xiong H, Mihalic K, Zhu H, Xiao W, Farrar WL. Transcriptional inactivation of STAT3 by PPARgamma suppresses IL-6-responsive multiple myeloma cells. Immunity. 2004;20:205–218. doi: 10.1016/s1074-7613(04)00030-5. [DOI] [PubMed] [Google Scholar]


